The Impact of Tumor Eco-Evolution in Renal Cell Carcinoma Sampling
Abstract
1. Introduction
2. Tumors as Dynamic Cell Communities Guided by Ecological Principles
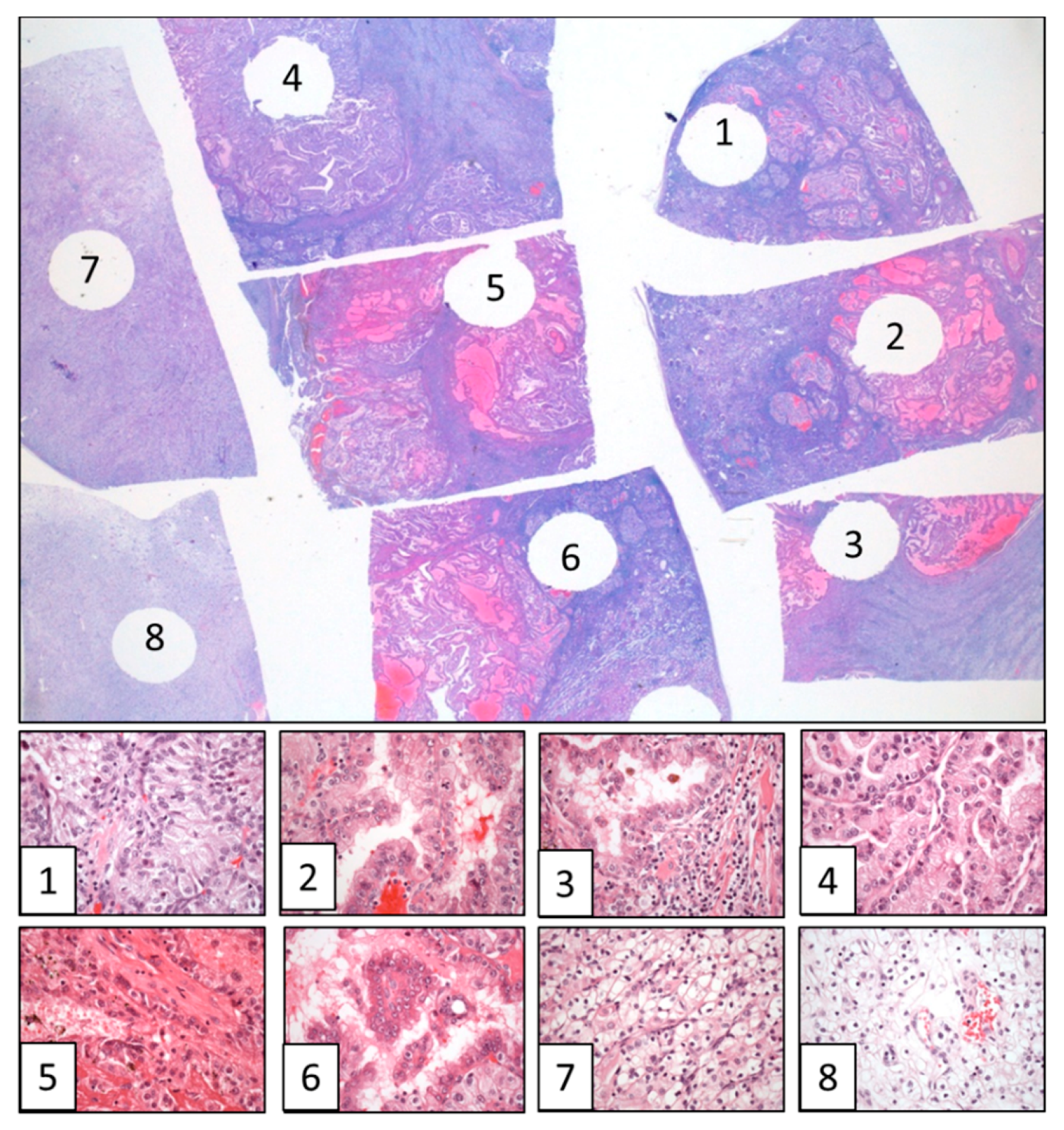
3. Spatial and Temporal Evolution in Clear Cell Renal Cell Carcinomas
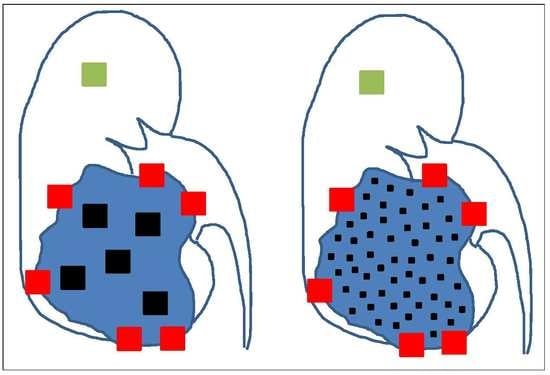
4. The Need for an Updated Tumor Sampling Adapted to Tumor Type
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Turajlic, S.; Swanton, C.; Boshoff, C. Kidney cancer: The next decade. J. Exp. Med. 2018, 215, 2477–2479. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidingen, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Primers 2017, 3, 17009. [Google Scholar] [CrossRef] [PubMed]
- Tannir, N.M.; Pal, S.K.; Atkins, M.B. Second-Line Treatment Landscape for Renal Cell Carcinoma: A Comprehensive Review. Oncologist 2018, 23, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Zhang, Q.; Sun, J. Cost-effectiveness of nivolumab plus ipilimumab as first-line therapy in advanced renal-cell carcinoma. J. Immunother. Cancer 2018, 6, 124. [Google Scholar] [CrossRef]
- Blumenfeld, A.J.; Guru, K.; Fuchs, G.J.; Kim, H.L. Percutaneous biopsy of renal cell carcinoma underestimates nuclear grade. Urology 2010, 76, 610–613. [Google Scholar] [CrossRef]
- Tomaszewski, J.J.; Uzzo, R.G.; Smaldone, M.C. Heterogeneity and renal mass biopsy: A review of its role and reliability. Cancer Biol. Med. 2014, 11, 162–172. [Google Scholar] [CrossRef]
- Beltrame, L.; di Martino, M.; Fruscio, R.; Calura, E.; Chapman, B.; Clivio, L.; Sina, F.; Mele, C.; Iatropoulos, P.; Grassi, T.; et al. Profiling cancer gene mutations in longitudinal epithelial ovarian cancer biopsies by targeted next-generation sequencing: A retrospective study. Ann. Oncol. 2015, 26, 1363–1371. [Google Scholar] [CrossRef]
- Bettoni, F.; Masotti, C.; Habr-Gama, A.; Correa, B.R.; Gama-Rodrigues, J.; Vianna, M.R.; Vailati, B.B.; Sao Juliao, G.P.; Fernandez, L.M.; Galante, P.A.; et al. Intratumoral genetic heterogeneity in rectal cancer. Are single biopsies representative of the entirety of the tumor? Ann. Surg. 2017, 265, e4–e6. [Google Scholar] [CrossRef]
- Ellsworth, R.E.; Blackburn, H.L.; Shriver, C.D.; Soon-Shiong, P.; Ellsworth, D.L. Molecular heterogeneity in breast cancer: State of the science and implications for patient care. Semin. Cell Dev. Biol. 2017, 64, 65–72. [Google Scholar] [CrossRef]
- Soultati, A.; Stares, M.; Swanton, C.; Larkin, J.; Turajlic, S. How should clinicians address intratumor heterogeneity in clear cell renal cell carcinoma? Curr. Opin. Urol. 2015, 25, 358–366. [Google Scholar] [CrossRef]
- Marusyk, A.; Almendro, V.; Polyak, K. Intra-tumour heterogeneity: A looking glass for cancer? Nat. Rev. Cancer 2012, 12, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Eng. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef]
- López, J.I.; Cortés, J.M. Multi-site tumor sampling (MSTS): A new tumor selection method to enhance intratumor heterogeneity detection. Hum. Pathol. 2017, 64, 1–6. [Google Scholar] [CrossRef] [PubMed]
- López, J.I.; Cortés, J.M. A divide and conquer strategy in tumor sampling enhances detection of intratumor heterogeneity in pathology routine: A modeling approach in clear cell renal cell carcinoma. F1000Research 2016, 5, 385. [Google Scholar] [CrossRef]
- De la Fuente, I.M. Elements of the cellular metabolic structure. Front. Mol. Biosci. 2015, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Diesboeck, T.S.; Couzin, I.D. Collective behavior in cancer cell populations. BioEssays 2009, 31, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Merlo, L.M.F.; Pepper, J.M.; Reid, B.J.; Maley, C.C. Cancer as an evolutionary and ecological process. Nat. Rev. Cancer 2006, 6, 924–935. [Google Scholar] [CrossRef]
- Horswell, S.; Matthews, N.; Swanton, C. Cancer heterogeneity and the “struggle for existence”: Diagnostic and analytical challenges. Cancer Lett. 2013, 340, 220–226. [Google Scholar] [CrossRef]
- Marcucci, F.; Stassi, G.; De Maria, R. Epithelial-mesenchymal transition: A new target in anticancer drug discovery. Nat. Rev. 2016, 15, 311–325. [Google Scholar] [CrossRef]
- Lopez, J.I.; Mosteiro, L.; Guarch, R.; Larrinaga, G.; Pulido, R.; Angulo, J.C. Low-grade metastases in high-grade clear cell renal cell carcinomas. A clinicopathologic study of 4 cases with an insight into the role of mesenchymal-to-epithelial transition process. Ann. Diagn. Pathol. 2016, 20, 13–18. [Google Scholar] [CrossRef]
- Nawaz, S.; Yuan, Y. Computational pathology: Exploring the spatial dimension of tumor ecology. Cancer Lett. 2016, 380, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Warsow, G.; Hübschmann, D.; Kleinheinz, K.; Nientiedt, C.; Heller, M.; Van Coile, L.; Tolstov, Y.; Trennheuser, L.; Wieczorek, K.; Pecqueux, C.; et al. Genomic features of renal cell carcinoma with venous tumor thrombus. Sci. Rep. 2018, 8, 7477. [Google Scholar] [CrossRef] [PubMed]
- López, J.I.; Pulido, R.; Lawrie, C.H.; Angulo, J.C. Loss of PD-L1 (SP-142) expression characterizes renal vein tumor thrombus microenvironment in clear cell renal cell carcinoma. Ann. Diagn. Pathol. 2018, 34, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Maley, C.C.; Koelble, K.; Natrajan, R.; Aktipis, A.; Yuan, Y. An ecological measure of immune-cancer colocalization as a prognostic factor for breast cancer. Breast Cancer Res. 2015, 17, 131. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J.Y.S.; Au, W.L.; Lo, K.Y.; Ni, Y.B.; Hlaing, T.; Hu, J.; Chan, S.K.; Chan, K.F.; Cheung, S.Y.; Tse, G.M. PD-L1 expression and tumor infiltrating PD-1+ lymphocytes associated with outcome in HER2+ breast cancer patients. Breast Cancer Res. Treat. 2017, 162, 19–30. [Google Scholar] [CrossRef]
- Chapman, A.; del Ama, L.F.; Ferguson, J.; Kamarashev, J.; Wellbrock, C.; Hurlstone, A. Heterogeneous tumor subpopulations cooperate to drive invasion. Cell Rep. 2014, 8, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Invest. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Hwang, R.F.; Moore, T.; Arumugam, T.; Ramachandran, V.; Amos, K.D.; Rivera, A.; Ji, B.; Evans, D.B.; Logsdon, C.D. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 2008, 68, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, H.; Cai, J.; Zhang, T.; Guo, J.; Feng, D.; Wang, Z. Ovarian cancer-associated fibroblasts contribute to epithelial ovarian carcinoma metastasis by promoting angiogenesis, lymphangiogenesis and tumor cell invasion. Cancer Lett. 2011, 303, 47–55. [Google Scholar] [CrossRef]
- López, J.I.; Errarte, P.; Erramuzpe, A.; Guarch, R.; Cortes, J.M.; Angulo, J.C.; Pulido, R.; Irazusta, J.; Llarena, R.; Larrinaga, G. Fibroblast activation protein predicts prognosis in clear cell renal cell carcinoma. Hum. Pathol. 2016, 54, 100–105. [Google Scholar] [CrossRef]
- Errarte, P.; Guarch, R.; Pulido, R.; Blanco, L.; Nunes-Xavier, C.E.; Beitia, M.; Gil, J.; Angulo, J.C.; Lopez, J.I.; Larrinaga, G. The expression of fibroblast activation protein in clear cell renal cell carcinomas is associated with synchronous lymph node metastases. PLoS ONE 2016, 11, e0169105. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Sanchez, A.; Memon, D.; Pourpe, S.; Veeraraghavan, H.; Li, Y.; Vargas, H.A.; Gill, M.B.; Park, K.J.; Zivanovic, O.; Konner, J.; et al. Heterogeneous tumor-immune microenvironments among differentially growing metastases in an ovarian cancer patient. Cell 2017, 170, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Pavlides, S.; Whitaker-Menezes, D.; Castello-Cros, H.; Flomenberg, N.; Vitkiewicz, A.K.; Frank, P.G.; Casimiro, M.C.; Wang, C.; Fortina, P.; Addya, S.; et al. The reverse Warburg effect: Aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle 2009, 8, 3984–4001. [Google Scholar] [CrossRef] [PubMed]
- Marusyk, A.; Polyak, K. Tumor heterogeneity: Causes and consequences. Biochim. Biophys. Acta 2010, 1805, 105–117. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Swanton, C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015, 27, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, T.J.; Turajlic, S.; Rowan, A.; Nicol, D.; Farmery, J.H.R.; O’Brien, T.; Martincorena, I.; Tarpey, P.; Angelopoulos, N.; Yates, L.; et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx Renal. Cell 2018, 173, 611–623. [Google Scholar] [CrossRef]
- Davis, A.; Gao, R.; Navin, N. Tumor evolution: Linear, branching, neutral or punctuated? Biochim. Biophys. Acta 2017, 1867, 151–161. [Google Scholar] [CrossRef]
- Turajlic, S.; Xu, H.; Litchfield, K.; Rowan, A.; Horswell, S.; Chambers, T.; O’Brien, T.; Lopez, J.I.; Watkins, T.B.K.; Nicol, N.; et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx Renal. Cell 2018, 173, 595–610. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Gerlinger, M.; Horswell, S.; Larkin, A.J.; Rowan, A.J.; Salm, M.P.; Varela, I.; Fisher, R.; McGranahan, N.; Matthews, N.; Santos, C.R.; et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014, 46, 225–233. [Google Scholar] [CrossRef]
- Sottoriva, A.; Kang, H.; Ma, Z.; Graham, T.A.; Salomon, M.P.; Zhao, J.; Marjoram, P.; Siegmund, K.; Press, M.F.; Shibata, D.; et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 2015, 47, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Turajlic, S.; Xu, H.; Litchfied, K.; Rowan, A.; Chambers, T.; Lopez, J.I.; Nicol, D.; O’Brien, T.; Larkin, J.; Horswell, S.; et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx Renal. Cell 2018, 173, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Arnal, A.; Ujvari, B.; Crespi, B.; Gatenby, R.A.; Tissot, T.; Vittecoq, M.; Ewald, P.W.; Casali, A.; Ducasse, H.; Jacqueline, C.; et al. Evolutionary perspective of cancer: Myth, metaphors, and reality. Evol. Appl. 2015, 8, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Pepper, J.W. Darwinian Strategies to Avoid the Evolution of Drug Resistance during Cancer Treatment. Evolutionary Thinking in Medicine; Alvergne, A., Ed.; Springer: Cham, Switzerland, 2016; pp. 167–176. [Google Scholar]
- Lloyd, M.C.; Cunningham, J.J.; Bui, M.M.; Gillies, R.J.; Brown, J.S.; Gatenby, R.A. Darwinian dynamics of intratumoral heterogeneity: Not solely random mutations but also variable environmental selection forces. Cancer Res. 2016, 76, 3136–3144. [Google Scholar] [CrossRef] [PubMed]
- Trpkov, K.; Grignon, D.J.; Bonsib, S.M.; Amin, M.B.; Billis, A.; Lopez-Beltran, A.; Samaratunga, H.; Tamboli, P.; Delahunt, B.; Egevad, L.; et al. Handling and staging of renal cell carcinoma: The International Society of Urological Pathology Consensus (ISUP) conference recommendations. Am. J. Surg. Pathol. 2013, 37, 1505–1517. [Google Scholar] [CrossRef] [PubMed]
- Cormen, T.H.; Leiserson, C.E.; Rivest, R.L.; Stein, C. Introduction to Algorithms, 2nd ed.; MIT Press: Cambridge, MA, USA, 2001. [Google Scholar]
- Ming, D.; Yang, W. A divide and conquer strategy to improve diffusion sampling in generalized ensemble simulators. J. Chem. Phys. 2008, 128, 094106. [Google Scholar] [CrossRef]
- Eisenstein, M. Cell sorting: Divide and conquer. Nature 2006, 441, 1179–1185. [Google Scholar] [CrossRef]
- Kristensen, V.N. Divide and conquer: The genetic basis of molecular subclassification of breast cancer. EMBO Mol. Med. 2011, 3, 183–185. [Google Scholar] [CrossRef]
- Guarch, R.; Cortés, J.M.; Lawrie, C.H.; Lopez, J.I. Multi-site tumor sampling (MSTS) significantly improves the performance of histological detection of intratumor heterogeneity in clear cell renal cell carcinoma. F1000Research 2016, 5, 2020. [Google Scholar] [CrossRef]
- López, J.I.; Cortés, J.M. A multi-site cutting device implements efficiently the divide-and-conquer strategy in tumor sampling. F1000Research 2016, 5, 1587. [Google Scholar] [CrossRef]
- Cortés, J.M.; de Petris, G.; López, J.I. Detection of intratumor heterogeneity in modern pathology: A multisite tumor sampling perspective. Front. Med. 2017, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Erramuzpe, A.; Cortés, J.M.; López, J.I. Multisite tumor sampling enhances the detection of intratumor heterogeneity at all different temporal stages of tumor evolution. Virchows Arch. 2018, 472, 187–194. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Swanton, C. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Fernández, E.; López, J.I. The Impact of Tumor Eco-Evolution in Renal Cell Carcinoma Sampling. Cancers 2018, 10, 485. https://doi.org/10.3390/cancers10120485
López-Fernández E, López JI. The Impact of Tumor Eco-Evolution in Renal Cell Carcinoma Sampling. Cancers. 2018; 10(12):485. https://doi.org/10.3390/cancers10120485
Chicago/Turabian StyleLópez-Fernández, Estíbaliz, and José I. López. 2018. "The Impact of Tumor Eco-Evolution in Renal Cell Carcinoma Sampling" Cancers 10, no. 12: 485. https://doi.org/10.3390/cancers10120485
APA StyleLópez-Fernández, E., & López, J. I. (2018). The Impact of Tumor Eco-Evolution in Renal Cell Carcinoma Sampling. Cancers, 10(12), 485. https://doi.org/10.3390/cancers10120485