Highly Sensitive Label-Free Detection of Small Molecules with an Optofluidic Microbubble Resonator
Abstract
:1. Introduction
2. Materials and Method
2.1. Materials
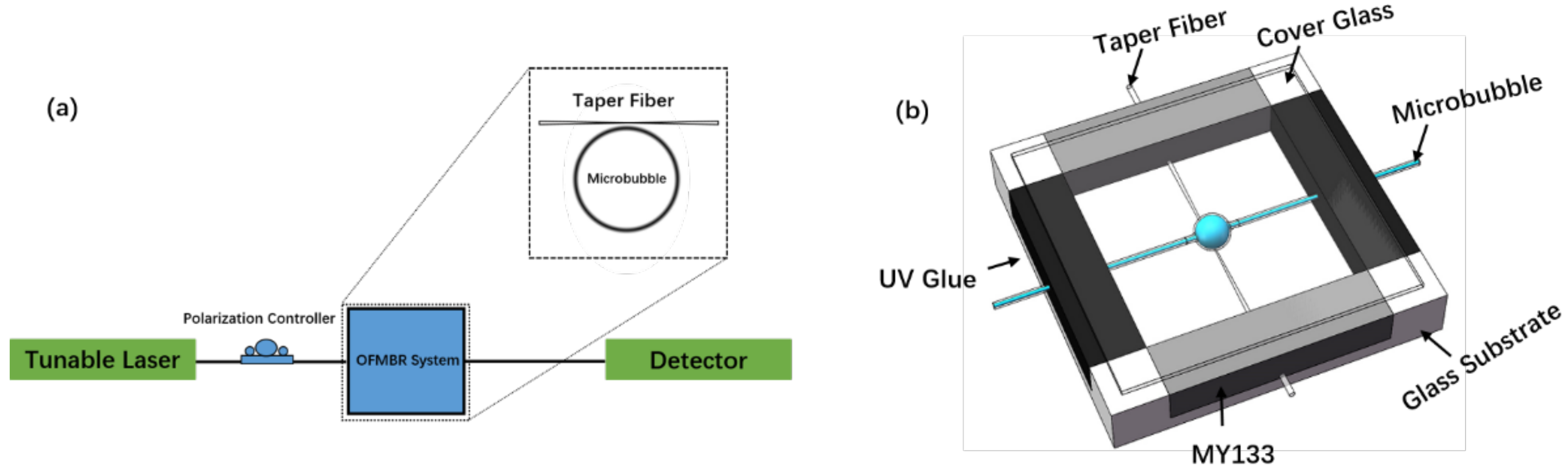
2.2. The Packaged OFMBR Sensor for Biomolecular Interaction Detection
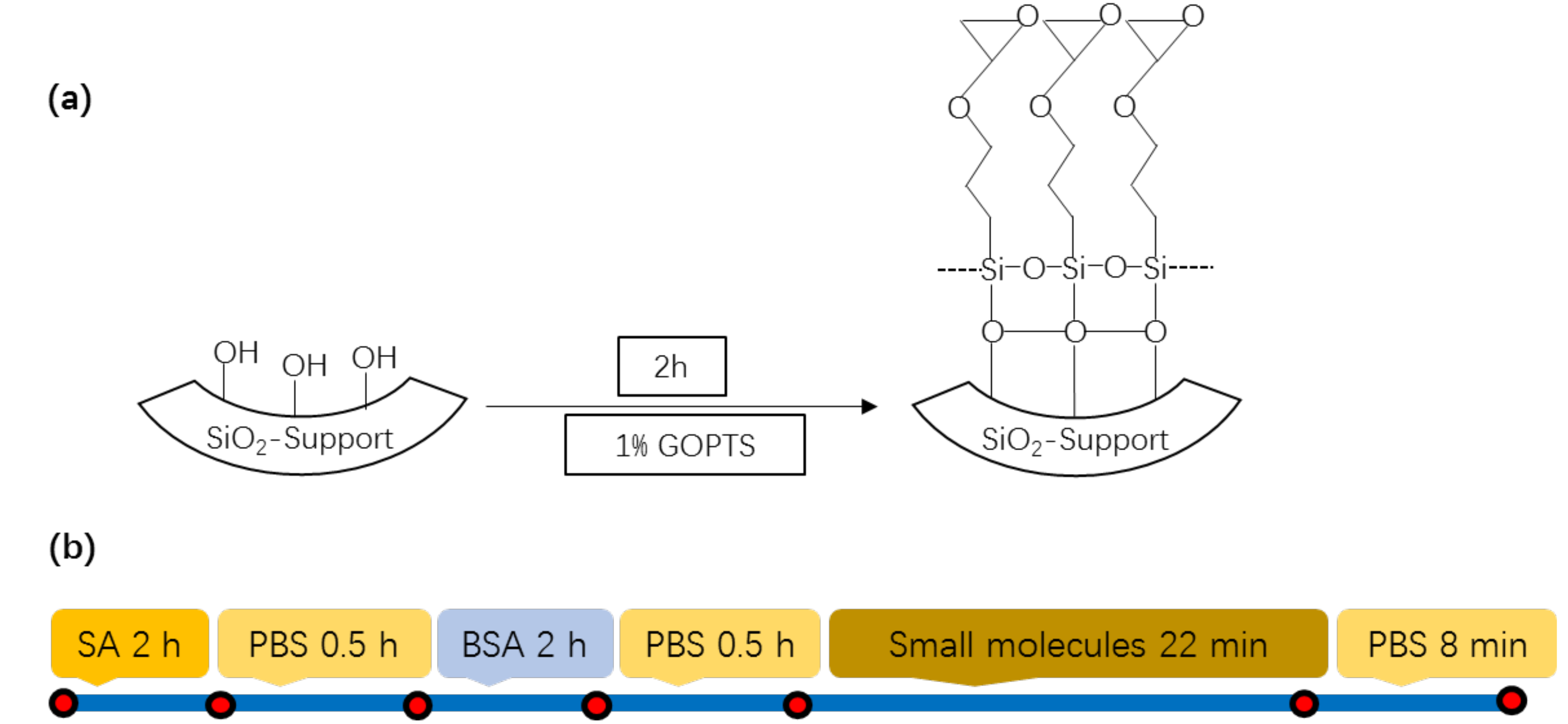
2.3. Detection of Small Molecule Binding to Surface-Immobilized Streptavidin
3. Results
3.1. Transmission Spectra and Q Factors of OFMBR Sensor
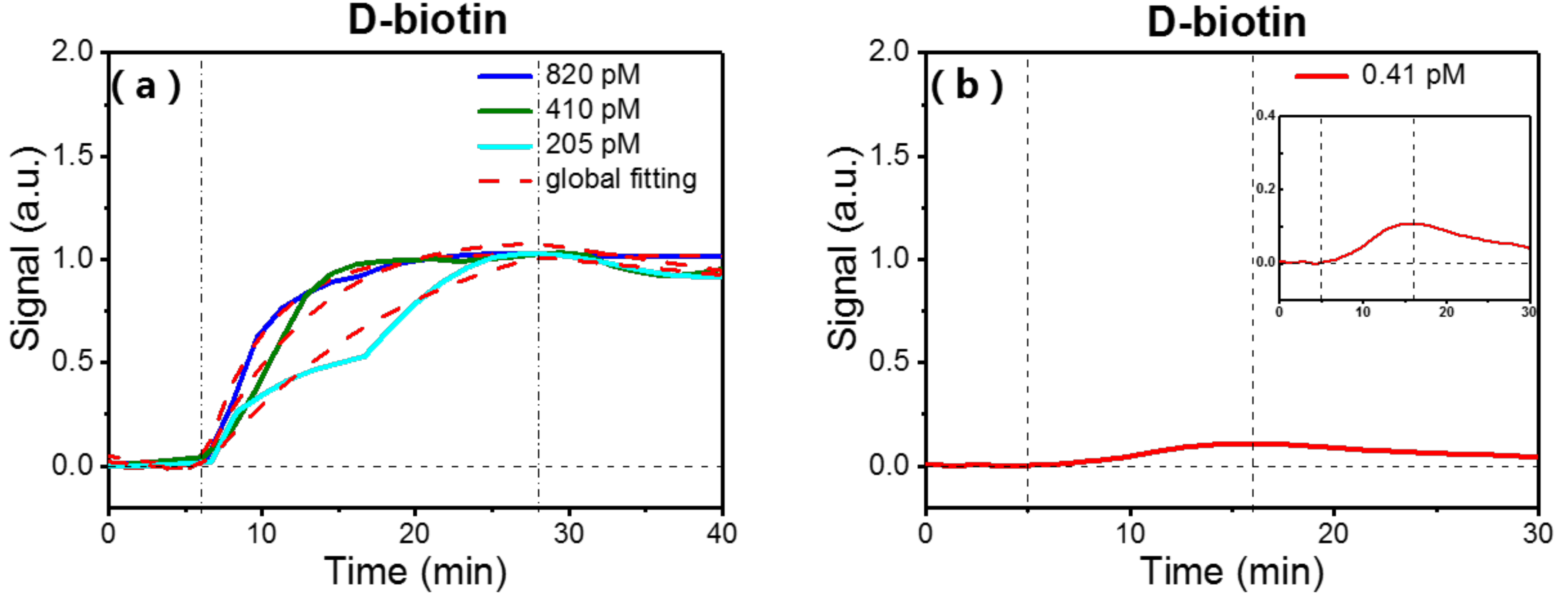
3.2. Binding Kinetics of Biotin to Surface-Immobilized Streptavidin
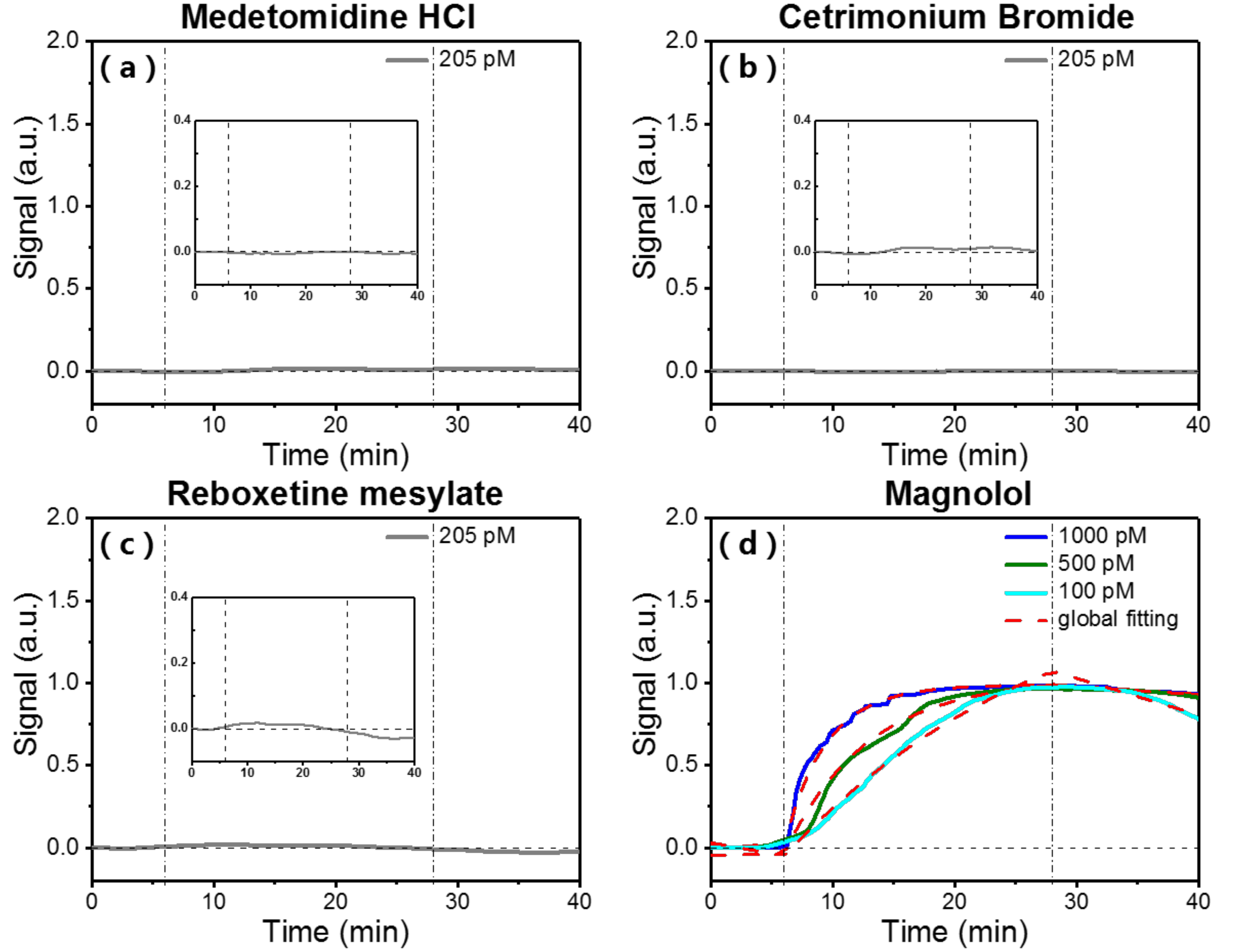
3.3. Specificity of Small Molecules Binding to Surface-Immobilized Streptavidin
4. Discussion and Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Zhang, J.; Yang, P.L.; Gray, N.S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 2009, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.U.; Chen, C.; Lin, K.H.; Fang, Y.; Lieber, C.M. Label-free detection of small-molecule—Protein interactions by using nanowire nanosensors. Proc. Natl. Acad. Sci. USA 2005, 102, 3208–3212. [Google Scholar] [CrossRef] [PubMed]
- Strausberg, R.L.; Schreiber, S.L. From knowing to controlling: A path from genomics to drugs using small molecule probes. Science 2003, 300, 294–295. [Google Scholar] [CrossRef] [PubMed]
- Kenakin, T.; Christopoulos, A. Signalling bias in new drug discovery: Detection, quantification and therapeutic impact. Nat. Rev. Drug Discov. 2013, 12, 205. [Google Scholar] [CrossRef] [PubMed]
- Holliger, P.; Hudson, P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 2005, 23, 1126. [Google Scholar] [CrossRef] [PubMed]
- Gooding, J.J. Biosensor technology for detecting biological warfare agents: Recent progress and future trends. Anal. Chim. Acta 2006, 559, 137–151. [Google Scholar] [CrossRef]
- Fitch, J.P.; Raber, E.; Imbro, D.R. Technology challenges in responding to biological or chemical attacks in the civilian sector. Science 2003, 302, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Anker, J.N.; Hall, W.P.; Lyandres, O.; Shah, N.C.; Zhao, J.; Van Duyne, R.P. Biosensing with plasmonic nanosensors. Nat. Mater. 2008, 7, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Ferrie, A.M.; Fontaine, N.H.; Mauro, J.; Balakrishnan, J. Resonant waveguide grating biosensor for living cell sensing. Biophys. J. 2006, 91, 1925–1940. [Google Scholar] [CrossRef] [PubMed]
- Owen, V. Real-time optical immunosensors—A commercial reality. Biosens. Bioelectron. 1997, 12, i–ii. [Google Scholar] [CrossRef]
- Daghestani, H.N.; Day, B.W. Theory and applications of surface plasmon resonance, resonant mirror, resonant waveguide grating, and dual polarization interferometry biosensors. Sensors 2010, 10, 9630–9646. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, F.; Yang, L. Review Label-free detection with high-Q microcavities: A review of biosensing mechanisms for integrated devices. Nanophotonics 2012, 1, 267–291. [Google Scholar] [CrossRef] [PubMed]
- Soria, S.; Berneschi, S.; Brenci, M.; Cosi, F.; Nunzi Conti, G.; Pelli, S.; Righini, G.C. Optical microspherical resonators for biomedical sensing. Sensors 2011, 11, 785–805. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wu, X.; Liu, L.; Fan, X.; Xu, L. Self-referencing optofluidic ring resonator sensor for highly sensitive biomolecular detection. Anal. Chem. 2013, 85, 9328–9332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, L.; Xu, L. Ultralow sensing limit in optofluidic micro-bottle resonator biosensor by self-referenced differential-mode detection scheme. Appl. Phys. Lett. 2014, 104, 033703. [Google Scholar] [CrossRef]
- Ren, L.; Wu, X.; Li, M.; Zhang, X.; Liu, L.; Xu, L. Ultrasensitive label-free coupled optofluidic ring laser sensor. Opt. Lett. 2012, 37, 3873–3875. [Google Scholar] [CrossRef] [PubMed]
- Arlett, J.L.; Myers, E.B.; Roukes, M.L. Comparative advantages of mechanical biosensors. Nat. Nanotechnol. 2011, 6, 203. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Dai, S.; Wang, L. Optical aptasensors for quantitative detection of small biomolecules: A review. Biosens. Bioelectron. 2014, 59, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Armani, A.M.; Kulkarni, R.P.; Fraser, S.E.; Flagan, R.C.; Vahala, K.J. Label-free, single-molecule detection with optical microcavities. Science 2007, 317, 783–787. [Google Scholar] [CrossRef] [PubMed]
- Berneschi, S.; Farnesi, D.; Cosi, F.; Conti, G.N.; Pelli, S.; Righini, G.C.; Soria, S. High Q silica microbubble resonators fabricated by arc discharge. Opt. Lett. 2011, 36, 3521–3523. [Google Scholar] [CrossRef] [PubMed]
- Cosci, A.; Quercioli, F.; Farnesi, D.; Berneschi, S.; Giannetti, A.; Cosi, F.; Barucci, A.; Conti, G.N.; Righini, G.; Pelli, S. Confocal reflectance microscopy for determination of microbubble resonator thickness. Opt. Express 2015, 23, 16693–16701. [Google Scholar] [CrossRef] [PubMed]
- Farnesi, D.; Barucci, A.; Righini, G.C.; Conti, G.N.; Soria, S. Generation of hyper-parametric oscillations in silica microbubbles. Opt. Lett. 2015, 40, 4508–4511. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.T.; Wu, X. Optical manipulation in optofluidic microbubble resonators. Sci. China Phys. Mech. Astron. 2015, 58, 114206. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, T.; Jiang, J.; Liu, K.; Yu, Z.; Chen, W.; Liu, W. Micro-bubble-based wavelength division multiplex optical fluidic sensing. In Proceedings of the International Society for Optics and Photonics, Beijing, China, 9–11 October 2014. [Google Scholar]
- Tang, T.; Wu, X.; Liu, L.; Xu, L. Packaged optofluidic microbubble resonators for optical sensing. Appl. Opt. 2016, 55, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; White, I.M.; Suter, J.D.; Dale, P.S.; Fan, X. Analysis of biomolecule detection with optofluidic ring resonator sensors. Opt. Express 2007, 15, 9139–9146. [Google Scholar]
- Srisa-Art, M.; Dyson, E.C.; de Mello, A.J.; Edel, J.B. Monitoring of real-time streptavidin—Biotin binding kinetics using droplet microfluidics. Anal. Chem. 2008, 80, 7063–7067. [Google Scholar] [CrossRef] [PubMed]
- Jung, L.S.; Nelson, K.E.; Stayton, P.S.; Campbell, C.T. Binding and dissociation kinetics of wild-type and mutant streptavidins on mixed biotin-containing alkylthiolate monolayers. Langmuir 2000, 16, 9421–9432. [Google Scholar] [CrossRef]
- Zhu, C.; Zhu, X.; Landry, J.P.; Cui, Z.; Li, Q.; Dang, Y.; Mi, L.; Zheng, F.; Fei, Y. Developing an efficient and general strategy for immobilization of small molecules onto microarrays using isocyanate chemistry. Sensors 2016, 16, 378. [Google Scholar] [CrossRef] [PubMed]

| Small Molecule | kon (min nM)−1 | koff (min)−1 | ka (M)−1 |
|---|---|---|---|
| Biotin | 0.30 | 4.5 × 10−7 | 6.7 × 1014 |
| Magnolol | 0.29 | 6.2 × 10−3 | 4.7 × 1010 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Zhu, C.; Guo, Z.; Wang, B.; Wu, X.; Fei, Y. Highly Sensitive Label-Free Detection of Small Molecules with an Optofluidic Microbubble Resonator. Micromachines 2018, 9, 274. https://doi.org/10.3390/mi9060274
Li Z, Zhu C, Guo Z, Wang B, Wu X, Fei Y. Highly Sensitive Label-Free Detection of Small Molecules with an Optofluidic Microbubble Resonator. Micromachines. 2018; 9(6):274. https://doi.org/10.3390/mi9060274
Chicago/Turabian StyleLi, Zihao, Chenggang Zhu, Zhihe Guo, Bowen Wang, Xiang Wu, and Yiyan Fei. 2018. "Highly Sensitive Label-Free Detection of Small Molecules with an Optofluidic Microbubble Resonator" Micromachines 9, no. 6: 274. https://doi.org/10.3390/mi9060274





