Any Place for Immunohistochemistry within the Predictive Biomarkers of Treatment in Lung Cancer Patients?
Abstract
:1. Introduction
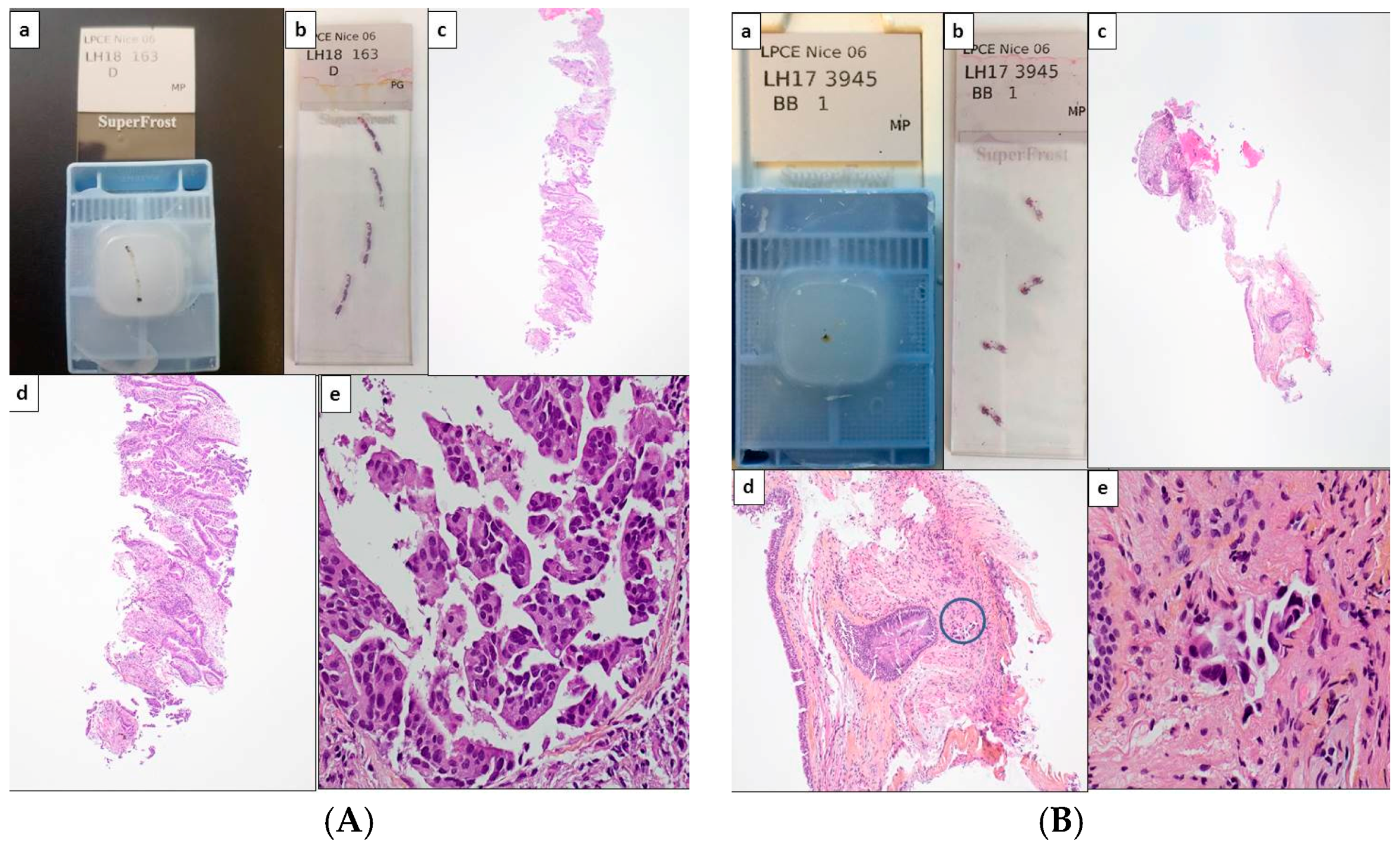
2. Tissue and Cell Samples and the Management of Daily Practice
3. The Therapeutic Targets Identified by Immunohistochemistry in Thoracic Oncology
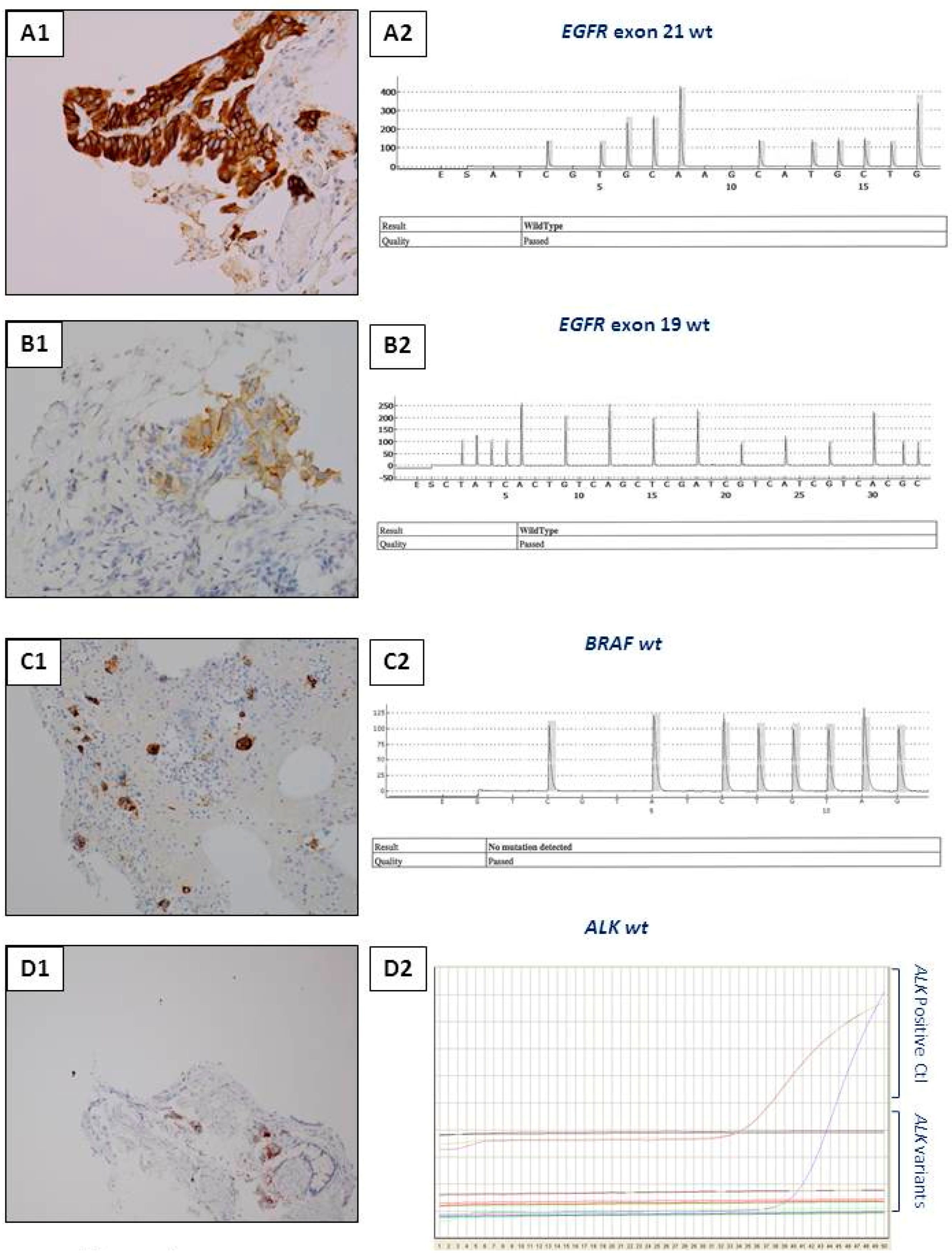
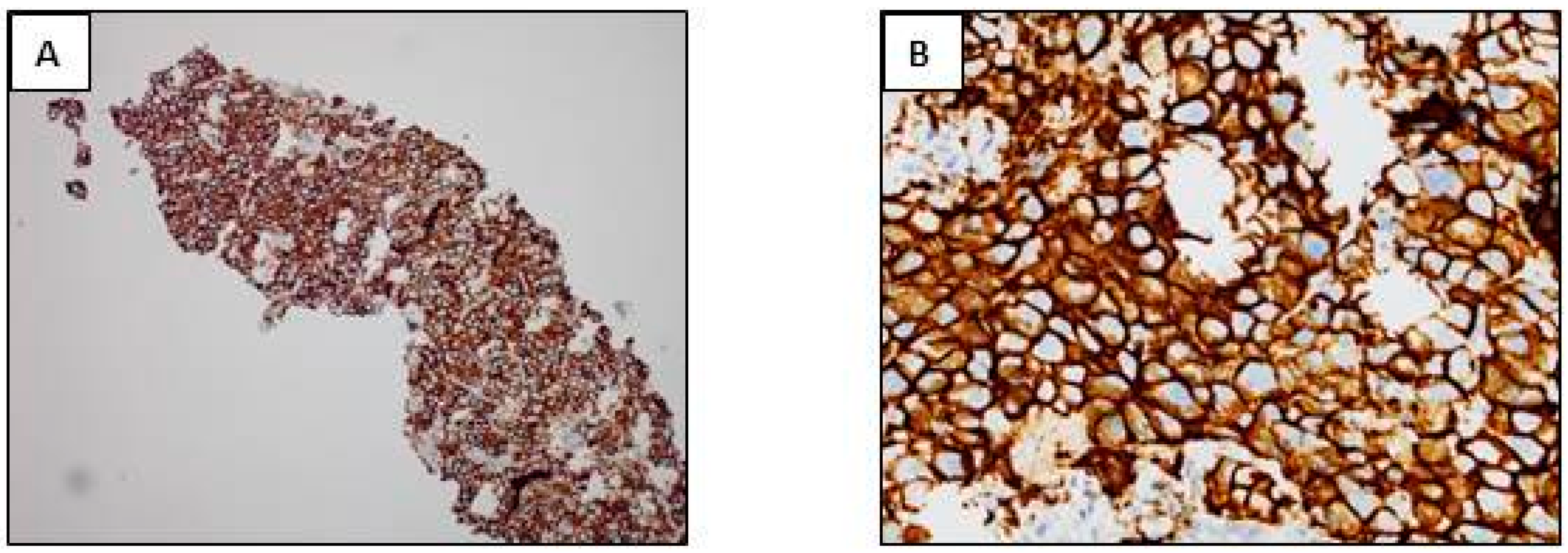
3.1. Anti-ALK Antibodies
3.2. Anti-ROS1 Antibodies
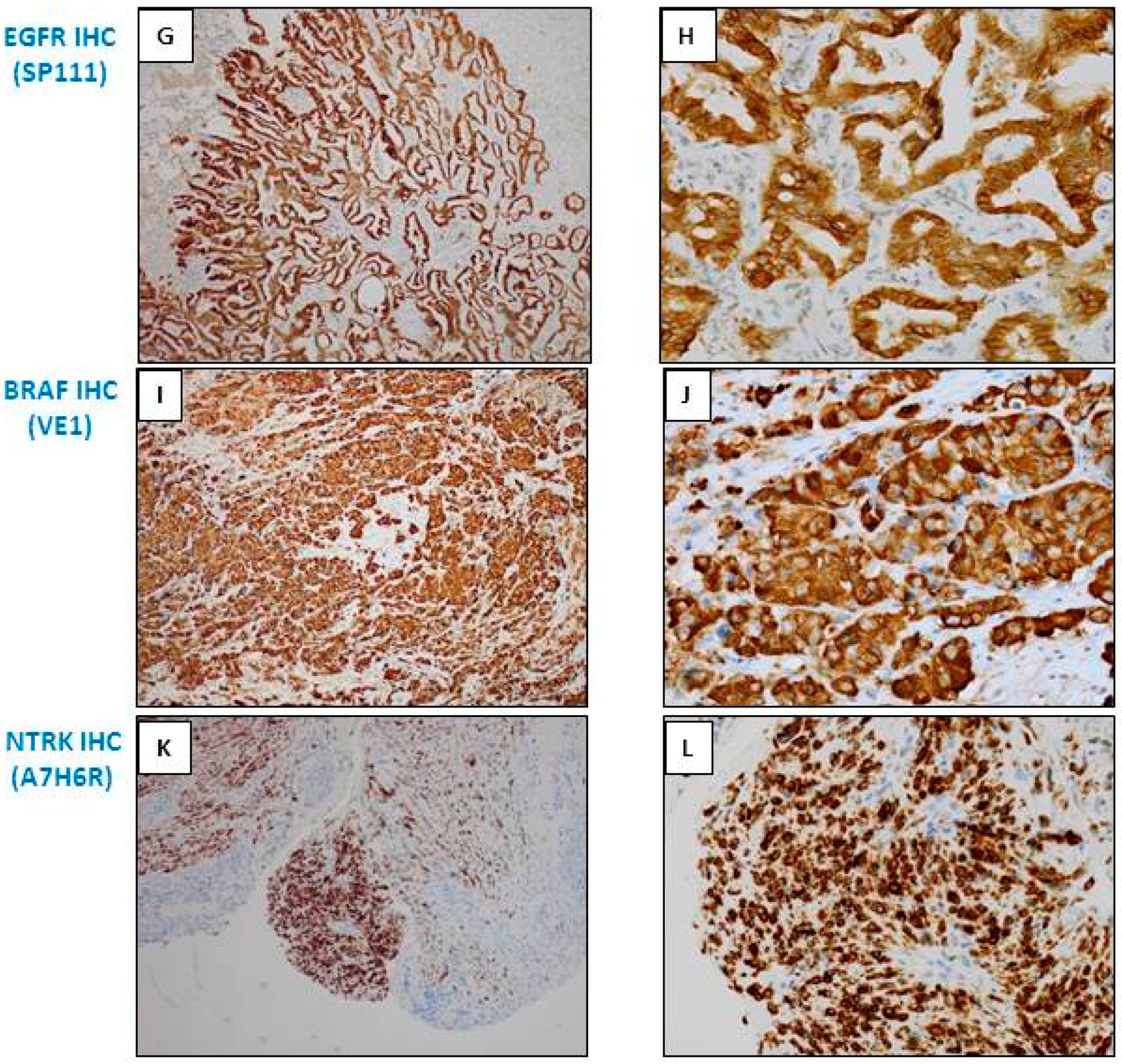
3.3. Anti-EGFR Mutated Antibodies
3.4. Anti-BRAF Antibodies
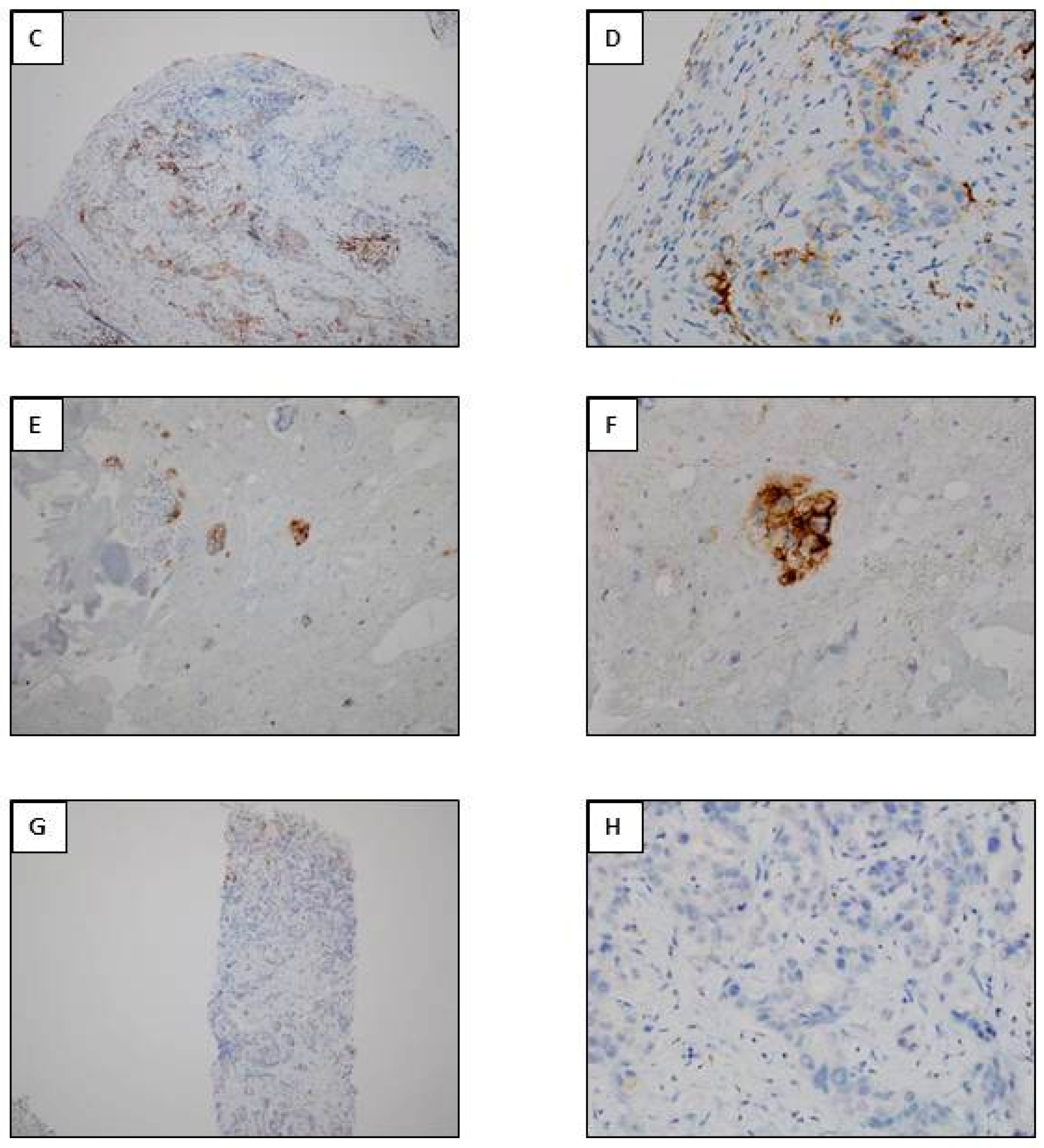
3.5. Anti-PD-L1 Antibodies
3.6. Anti-NTRK Antibodies
3.7. Anti-RET Antibodies
3.8. Anti-MET Antibodies
3.9. Anti-ERCC1 Antibodies
3.10. Anti-DLL3 Antibodies
3.11. Other Antibodies
4. Integration of Molecular and Immunohistochemical Approaches: What Is the Future?
5. Which Algorithms? For Which Patients? For Which Samples?
6. Perspectives and Conclusions
Acknowledgments
Conflicts of Interest
References
- Hoseok, I.; Cho, J.Y. Lung Cancer Biomarkers. Adv. Clin. Chem. 2015, 72, 107–170. [Google Scholar]
- Lewis, C.; McQuaid, S.; Hamilton, P.W.; Salto-Tellez, M.; McArt, D.; James, J.A. Building a ‘Repository of Science’: The importance of integrating biobanks within molecular pathology programmes. Eur. J. Cancer 2016, 67, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Oberndorfer, F.; Müllauer, L. Molecular pathology of lung cancer: Current status and perspectives. Curr. Opin. Oncol. 2017, 30, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L. Molecular diagnostics of lung cancer in the clinic. Transl. Lung Cancer Res. 2017, 6, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Aisner, D.L.; Sholl, L.M.; Berry, L.; Rossi, M.; Chen, H.; Fujimoto, J.; Moreira, A.L.; Ramalingam, S.; Villaruz, L.C.; Otterson, G.A.; et al. The Impact of Smoking and TP53 mutations in lung adenocarcinoma patients with targetable mutations—The Lung Cancer Mutation Consortium (LCMC2). Clin. Cancer Res. 2018, 24, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Barlesi, F.; Mazieres, J.; Merlio, J.P.; Debieuvre, D.; Mosser, J.; Lena, H.; Ouafik, L.; Besse, B.; Rouquette, I.; Westeel, V.; et al. Biomarkers France contributors. Routine molecular profiling of patients with advanced non-small-cell lung cancer: Results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016, 387, 1415–1426. [Google Scholar] [CrossRef]
- Mascaux, C.; Tomasini, P.; Greillier, L.; Barlesi, F. Personalised medicine for nonsmall cell lung cancer. Eur. Respir. Rev. 2017, 26, 170066. [Google Scholar] [PubMed]
- Osmani, L.; Askin, F.; Gabrielson, E.; Li, Q.K. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): Moving from targeted therapy to immunotherapy. Semin. Cancer Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Cree, I.A.; Deans, Z.; Ligtenberg, M.J.; Normanno, N.; Edsjö, A.; Rouleau, E.; Solé, F.; Thunnissen, E.; Timens, W.; Schuuring, E.; et al. European Society of Pathology Task Force on Quality Assurance in Molecular Pathology; Royal College of Pathologists. Guidance for laboratories performing molecular pathology for cancer patients. J. Clin. Pathol. 2014, 67, 923–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, D.; Ames, D.; Lopez, B.; Still, R.; Simpson, W.; Twomey, P. External quality assessment: Best practice. J. Clin. Pathol. 2014, 67, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Tembuyser, L.; Dequeker, E.M. Endorsing good quality assurance practices in molecular pathology: Risks and recommendations for diagnostic laboratories and external quality assessment providers. Virchows Arch. 2016, 468, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Vermaelen, K.; Waeytens, A.; Kholmanskikh, O.; van den Bulcke, M.; van Valckenborgh, E. Perspectives on the integration of Immuno-Oncology Biomarkers and drugs in a Health Care setting. Semin. Cancer Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S.S.; Harris, K. Bronchoscopy for the diagnosis of peripheral lung lesions. J. Thorac. Dis. 2017, 9, S1047–S1058. [Google Scholar] [CrossRef] [PubMed]
- Labarca, G.; Aravena, C.; Ortega, F.; Arenas, A.; Majid, A.; Folch, E.; Mehta, H.J.; Jantz, M.A.; Fernandez-Bussy, S. Minimally invasive methods for staging in lung cancer: Systematic review and meta-analysis. Pulm. Med. 2016, 2016, 1024709. [Google Scholar] [CrossRef] [PubMed]
- Mudambi, L.; Ost, D.E. Advanced bronchoscopic techniques for the diagnosis of peripheral pulmonary lesions. Curr. Opin. Pulm. Med. 2016, 22, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, R.W. Bronchoscopic pursuit of the peripheral pulmonary lesion: Navigational bronchoscopy, radial endobronchial ultrasound, and ultrathin bronchoscopy. Curr. Opin. Pulm. Med. 2016, 22, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Bubendorf, L.; Lantuejoul, S.; de Langen, A.J.; Thunnissen, E. Nonsmall cell lung carcinoma: Diagnostic difficulties in small biopsies and cytological specimens: Number 2 in the Series “Pathology for the clinician” Edited by Peter Dorfmüller and Alberto Cavazza. Eur. Respir. Rev. 2017, 26, 170007. [Google Scholar] [CrossRef] [PubMed]
- Long, É.; Ilie, M.; Hofman, V.; Lassalle, S.; Butori, C.; Alsubaie, S.; Hofman, P. Role of the surgical pathologist for tissue management in oncology. Bull. Cancer 2013, 100, 837–845. [Google Scholar] [PubMed]
- Bussolati, G.; Annaratone, L.; Maletta, F. The pre-analytical phase in surgical pathology. Recent Results Cancer Res. 2015, 199, 1–13. [Google Scholar] [PubMed]
- Hofman, V.; Ilie, M.; Gavric-Tanga, V.; Lespinet, V.; Mari, M.; Lassalle, S.; Butori, C.; Coelle, C.; Bordone, O.; Selva, E.; et al. Role of the surgical pathology laboratory in the pre-analytical approach of molecular biology techniques. Ann. Pathol. 2010, 30, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Engel, K.B.; Moore, H.M. Effects of preanalytical variables on the detection of proteins by immunohistochemistry in formalin-fixed, paraffin-embedded tissue. Arch. Pathol. Lab. Med. 2011, 135, 537–543. [Google Scholar] [PubMed]
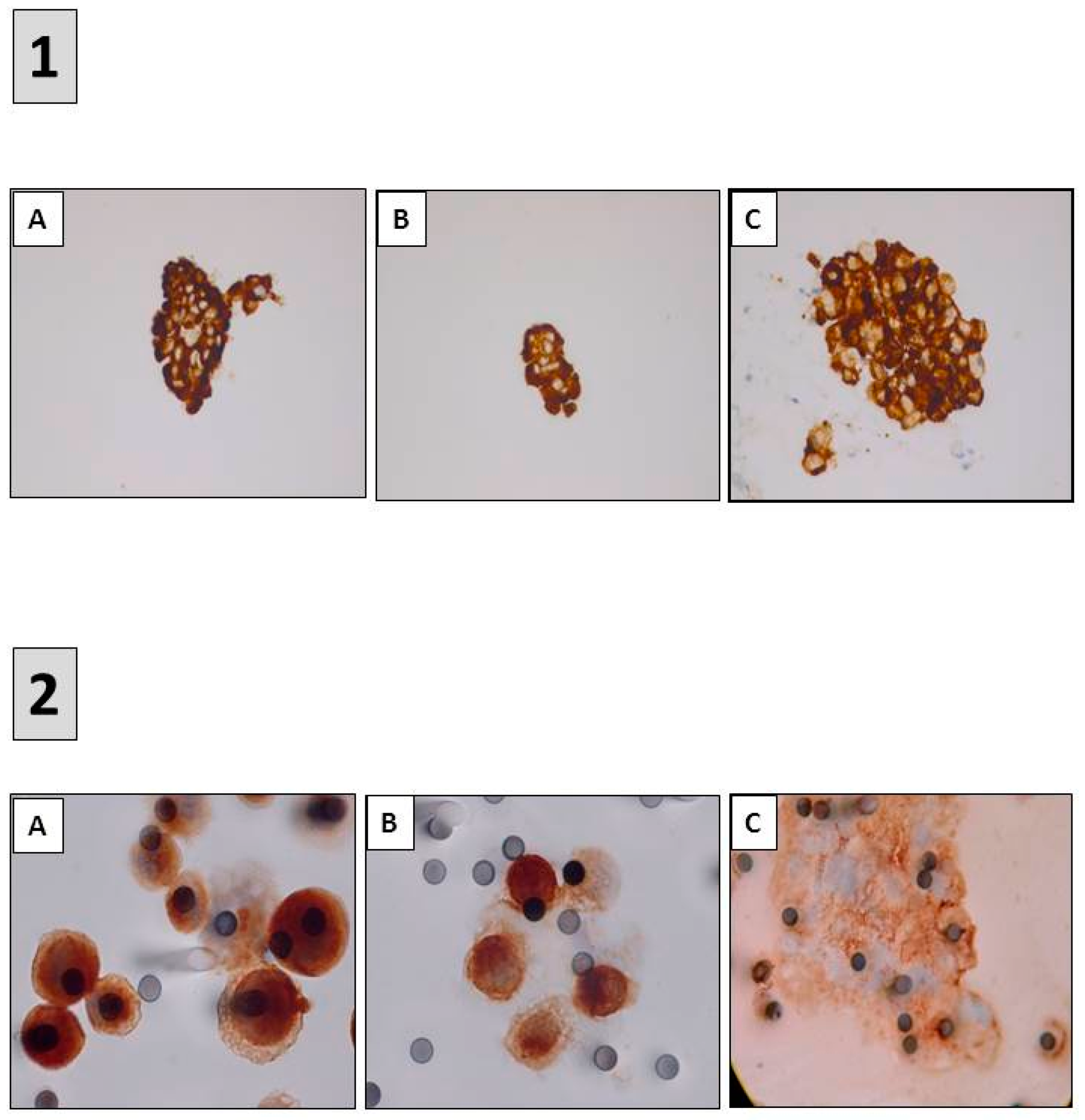
- Roy-Chowdhuri, S.; Aisner, D.L.; Allen, T.C.; Beasley, M.B.; Borczuk, A.; Cagle, P.T.; Capelozzi, V.; Dacic, S.; da Cunha Santos, G.; Hariri, L.P.; et al. Biomarker Testing in Lung Carcinoma Cytology Specimens: A Perspective From Members of the Pulmonary Pathology Society. Arch. Pathol. Lab. Med. 2016, 140, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Hofman, P. Pitfalls in lung cancer molecular pathology: How to limit them in routine practice? Curr. Med. Chem. 2012, 19, 2638–2651. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Li, H.; Zhou, J.; Zhang, W.; Wu, C. The value of cell block based on fine needle aspiration for lung cancer diagnosis. J. Thorac. Dis. 2017, 9, 2375–2382. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Juco, J.; Huang, L.; Hofman, V.; Khambata-Ford, S.; Hofman, P. Use of the 22C3 anti-programmed death ligand 1 antibody to determine programmed death ligand 1 expression in cytology samples obtained from non-small cell lung cancer patients. Cancer Cytopathol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Russell-Goldman, E.; Kravets, S.; Dahlberg, S.E.; Sholl, L.M.; Vivero, M. Cytologic-histologic correlation of programmed death-ligand 1 immunohistochemistry in lung carcinomas. Cancer Cytopathol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yao, H.; Zhao, J.; Zhang, S.; You, Q.; Sun, K.; Zou, Y.; Zhou, C.; Zhou, J. Cell block samples from malignant pleural effusion might be valid alternative samples for anaplastic lymphoma kinase detection in patients with advanced non-small-cell lung cancer. Histopathology 2015, 66, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P. PD-L1 immunohistochemistry for non-small cell lung carcinoma: Which strategy should be adopted? Expert Rev. Mol. Diagn. 2017. [Google Scholar] [CrossRef] [PubMed]
- Zhiwei, W.; Yuan, J.; Yihui, Y.; Xin, H.; Jingtao, C.; Lei, S.; Yongjian, D. Ventana immunohistochemistry assay for anaplastic lymphoma kinase gene rearrangement detection in patients with non-small cell lung cancer: A meta-analysis. Thorac. Cancer 2017, 8, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Conde, E.; Hernandez, S.; Prieto, M.; Martinez, R.; Lopez-Rios, F. Profile of Ventana ALK (D5F3) companion diagnostic assay for non-small-cell lung carcinomas. Expert Rev. Mol. Diagn. 2016, 16, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Thorne-Nuzzo, T.; Williams, C.; Catallini, A.; Clements, J.; Singh, S.; Amberson, J.; Dickinson, K.; Gatalica, Z.; Ho, S.N.; Loftin, I.; et al. A Sensitive ALK Immunohistochemistry Companion Diagnostic Test Identifies Patients Eligible for Treatment with Crizotinib. J. Thorac. Oncol. 2017, 12, 804–813. [Google Scholar] [CrossRef] [PubMed]
- Vinciguerra, G.L.R.; Scarpino, S.; Pini, B.; Cippitelli, C.; Fochetti, F.; Ruco, L. Optimized immunohistochemistry using the D5F3 antibody provides a reliable test for identification of ALK-positive lung adenocarcinomas. Virchows Arch. 2017, 47, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P.; Ilie, M.; Hofman, V.; Roux, S.; Valent, A.; Bernheim, A.; Alifano, M.; Leroy-Ladurie, F.; Vaylet, F.; Rouquette, I.; et al. Immunohistochemistry to identify EGFR mutations or ALK rearrangements in patients with lung adenocarcinoma. Ann. Oncol. 2012, 23, 1738–1743. [Google Scholar] [CrossRef] [PubMed]
- Von Laffert, M.; Schirmacher, P.; Warth, A.; Weichert, W.; Büttner, R.; Huber, R.M.; Wolf, J.; Griesinger, F.; Dietel, M.; Grohé, C. ALK-Testing in non-small cell lung cancer (NSCLC): Immunohistochemistry (IHC) and/or fluorescence in-situ Hybridisation (FISH)?: Statement of the Germany Society for Pathology (DGP) and the Working Group Thoracic Oncology (AIO) of the German Cancer Society e.V. (Stellungnahme der Deutschen Gesellschaft für Pathologie und der AG Thorakale Onkologie der Arbeitsgemeinschaft Onkologie/Deutsche Krebsgesellschaft e.V.). Lung Cancer 2017, 103, 1–5. [Google Scholar] [PubMed]
- Ibrahim, M.; Parry, S.; Wilkinson, D.; Bilbe, N.; Allen, D.; Forrest, S.; Maxwell, P.; O’Grady, A.; Starczynski, J.; Tanier, P.; et al. ALK Immunohistochemistry in NSCLC: Discordant staining can impact patient treatment regimen. J. Thorac. Oncol. 2016, 11, 2241–2247. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.I.; Bence, C.; Hofman, V.; Long-Mira, E.; Butori, C.; Bouhlel, L.; Lalvée, S.; Mouroux, J.; Poudenx, M.; Otto, J.; et al. Discrepancies between FISH and immunohistochemistry for assessment of the ALK status are associated with ALK ‘borderline’-positive rearrangements or a high copy number: A potential major issue for anti-ALK therapeutic strategies. Ann. Oncol. 2015, 26, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Rosoux, A.; Pauwels, P.; Duplaquet, F.; D’Haene, N.; Weynand, B.; Delos, M.; Menon, R.; Heukamp, L.C.; Thunnissen, E.; Ocak, S. Effectiveness of crizotinib in a patient with ALK IHC-positive/FISH-negative metastatic lung adenocarcinoma. Lung Cancer 2016, 98, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Trejo Bittar, H.E.; Luvison, A.; Miller, C.; Dacic, S. A comparison of ALK gene rearrangement and ALK protein expression in primary lung carcinoma and matched metastasis. Histopathology 2017, 71, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Rosenblum, F.; Hutchinson, L.M.; Garver, J.; Woda, B.; Cosar, E.; Kurian, E.M. Cytology specimens offer an effective alternative to formalin-fixed tissue as demonstrated by novel automated detection for ALK break-apart FISH testing and immunohistochemistry in lung adenocarcinoma. Cancer Cytopathol. 2014, 122, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Pisapia, P.; Lozano, M.D.; Vigliar, E.; Bellevicine, C.; Pepe, F.; Malapelle, U.; Troncone, G. ALK and ROS1 testing on lung cancer cytologic samples: Perspectives. Cancer Cytopathol. 2017, 125, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Bubendorf, L.; Büttner, R.; Al-Dayel, F.; Dietel, M.; Elmberger, G.; Kerr, K.; López-Ríos, F.; Marchetti, A.; Öz, B.; Pauwels, P.; et al. Testing for ROS1 in non-small cell lung cancer: A review with recommendations. Virchows Arch. 2016, 469, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Pal, P.; Khan, Z. ROS1-1. J. Clin. Pathol. 2017, 70, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Goncalves, T.; Dias-Santagata, D.; Hoang, M.P. Immunohistochemical Detection of ROS1 Fusion. Am. J. Clin. Pathol. 2017, 147, 77–82. [Google Scholar] [PubMed]
- Viola, P.; Maurya, M.; Croud, J.; Gazdova, J.; Suleman, N.; Lim, E.; Newsom-Davis, T.; Plowman, N.; Rice, A.; Montero, M.A.; et al. A Validation Study for the Use of ROS1 Immunohistochemical Staining in Screening for ROS1 Translocations in Lung Cancer. J. Thorac. Oncol. 2016, 11, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Selinger, C.I.; Li, B.T.; Pavlakis, N.; Links, M.; Gill, A.J.; Lee, A.; Clarke, S.; Tran, T.N.; Lum, T.; Yip, P.Y.; et al. Screening for ROS1 gene rearrangements in non-small-cell lung cancers using immunohistochemistry with FISH confirmation is an effective method to identify this rare target. Histopathology 2017, 70, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Allo, G.; Bandarchi, B.; Yanagawa, N.; Wang, A.; Shih, W.; Xu, J.; Dalby, M.; Nitta, H.; To, C.; Liu, N.; et al. Epidermal growth factor receptor mutation-specific immunohistochemical antibodies in lung adenocarcinoma. Histopathology 2014, 64, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Bondgaard, A.L.; Høgdall, E.; Mellemgaard, A.; Skov, B.G. High specificity but low sensitivity of mutation-specific antibodies against EGFR mutations in non-small-cell lung cancer. Mod. Pathol. 2014, 27, 1590–1598. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Liu, B.; Xu, H.; Yu, B.; Shi, S.; Zhang, J.; Wang, X.; Wang, J.; Lu, Z.; Ma, H.; et al. Immunostaining with EGFR mutation-specific antibodies: A reliable screening method for lung adenocarcinomas harboring EGFR mutation in biopsy and resection samples. Hum. Pathol. 2013, 44, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Houang, M.; Sioson, L.; Clarkson, A.; Watson, N.; Farzin, M.; Toon, C.W.; Raut, A.; O’Toole, S.A.; Cooper, W.A.; Pavlakis, N.; et al. EGFR mutation specific immunohistochemistry is a useful adjunct which helps to identify false negative mutation testing in lung cancer. Pathology 2014, 46, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H.; Kim, S.H.; Park, S.Y.; Yoo, J.; Kim, S.K.; Kim, H.K. Identification of EGFR Mutations by Immunohistochemistry with EGFR Mutation-Specific Antibodies in Biopsy and Resection Specimens from Pulmonary Adenocarcinoma. Cancer Res. Treat. 2015, 47, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Seo, A.N.; Park, T.I.; Jin, Y.; Sun, P.L.; Kim, H.; Chang, H.; Chung, J.H. Novel EGFR mutation-specific antibodies for lung adenocarcinoma: Highly specific but not sensitive detection of an E746_A750 deletion in exon 19 and an L858R mutation in exon 21 by immunohistochemistry. Lung Cancer 2014, 83, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.H.; Brogi, E.; Hasanovic, A.; Ladanyi, M.; Soslow, R.A.; Chitale, D.; Shia, J.; Moreira, A.L. Immunohistochemical staining with EGFR mutation-specific antibodies: High specificity as a diagnostic marker for lung adenocarcinoma. Mod. Pathol. 2013, 26, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Verdu, M.; Trias, I.; Roman, R.; Rodon, N.; Pubill, C.; Arraiza, N.; Martinez, B.; Garcia-Pelaez, B.; Serrano, T.; Puig, X. Cross-reactivity of EGFR mutation-specific immunohistochemistry assay in HER2-positive tumors. Appl. Immunohistochem. Mol. Morphol. 2015, 23, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Butori, C.; Lassalle, S.; Heeke, S.; Piton, N.; Sabourin, J.C.; Tanga, V.; Washetine, K.; Long-Mira, E.; Maitre, P.; et al. Optimization of EGFR mutation detection by the fully-automated qPCR-based Idylla system on tumor tissue from patients with non-small cell lung cancer. Oncotarget 2017, 8, 103055–103062. [Google Scholar] [CrossRef] [PubMed]
- Kinno, T.; Tsuta, K.; Shiraishi, K.; Mizukami, T.; Suzuki, M.; Yoshida, A.; Suzuki, K.; Asamura, H.; Furuta, K.; Kohno, T.; et al. Clinicopathological features of nonsmall cell lung carcinomas with BRAF mutations. Ann. Oncol. 2014, 25, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Luk, P.P.; Yu, B.; Ng, C.C.; Mercorella, B.; Selinger, C.; Lum, T.; Kao, S.; O’Toole, S.A.; Cooper, W.A. BRAF mutations in non-small cell lung cancer. Transl. Lung Cancer Res. 2015, 4, 142–148. [Google Scholar] [PubMed]
- Marchetti, A.; Felicioni, L.; Malatesta, S.; Grazia Sciarrotta, M.; Guetti, L.; Chella, A.; Viola, P.; Pullara, C.; Mucilli, F.; Buttitta, F. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J. Clin. Oncol. 2011, 29, 3574–3579. [Google Scholar] [CrossRef] [PubMed]
- Tissot, C.; Couraud, S.; Tanguy, R.; Bringuier, P.P.; Girard, N.; Souquet, P.J. Clinical characteristics and outcome of patients with lung cancer harboring BRAF mutations. Lung Cancer 2016, 91, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Long, E.; Hofman, V.; Dadone, B.; Marquette, C.H.; Mouroux, J.; Vignaud, J.M.; Begueret, H.; Merlio, J.P.; Capper, D.; et al. Diagnostic value of immunohistochemistry for the detection of the BRAFV600E mutation in primary lung adenocarcinoma Caucasian patients. Ann. Oncol. 2013, 24, 742–748. [Google Scholar] [CrossRef] [PubMed]
- Ritterhouse, L.L.; Barletta, J.A. BRAF V600E mutation-specific antibody: A review. Semin. Diagn. Pathol. 2015, 32, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.T.; Abedalthagafi, M.S.; Brahmandam, M.; Greenfield, E.A.; Hoang, M.P.; Louis, D.N.; Hornick, J.L.; Santagata, S. Cross-reactivity of the BRAF VE1 antibody with epitopes in axonemal dyneins leads to staining of cilia. Mod. Pathol. 2015, 28, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Hofman, P. Immunotherapy supplanting chemotherapy for upfront treatment of advanced non-small cell lung cancer: What’s next? J. Thorac. Dis. 2017, 9, E519–E521. [Google Scholar] [CrossRef] [PubMed]
- Reck, M. Pembrolizumab as first-line therapy for metastatic non-small-cell lung cancer. Immunotherapy 2018, 10, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Cree, I.A.; Booton, R.; Cane, P.; Gosney, J.; Ibrahim, M.; Kerr, K.; Lal, R.; Lewanski, C.; Navani, N.; Nicholson, A.G.; et al. PD-L1 testing for lung cancer in the UK: Recognizing the challenges for implementation. Histopathology 2016, 69, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Hofman, V.; Dietel, M.; Soria, J.C.; Hofman, P. Assessment of the PD-L1 status by immunohistochemistry: Challenges and perspectives for therapeutic strategies in lung cancer patients. Virchows Arch. 2016, 468, 511–525. [Google Scholar] [CrossRef] [PubMed]
- Thunnissen, E.; de Langen, A.J.; Smit, E.F. PD-L1 IHC in NSCLC with a global and methodological perspective. Lung Cancer 2017, 113, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Boyle, T.A.; Zhou, C.; Rimm, D.L.; Hirsch, F.R. PD-L1 Expression in Lung Cancer. J. Thorac. Oncol. 2016, 11, 964–975. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.K.; Pan, A.; Davis, A.A.; Raparia, K.; Mohindra, N.A.; Matsangou, M.; Giles, F.J. Biomarkers for PD-1/PD-L1 Blockade Therapy in Non-Small-cell Lung Cancer: Is PD-L1 Expression a Good Marker for Patient Selection? Clin. Lung Cancer 2016, 17, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Long-Mira, E.; Bence, C.; Butori, C.; Lassalle, S.; Bouhlel, L.; Fazzalari, L.; Zahaf, K.; Lalvée, S.; Washetine, K.; et al. Comparative study of the PD-L1 status between surgically resected specimens and matched biopsies of NSCLC patients reveal major discordances: A potential issue for anti-PD-L1 therapeutic strategies. Ann. Oncol. 2016, 27, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Ng Kee Kwong, F.; Laggner, U.; Mc Kinney, O.; Croud, J.; Rice, A.; Nicholson, A.G. Expression of PD-L1 correlates with pleomorphic morphology and histological patterns of non-small cell lung carcinomas. Histopathology 2018. [Google Scholar] [CrossRef] [PubMed]
- Kerr, K.M.; Nicolson, M.C. Non-Small Cell Lung Cancer, PD-L1, and the Pathologist. Arch. Pathol. Lab. Med. 2016, 140, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Adam, J.; Le Stang, N.; Rouquette, I.; Cazes, A.; Badoual, C.; Pinot-Roussel, H.; Tixier, L.; Danel, C.; Damiola, F.; Damotte, D.; et al. Multicenter French harmonization study for PD-L1 IHC testing in non-small cell lung cancer. Ann. Oncol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Brody, R.; Zhang, Y.; Ballas, M.; Siddiqui, M.K.; Gupta, P.; Barker, C.; Midha, A.; Walker, J. PD-L1 expression in advanced NSCLC: Insights into risk stratification and treatment selection from a systematic literature review. Lung Cancer 2017, 112, 200–215. [Google Scholar] [CrossRef] [PubMed]
- Hendry, S.; Byrne, D.J.; Wright, G.M.; Young, R.J.; Sturrock, S.; Cooper, W.A.; Fox, S.B. Comparison of Four PD-L1 Immunohistochemical Assays in Lung Cancer. J. Thorac. Oncol. 2018, 13, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Khambata-Ford, S.; Copie-Bergman, C.; Huang, L.; Juco, J.; Hofman, V.; Hofman, P. Use of the 22C3 anti-PD-L1 antibody to determine PD-L1 expression in multiple automated immunohistochemistry platforms. PLoS ONE 2017, 12, e0183023. [Google Scholar]
- Rolfo, C.; Raez, L. New targets bring hope in squamous cell lung cancer: Neurotrophic tyrosine kinase gene fusions. Lab. Investig. 2017, 97, 1268–1270. [Google Scholar] [CrossRef] [PubMed]
- Hechtman, J.F.; Benayed, R.; Hyman, D.M.; Drilon, A.; Zehir, A.; Frosina, D.; Arcila, M.E.; Dogan, S.; Klimstra, D.S.; Ladanyi, M.; et al. Pan-Trk Immunohistochemistry Is an Efficient and Reliable Screen for the Detection of NTRK Fusions. Am. J. Surg. Pathol. 2017, 41, 1547–1551. [Google Scholar] [CrossRef] [PubMed]
- Farago, A.F.; Azzoli, C.G. Beyond ALK and ROS1: RET, NTRK, EGFR and BRAF gene rearrangements in non-small cell lung cancer. Transl. Lung Cancer Res. 2017, 6, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, R.; Auger, N.; Auclin, E.; Besse, B. Clinical and translational implications of RET rearrangements in non-small cell lung cancer. J. Thorac. Oncol. 2018, 13, 27–45. [Google Scholar] [CrossRef] [PubMed]
- Gautschi, O.; Milia, J.; Filleron, T.; Wolf, J.; Carbone, D.P.; Owen, D.; Camidge, R.; Narayanan, V.; Doebele, R.C.; Besse, B.; et al. Targeting RET in patients with RET-rearranged lung cancers: Results from the global, multicenter RET registry. J. Clin. Oncol. 2017, 35, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
- Platt, A.; Morten, J.; Ji, Q.; Elvin, P.; Womack, C.; Su, X.; Donald, E.; Gray, N.; Read, J.; Bigley, G.; et al. A retrospective analysis of RET translocation, gene copy number gain and expression in NSCLC patients treated with vandetanib in four randomized Phase III studies. BMC Cancer 2015, 15, 171. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Lee, B.; Hong, M.; Song, J.Y.; Jung, K.; Lira, M.E.; Mao, M.; Han, J.; Kim, J.; Choi, Y.L. Comprehensive analysis of RET and ROS1 rearrangement in lung adenocarcinoma. Mod. Pathol. 2015, 28, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Koeppen, H.; Yu, W.; Zha, J.; Pandita, A.; Penuel, E.; Rangell, L.; Raja, R.; Mohan, S.; Patel, R.; Desai, R.; et al. Biomarker analyses from a placebo-controlled phase II study evaluating erlotinib±onartuzumab in advanced non-small cell lung cancer: MET expression levels are predictive of patient benefit. Clin. Cancer Res. 2014, 20, 4488–4498. [Google Scholar] [CrossRef] [PubMed]
- Spigel, D.R.; Edelman, M.J.; O’Byrne, K.; Paz-Ares, L.; Mocci, S.; Phan, S.; Shames, D.S.; Smith, D.; Yu, W.; Paton, V.E.; et al. Results from the Phase III Randomized Trial of Onartuzumab Plus Erlotinib Versus Erlotinib in Previously Treated Stage IIIB or IV Non-Small-Cell Lung Cancer: METLung. J. Clin. Oncol. 2017, 35, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Koh, J.; Kim, D.W.; Kim, M.; Keam, B.; Kim, T.M.; Jeon, Y.K.; Chung, D.H.; Heo, D.S. MET amplification, protein expression, and mutations in pulmonary adenocarcinoma. Lung Cancer 2015, 90, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Sterlacci, W.; Fiegl, M.; Gugger, M.; Bubendorf, L.; Savic, S.; Tzankov, A. MET overexpression and gene amplification: Prevalence, clinico-pathological characteristics and prognostic significance in a large cohort of patients with surgically resected NSCLC. Virchows Arch. 2017, 471, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Watermann, I.; Schmitt, B.; Stellmacher, F.; Müller, J.; Gaber, R.; Kugler, C.H.; Reinmuth, N.; Huber, R.M.; Thomas, M.; Zabel, P.; et al. Improved diagnostics targeting c-MET in non-small cell lung cancer: Expression, amplification and activation? Diagn. Pathol. 2015, 10, 130. [Google Scholar] [CrossRef] [PubMed]
- Weingertner, N.; Meyer, N.; Voegeli, A.C.; Guenot, D.; Renaud, S.; Massard, G.; Falcoz, P.E.; Olland, A.; Mennecier, B.; Gaub, M.P.; et al. Correlation between MET protein expression and MET gene copy number in a Caucasian cohort of non-small cell lung cancers according to the new IASLC/ATS/ERS classification. Pathology 2015, 47, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Friboulet, L.; Olaussen, K.A.; Pignon, J.P.; Shepherd, F.A.; Tsao, M.S.; Graziano, S.; Kratzke, R.; Douillard, J.Y.; Seymour, L.; Pirker, R.; et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N. Engl. J. Med. 2013, 368, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Olaussen, K.A.; Dunant, A.; Fouret, P.; Brambilla, E.; André, F.; Haddad, V.; Taranchon, E.; Filipits, M.; Pirker, R.; Popper, H.H.; et al. IALT Bio Investigators. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N. Engl. J. Med. 2006, 355, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Pierceall, W.E.; Olaussen, K.A.; Rousseau, V.; Brambilla, E.; Sprott, K.M.; Andre, F.; Pignon, J.P.; Le Chevalier, T.; Pirker, R.; Jiang, C.; et al. Cisplatin benefit is predicted by immunohistochemical analysis of DNA repair proteins in squamous cell carcinoma but not adenocarcinoma: Theranostic modeling by NSCLC constituent histological subclasses. Ann. Oncol. 2012, 23, 2245–2252. [Google Scholar] [CrossRef] [PubMed]
- Handra-Luca, A.; Hernandez, J.; Mountzios, G.; Taranchon, E.; Lacau-St-Guily, J.; Soria, J.C.; Fouret, P. Excision repair cross complementation group 1 immunohistochemical expression predicts objective response and cancer-specific survival in patients treated by Cisplatin-based induction chemotherapy for locally advanced head and neck squamous cell carcinoma. Clin. Cancer Res. 2007, 13, 3855–3859. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Falzon, M.; Blackhall, F.; Spicer, J.; Nicolson, M.; Chaudhuri, A.; Middleton, G.; Ahmed, S.; Hicks, J.; Crosse, B.; et al. Randomized prospective biomarker trial of ERCC1 for comparing platinum and nonplatinum therapy in advanced non-small-cell lung cancer: ERCC1 Trial (ET). J. Clin. Oncol. 2017, 35, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Postel-Vinay, S.; Soria, J.C. ERCC1 as predictor of platinum benefit in Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2017, 35, 384–386. [Google Scholar] [CrossRef] [PubMed]
- Cardnell, R.J.; Li, L.; Sen, T.; Bara, R.; Tong, P.; Fujimoto, J.; Ireland, A.S.; Guthrie, M.R.; Bheddah, S.; Banerjee, U.; et al. Protein expression of TTF1 and cMYC define distinct molecular subgroups of small cell lung cancer with unique vulnerabilities to aurora kinase inhibition, DLL3 targeting, and other targeted therapies. Oncotarget 2017, 8, 73419–73432. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C.M.; Pietanza, M.C.; Bauer, T.M.; Ready, N.; Morgensztern, D.; Glisson, B.S.; Byers, L.A.; Johnson, M.L.; Burris, H.A., 3rd; Robert, F.; et al. SCRX16-001 investigators. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: A first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol. 2017, 18, 42–51. [Google Scholar] [CrossRef]
- Sharma, S.K.; Pourat, J.; Abdel-Atti, D.; Carlin, S.D.; Piersigilli, A.; Bankovich, A.J.; Gardner, E.E.; Hamdy, O.; Isse, K.; Bheddah, S.; et al. Noninvasive Interrogation of DLL3 expression in metastatic small cell lung cancer. Cancer Res. 2017, 77, 3931–3941. [Google Scholar] [CrossRef] [PubMed]
- Calles, A.; Sholl, L.M.; Rodig, S.J.; Pelton, A.K.; Hornick, J.L.; Butaney, M.; Lydon, C.; Dahlberg, S.E.; Oxnard, G.R.; Jackman, D.M.; et al. Immunohistochemical loss of LKB1 is a biomarker for more aggressive biology in KRAS-mutant lung adenocarcinoma. Clin. Cancer Res. 2015, 21, 2851–2860. [Google Scholar] [CrossRef] [PubMed]
- Cumberbatch, M.; Tang, X.; Beran, G.; Eckersley, S.; Wang, X.; Ellston, R.P.; Dearden, S.; Cosulich, S.; Smith, P.D.; Behrens, C.; et al. Identification of a subset of human non-small cell lung cancer patients with high PI3Kβ and low PTEN expression, more prevalent in squamous cell carcinoma. Clin. Cancer Res. 2014, 20, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Long-Mira, E.; Funck-Brentano, E.; Lassalle, S.; Butori, C.; Lespinet-Fabre, V.; Bordone, O.; Gay, A.; Zahaf, K.; Poissonnet, G.; et al. Immunohistochemistry as a potential tool for routine detection of the NRAS Q61R mutation in patients with metastatic melanoma. J. Am. Acad. Dermatol. 2015, 72, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Shin, E.; Choi, C.M.; Kim, H.R.; Jang, S.J.; Park, Y.S. Immunohistochemical characterization of the mTOR pathway in stage-I non-small-cell lung carcinoma. Lung Cancer 2015, 89, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, V.; Steinmetz, H.B.; Rizzo, E.J.; Erskine, A.J.; Fairbank, T.L.; de Abreu, F.B.; Tsongalis, G.J.; Tafe, L.J. Improving Adequacy of Small Biopsy and Fine-Needle Aspiration Specimens for Molecular Testing by Next-Generation Sequencing in Patients with Lung Cancer: A Quality Improvement Study at Dartmouth-Hitchcock Medical Center. Arch. Pathol. Lab. Med. 2017, 141, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Tafe, L.J.; Pierce, K.J.; Peterson, J.D.; de Abreu, F.; Memoli, V.A.; Black, C.C.; Pettus, J.R.; Marotti, J.D.; Gutmann, E.J.; Liu, X.; et al. Clinical Genotyping of Non-Small Cell Lung Cancers Using Targeted Next-Generation Sequencing: Utility of Identifying Rare and Co-mutations in Oncogenic Driver Genes. Neoplasia 2016, 18, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Letovanec, I.; Finn, S.; Zygoura, P.; Smyth, P.; Soltermann, A.; Bubendorf, L.; Speel, E.J.; Marchetti, A.; Nonaka, D.; Monkhorst, K.; et al. ETOP Lungscape Consortium. Evaluation of NGS and RT-PCR methods for ALK rearrangement in European NSCLC patients: Results from the ETOP Lungscape Project. J. Thorac. Oncol. 2018, 13, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Jocollé, G.; Conti, A.; Tiseo, M.; Zito Marino, F.; Donati, G.; Franco, R.; Bono, F.; Barbisan, F.; Facchinetti, F. Detection of ROS1 rearrangement in non-small cell lung cancer: Current and future perspectives. Lung Cancer 2017, 8, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Rapisuwon, S.; Busam, K.J.; Parks, K.; Chapman, P.B.; Lee, E.; Atkins, M.B. Discordance Between Cobas BRAF V600 Testing and VE1 Immunohistochemistry in a Melanoma Patient with Bone Marrow Metastases. Am. J. Dermatopathol. 2016, 38, 687–689. [Google Scholar] [CrossRef] [PubMed]
- Kao, H.L.; Yeh, Y.C.; Lin, C.H.; Hsu, W.F.; Hsieh, W.Y.; Ho, H.L.; Chou, T.Y. Diagnostic algorithm for detection of targetable driver mutations in lung adenocarcinomas: Comprehensive analyses of 205 cases with immunohistochemistry, real-time PCR and fluorescence in situ hybridization methods. Lung Cancer 2016, 101, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Vincenten, J.P.; Smit, E.F.; Grünberg, K.; Postmus, P.E.; Snijders, P.J.; Witte, B.I.; Heideman, D.A.; Thunnissen, E. Is the current diagnostic algorithm reliable for selecting cases for EGFR- and KRAS-mutation analysis in lung cancer? Lung Cancer 2015, 89, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.S.; Greer, W.; Bethune, D.; Craddock, K.J.; Flowerdew, G.; Xu, Z. ALK+ lung adenocarcinoma in never smokers and long-term ex-smokers: Prevalence and detection by immunohistochemistry and fluorescence in situ hybridization. Virchows Arch. 2016, 469, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Campesato, L.F.; Barroso-Sousa, R.; Jimenez, L.; Correa, B.R.; Sabbaga, J.; Hoff, P.M.; Reis, L.F.; Galante, P.A.; Camargo, A.A. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to PD-1 blockade in clinical practice. Oncotarget 2015, 6, 34221–34227. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Soo, R.A.; Lim, S.M.; Syn, N.L.; Teng, R.; Soong, R.; Mok, T.S.K.; Cho, B.C. Immune checkpoint inhibitors in epidermal growth factor receptor mutant non-small cell lung cancer: Current controversies and future directions. Lung Cancer 2018, 115, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Gorris, M.A.J.; Halilovic, A.; Rabold, K.; van Duffelen, A.; Wickramasinghe, I.N.; Verweij, D.; Wortel, I.M.N.; Textor, J.C.; de Vries, I.J.M.; Figdor, C.G. Eight-color multiplex immunohistochemistry for simultaneous detection of multiple immune checkpoint molecules within the tumor microenvironment. J. Immunol. 2018, 200, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Mezheyeuski, A.; Bergsland, C.H.; Backman, M.; Djureinovic, D.; Sjöblom, T.; Bruun, J.; Micke, P. Multispectral imaging for quantitative and compartment-specific immune infiltrates reveals distinct immune profiles that classify lung cancer patients. J. Pathol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Parra, E.R.; Uraoka, N.; Jiang, M.; Cook, P.; Gibbons, D.; Forget, M.A.; Bernatchez, C.; Haymaker, C.; Wistuba, II.; Rodriguez-Canales, J. Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Sci. Rep. 2017, 7, 13380. [Google Scholar] [CrossRef] [PubMed]
- Roussel, H.; De Guillebon, E.; Biard, L.; Mandavit, M.; Gibault, L.; Fabre, E.; Antoine, M.; Hofman, P.; Beau-Faller, M.; Blons, H.; et al. Composite biomarkers defined by multiparametric immunofluorescence analysis identify ALK-positive adenocarcinoma as a potential target for immunotherapy. Oncoimmunology 2017, 6, e1286437. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Hofman, V.; Long, E.; Bordone, O.; Selva, E.; Washetine, K.; Marquette, C.H.; Hofman, P. Current challenges for detection of circulating tumor cells and cell-free circulating nucleic acids, and their characterization in non-small cell lung carcinoma patients. What is the best blood substrate for personalized medicine? Ann. Transl. Med. 2014, 2, 107. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Szafer-Glusman, E.; Hofman, V.; Long-Mira, E.; Suttmann, R.; Darbonne, W.; Butori, C.; Lalvée, S.; Fayada, J.; Selva, E.; et al. Expression of MET in circulating tumor cells correlates with expression in tumor tissue from advanced-stage lung cancer patients. Oncotarget 2017, 8, 26112–26121. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Szafer-Glusman, E.; Hofman, V.; Chamorey, E.; Lalvée, S.; Selva, E.; Leroy, S.; Marquette, C.H.; Kowanetz, M.; Hedge, P.; et al. Detection of PD-L1 in circulating tumor cells and white blood cells from patients with advanced non-small-cell lung cancer. Ann. Oncol. 2017, 29, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P. Liquid Biopsy and Therapeutic Targets: Present and future issues in thoracic oncology. Cancers 2017, 9, 154. [Google Scholar] [CrossRef] [PubMed]
- Ilié, M.; Hofman, P. Pros: Can tissue biopsy be replaced by liquid biopsy? Transl. Lung Cancer Res. 2016, 5, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Mayo-de-Las-Casas, C.; Garzón Ibáñez, M.; Jordana-Ariza, N.; García-Peláez, B.; Balada-Bel, A.; Villatoro, S.; Malapelle, U.; Karachaliou, N.; Troncone, G.; Rosell, R.; et al. An update on liquid biopsy analysis for diagnostic and monitoring applications in non-small cell lung cancer. Expert. Rev. Mol. Diagn. 2018, 18, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Hofman, V.J.; Ilie, M.; Hofman, P.M. Detection and characterization of circulating tumor cells in lung cancer: Why and how? Cancer Cytopathol. 2016, 124, 380–387. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hofman, V.; Lassalle, S.; Bence, C.; Long-Mira, E.; Nahon-Estève, S.; Heeke, S.; Lespinet-Fabre, V.; Butori, C.; Ilié, M.; Hofman, P. Any Place for Immunohistochemistry within the Predictive Biomarkers of Treatment in Lung Cancer Patients? Cancers 2018, 10, 70. https://doi.org/10.3390/cancers10030070
Hofman V, Lassalle S, Bence C, Long-Mira E, Nahon-Estève S, Heeke S, Lespinet-Fabre V, Butori C, Ilié M, Hofman P. Any Place for Immunohistochemistry within the Predictive Biomarkers of Treatment in Lung Cancer Patients? Cancers. 2018; 10(3):70. https://doi.org/10.3390/cancers10030070
Chicago/Turabian StyleHofman, Véronique, Sandra Lassalle, Coraline Bence, Elodie Long-Mira, Sacha Nahon-Estève, Simon Heeke, Virginie Lespinet-Fabre, Catherine Butori, Marius Ilié, and Paul Hofman. 2018. "Any Place for Immunohistochemistry within the Predictive Biomarkers of Treatment in Lung Cancer Patients?" Cancers 10, no. 3: 70. https://doi.org/10.3390/cancers10030070




