Epstein–Barr Virus Gene BARF1 Expression is Regulated by the Epithelial Differentiation Factor ΔNp63α in Undifferentiated Nasopharyngeal Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmids
2.3. In Vitro DNA Methylation
2.4. Luciferase Reporter Assays
2.5. Chromatin Immunoprecipitation (ChIP) Assay
2.6. Quantitative RT-PCR
2.7. SDS-Page and Western Blot
3. Results
3.1. Potential p53 Family Response Elements Found in the BARF1 Promoter
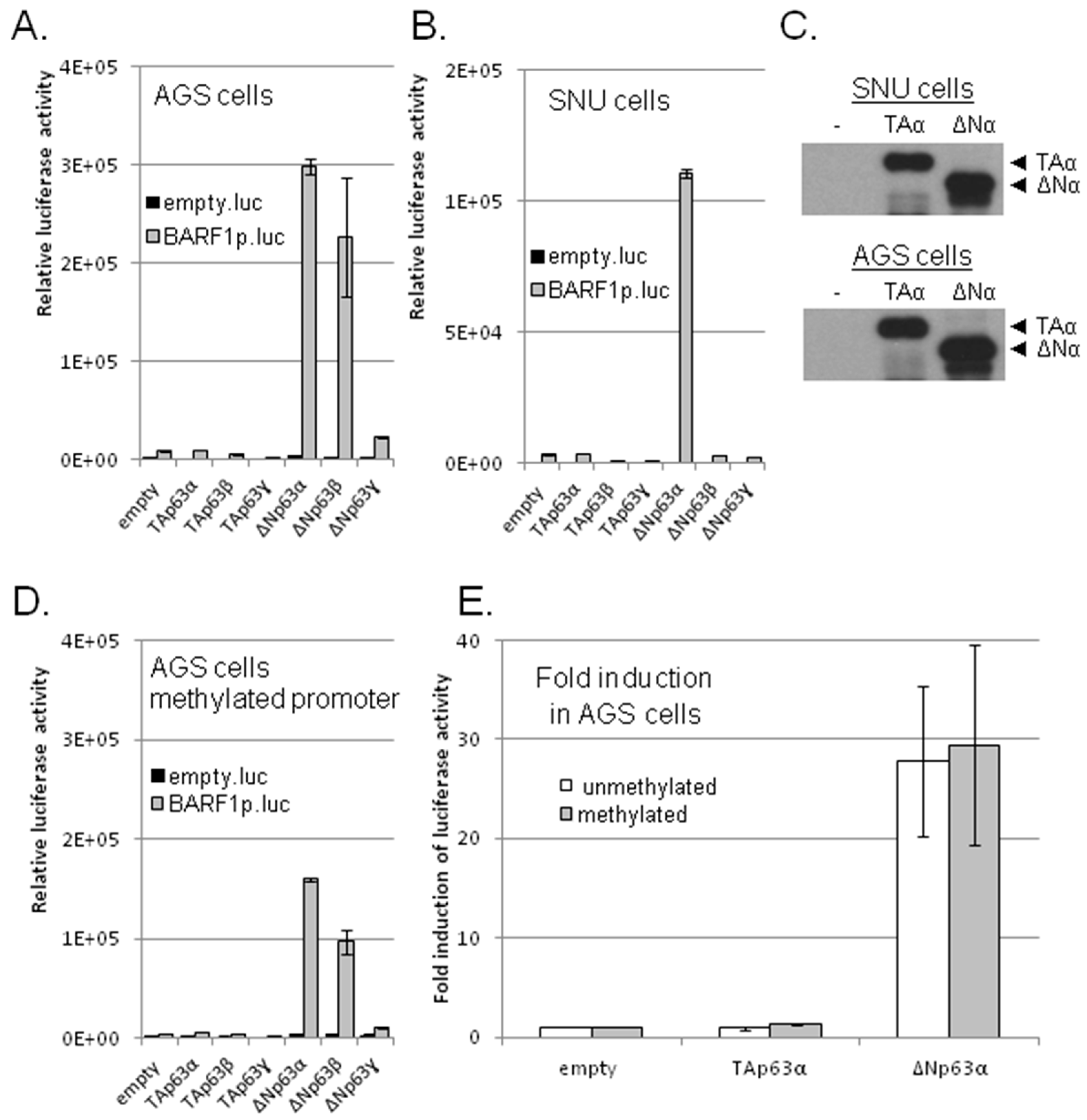
3.2. ΔNp63α, But No Other p53 Family Members, Transactivates BARF1 Promoter in Luciferase Reporter Assays
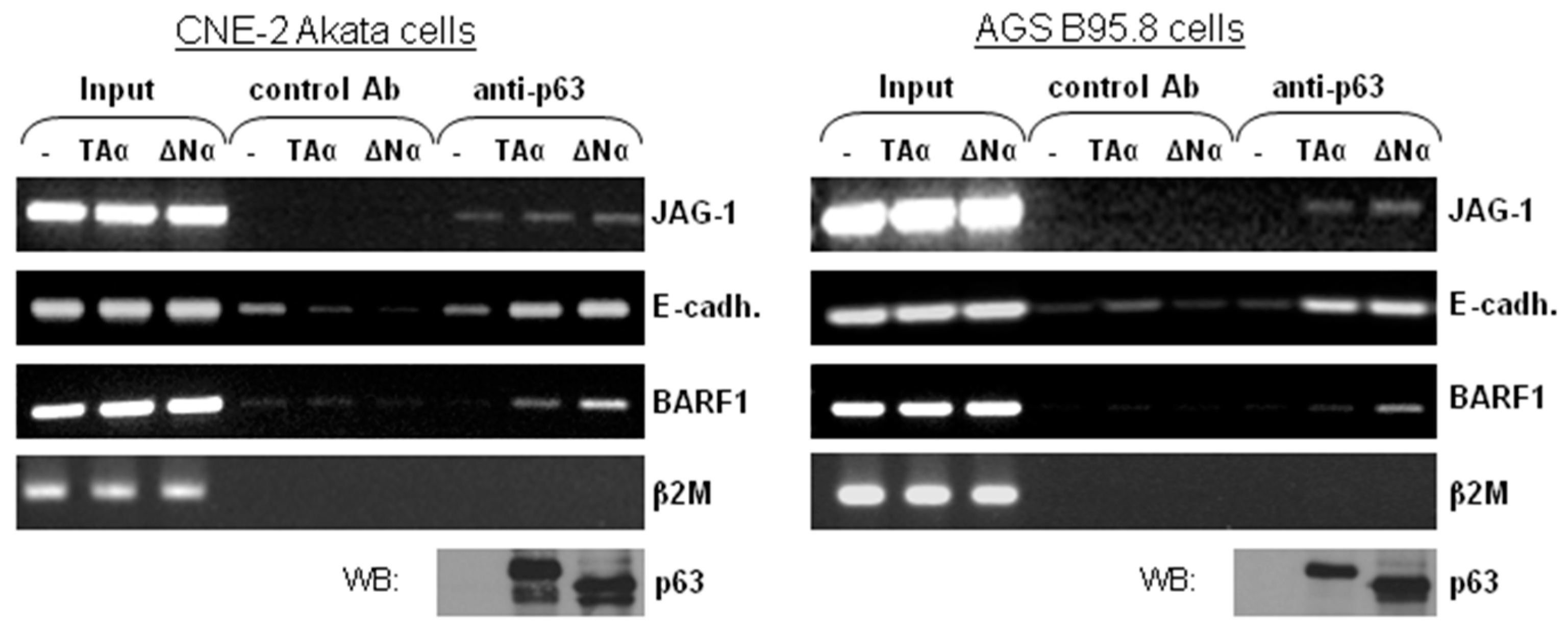
3.3. ΔNp63α Is Complexed with the BARF1 Promoter In Vivo
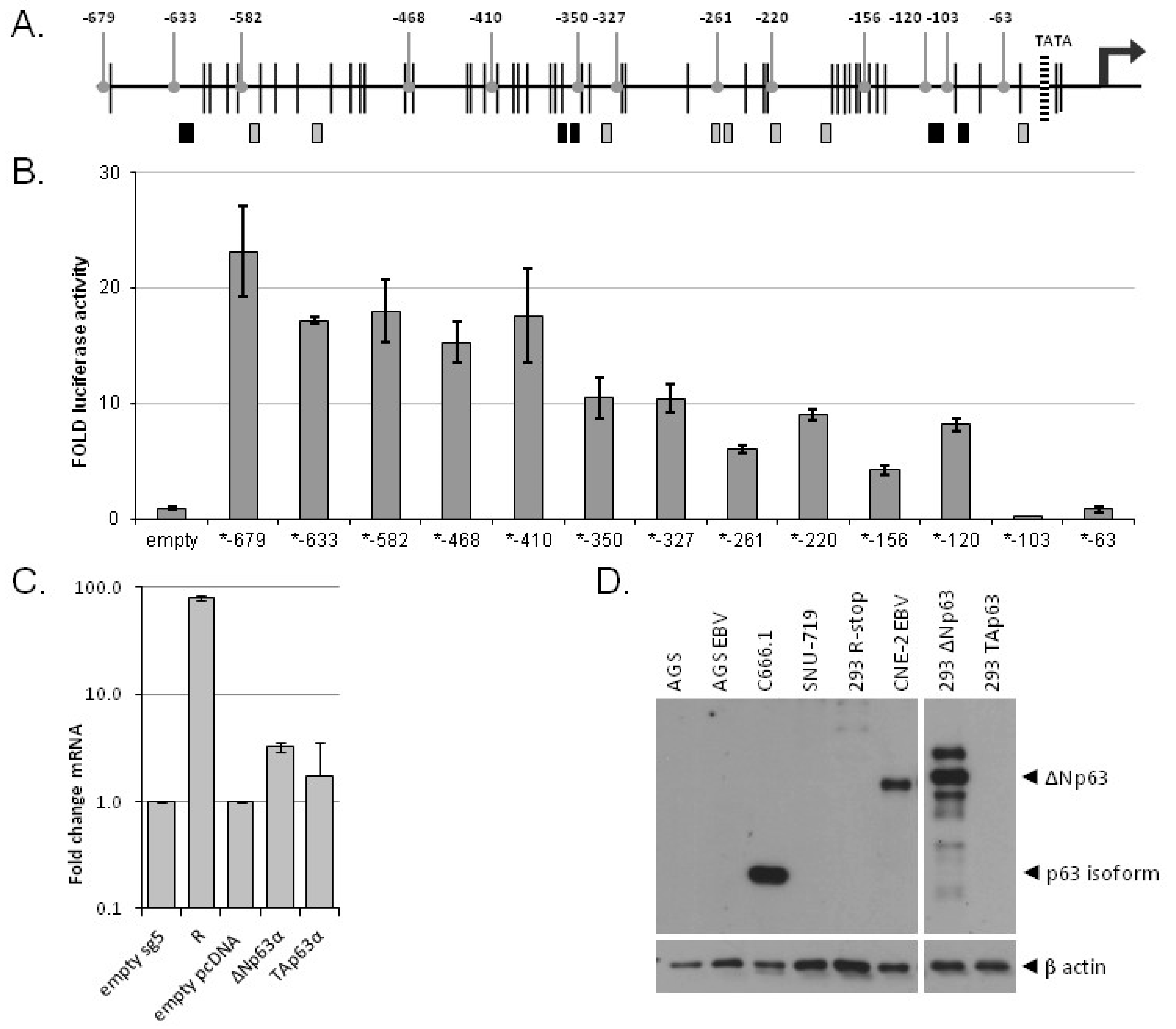
3.4. Multiple p63 Response Elements Are Responsible for BARF1 Promoter Transactivation
3.5. ΔNp63α Alone Is Not Sufficient to Induce BARF1 in Context of the Viral Genome
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Middeldorp, J.M.; Brink, A.A.; van den Brule, A.J.; Meijer, C.J. Pathogenic roles for Epstein-Barr virus (EBV) gene products in EBV-associated proliferative disorders. Crit. Rev. Oncol. Hematol. 2003, 45, 1–36. [Google Scholar] [CrossRef]
- Young, L.S.; Rickinson, A.B. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 2004, 4, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Frappier, L. Role of EBNA1 in NPC tumourigenesis. Semin. Cancer Biol. 2012, 22, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Hausen zur, A.; Brink, A.A.; Craanen, M.E.; Middeldorp, J.M.; Meijer, C.J.; van den Brule, A.J. Unique transcription pattern of Epstein-Barr virus (EBV) in EBV-carrying gastric adenocarcinomas: Expression of the transforming BARF1 gene. Cancer Res. 2000, 60, 2745–2748. [Google Scholar]
- Seto, E.; Yang, L.; Middeldorp, J.; Sheen, T.S.; Chen, J.Y.; Fukayama, M.; Eizuru, Y.; Ooka, T.; Takada, K. Epstein-Barr virus (EBV)-encoded BARF1 gene is expressed in nasopharyngeal carcinoma and EBV-associated gastric carcinoma tissues in the absence of lytic gene expression. J. Med. Virol. 2005, 76, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Stevens, S.J.C.; Verkuijlen, S.A.W.M.; Hariwiyanto, B.; Harijadi; Paramita, D.K.; Fachiroh, J.; Adham, M.; Tan, I.B.; Haryana, S.M.; Middeldorp, J.M. Noninvasive diagnosis of nasopharyngeal carcinoma: Nasopharyngeal brushings reveal high Epstein-Barr virus DNA load and carcinoma-specific viral BARF1 mRNA. Int. J. Cancer 2006, 119, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Tarbouriech, N.; Ruggiero, F.; de Turenne-Tessier, M.; Ooka, T.; Burmeister, W.P. Structure of the Epstein-Barr virus oncogene BARF1. J. Mol. Biol. 2006, 359, 667–678. [Google Scholar] [CrossRef] [PubMed]
- De Turenne-Tessier, M.; Jolicoeur, P.; Ooka, T. Expression of the protein encoded by Epstein-Barr virus (EBV) BARF1 open reading frame from a recombinant adenovirus system. Virus Res. 1997, 52, 73–85. [Google Scholar] [CrossRef]
- Hoebe, E.K.; Hutajulu, S.H.; van, B.J.; Stevens, S.J.; Paramita, D.K.; Greijer, A.E.; Middeldorp, J.M. Purified hexameric Epstein-Barr virus-encoded BARF1 protein for measuring anti-BARF1 antibody responses in nasopharyngeal carcinoma patients. Clin. Vaccine Immunol. 2011, 18, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Ooka, T. Biological Role of the BARF1 Gene Encoded by Epstein-Barr Virus. In Epstein-Barr Virus; Robertson, E.S., Ed.; Caister Academic Press: Poole, UK, 2005; p. 613. [Google Scholar]
- Strockbine, L.D.; Cohen, J.I.; Farrah, T.; Lyman, S.D.; Wagener, F.; DuBose, R.F.; Armitage, R.J.; Spriggs, M.K. The Epstein-Barr virus BARF1 gene encodes a novel, soluble colony-stimulating factor-1 receptor. J. Virol. 1998, 72, 4015–4021. [Google Scholar] [PubMed]
- Ohashi, M.; Orlova, N.; Quink, C.; Wang, F. Cloning of the Epstein-Barr virus-related rhesus lymphocryptovirus as a bacterial artificial chromosome: A loss-of-function mutation of the rhBARF1 immune evasion gene. J. Virol. 2011, 85, 1330–1339. [Google Scholar] [CrossRef] [PubMed]
- Hoebe, E.K.; Le Large, T.Y.; Tarbouriech, N.; Oosterhoff, D.; de Gruijl, T.D.; Middeldorp, J.M.; Greijer, A.E. Epstein-Barr Virus-Encoded BARF1 Protein is a Decoy Receptor for Macrophage Colony Stimulating Factor and Interferes with Macrophage Differentiation and Activation. Viral Immunol. 2012, 25, 461–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoebe, E.K.; Wille, C.; Hopmans, E.S.; Robinson, A.R.; Middeldorp, J.M.; Kenney, S.C.; Greijer, A.E. Epstein-Barr virus transcription activator R upregulates BARF1 expression by direct binding to its promoter, independent of methylation. J. Virol. 2012, 86, 11322–11332. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, B.; Zhao, P.; Huang, B.H. Expression of Epstein-Barr virus genes in EBV-associated gastric carcinoma. Ai Zheng 2004, 23, 782–787. (In Chinese) [Google Scholar] [PubMed]
- Jiang, R.; Cabras, G.; Sheng, W.; Zeng, Y.; Ooka, T. Synergism of BARF1 with Ras induces malignant transformation in primary primate epithelial cells and human nasopharyngeal epithelial cells. Neoplasia 2009, 11, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Hoebe, E.K.; Le Large, T.Y.; Greijer, A.E.; Middeldorp, J.M. BamHI-A rightward frame 1, an Epstein-Barr virus-encoded oncogene and immune modulator. Rev. Med. Virol. 2013, 23, 367–383. [Google Scholar] [CrossRef] [PubMed]
- Bourne, T.D.; Bellizzi, A.M.; Stelow, E.B.; Loy, A.H.; Levine, P.A.; Wick, M.R.; Mills, S.E. p63 Expression in olfactory neuroblastoma and other small cell tumors of the sinonasal tract. Am. J. Clin. Pathol. 2008, 130, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Pan, Z.G.; Li, D.J.; Yun, J.P.; Zheng, M.Z.; Hu, Z.Y.; Cheng, L.Z.; Zeng, Y.X. The expression of p63 is associated with the differential stage in nasopharyngeal carcinoma and EBV infection. J. Transl. Med. 2006, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Tannapfel, A.; Schmelzer, S.; Benicke, M.; Klimpfinger, M.; Kohlhaw, K.; Mossner, J.; Engeland, K.; Wittekind, C. Expression of the p53 homologues p63 and p73 in multiple simultaneous gastric cancer. J. Pathol. 2001, 195, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, Z.; Li, J.; Zhao, X.; Wang, Z.; Xu, H. DeltaNp63alpha mediates proliferation and apoptosis in human gastric cancer cells by the regulation of GATA-6. Neoplasma 2012, 59, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Kaghad, M.; Wang, Y.; Gillett, E.; Fleming, M.D.; Dotsch, V.; Andrews, N.C.; Caput, D.; McKeon, F. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 1998, 2, 305–316. [Google Scholar] [CrossRef]
- Yang, A.; Kaghad, M.; Caput, D.; McKeon, F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 2002, 18, 90–95. [Google Scholar] [CrossRef]
- Osada, M.; Park, H.L.; Nagakawa, Y.; Yamashita, K.; Fomenkov, A.; Kim, M.S.; Wu, G.; Nomoto, S.; Trink, B.; Sidransky, D. Differential recognition of response elements determines target gene specificity for p53 and p63. Mol. Cell. Biol. 2005, 25, 6077–6089. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Chakravarti, D.; Flores, E.R. p63 steps into the limelight: Crucial roles in the suppression of tumorigenesis and metastasis. Nat. Rev. Cancer 2012, 13, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Ghioni, P.; Bolognese, F.; Duijf, P.H.; Van, B.H.; Mantovani, R.; Guerrini, L. Complex transcriptional effects of p63 isoforms: Identification of novel activation and repression domains. Mol. Cell. Biol. 2002, 22, 8659–8668. [Google Scholar] [CrossRef] [PubMed]
- Perez, C.A.; Pietenpol, J.A. Transcriptional programs regulated by p63 in normal epithelium and tumors. Cell Cycle 2007, 6, 246–254. [Google Scholar] [CrossRef] [PubMed]
- King, K.E.; Ponnamperuma, R.M.; Yamashita, T.; Tokino, T.; Lee, L.A.; Young, M.F.; Weinberg, W.C. Deltanp63alpha functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene 2003, 22, 3635–3644. [Google Scholar] [CrossRef] [PubMed]
- Helton, E.S.; Zhu, J.; Chen, X. The unique NH2-terminally deleted (DeltaN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the DeltaN variant of p63. J. Biol. Chem. 2006, 281, 2533–2542. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Osada, M.; Guo, Z.; Fomenkov, A.; Begum, S.; Zhao, M.; Upadhyay, S.; Xing, M.; Wu, F.; Moon, C.; et al. DeltaNp63alpha up-regulates the Hsp70 gene in human cancer. Cancer Res. 2005, 65, 758–766. [Google Scholar] [PubMed]
- Graziano, V.; De Laurenzi, V. Role of p63 in cancer development. Biochim. Biophys. Acta 2011, 1816, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Fotheringham, J.A.; Mazzucca, S.; Raab-Traub, N. Epstein-Barr virus latent membrane protein-2A-induced DeltaNp63alpha expression is associated with impaired epithelial-cell differentiation. Oncogene 2010, 29, 4287–4296. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lu, J.; Zuo, L.; Yan, Q.; Yu, Z.; Li, X.; Huang, J.; Zhao, L.; Tang, H.; Luo, Z.; et al. Epstein-Barr virus downregulates microRNA 203 through the oncoprotein latent membrane protein 1: A contribution to increased tumor incidence in epithelial cells. J. Virol. 2012, 86, 3088–3099. [Google Scholar] [CrossRef] [PubMed]
- Hau, P.M.; Yip, Y.L.; Huen, M.S.Y.; Tsao, S.W. Loss of ΔNp63α promotes mitotic exit in epithelial cells. FEBS Lett. 2011, 585, 2710–2726. [Google Scholar] [CrossRef] [PubMed]
- Nekulova, M.; Holcakova, J.; Coates, P.; Vojtesek, B. The role of p63 in cancer, stem cells and cancer stem cells. Cell. Mol. Biol. Lett. 2011, 16, 296–327. [Google Scholar] [CrossRef] [PubMed]
- Yip, Y.L.; Tsao, S.W. Regulation of p63 expression in primary and immortalized nasopharyngeal epithelial cells. Int. J. Oncol. 2008, 33, 713–724. [Google Scholar] [PubMed]
- Park, J.G.; Yang, H.K.; Kim, W.H.; Chung, J.K.; Kang, M.S.; Lee, J.H.; Oh, J.H.; Park, H.S.; Yeo, K.S.; Kang, S.H.; et al. Establishment and characterization of human gastric carcinoma cell lines. Int. J. Cancer 1997, 70, 443–449. [Google Scholar] [CrossRef]
- Feederle, R.; Kost, M.; Baumann, M.; Janz, A.; Drouet, E.; Hammerschmidt, W.; Delecluse, H.J. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 2000, 19, 3080–3089. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.R.; Kwek, S.S.; Hagemeier, S.R.; Wille, C.K.; Kenney, S.C. Cellular transcription factor Oct-1 interacts with the Epstein-Barr virus BRLF1 protein to promote disruption of viral latency. J. Virol. 2011, 85, 8940–8953. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, J.M.; Lieberman, P.M.; Hayward, S.D. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 1988, 62, 2274–2284. [Google Scholar] [PubMed]
- Flores, E.R.; Tsai, K.Y.; Crowley, D.; Sengupta, S.; Yang, A.; McKeon, F.; Jacks, T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 2002, 416, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Klug, M.; Rehli, M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics 2006, 1, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Testoni, B.; Borrelli, S.; Tenedini, E.; Alotto, D.; Castagnoli, C.; Piccolo, S.; Tagliafico, E.; Ferrari, S.; Vigano, M.A.; Mantovani, R. Identification of new p63 targets in human keratinocytes. Cell Cycle 2006, 5, 2805–2811. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Munoz, A.; Halvorsen, K.; Rai, P. Creation and validation of a ligation-independent cloning (LIC) retroviral vector for stable gene transduction in mammalian cells. BMC Biotechnol. 2012, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Kel, A.E.; Gossling, E.; Reuter, I.; Cheremushkin, E.; Kel-Margoulis, O.V.; Wingender, E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003, 31, 3576–3579. [Google Scholar] [CrossRef] [PubMed]
- Ambinder, R.F.; Robertson, K.D.; Tao, Q. DNA methylation and the Epstein-Barr virus. Semin. Cancer Biol. 1999, 9, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Craig, A.L.; Holcakova, J.; Finlan, L.E.; Nekulova, M.; Hrstka, R.; Gueven, N.; DiRenzo, J.; Smith, G.; Hupp, T.R.; Vojtesek, B. DeltaNp63 transcriptionally regulates ATM to control p53 Serine-15 phosphorylation. Mol. Cancer 2010, 9, 195. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.S.; Maronian, N.; Vieira, J. Activation of Kaposi’s sarcoma-associated herpesvirus lytic gene expression during epithelial differentiation. J. Virol. 2005, 79, 13769–13777. [Google Scholar] [CrossRef] [PubMed]
- Moody, C.A.; Laimins, L.A. Human papillomavirus oncoproteins: Pathways to transformation. Nat. Rev. Cancer 2010, 10, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Flemington, E.K. Herpesvirus lytic replication and the cell cycle: Arresting new developments. J. Virol. 2001, 75, 4475–4481. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Leser, U.; Marschall, M.; Langford, A.; Jilg, W.; Gelderblom, H.; Reichart, P.; Wolf, H. Expression of proteins encoded by Epstein-Barr virus trans-activator genes depends on the differentiation of epithelial cells in oral hairy leukoplakia. Proc. Natl. Acad. Sci. USA 1991, 88, 8332–8336. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.X.; Young, L.S.; Niedobitek, G.; Dawson, C.W.; Birkenbach, M.; Wang, F.; Rickinson, A.B. Epstein-Barr virus infection and replication in a human epithelial cell system. Nature 1992, 356, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Kajitani, N.; Satsuka, A.; Kawate, A.; Sakai, H. Productive Lifecycle of Human Papillomaviruses that Depends Upon Squamous Epithelial Differentiation. Front. Microbiol. 2012, 3, 152. [Google Scholar] [CrossRef] [PubMed]
- Mighty, K.K.; Laimins, L.A. p63 is necessary for the activation of human papillomavirus late viral functions upon epithelial differentiation. J. Virol. 2011, 85, 8863–8869. [Google Scholar] [CrossRef] [PubMed]
- Yugawa, T.; Kiyono, T. Molecular mechanisms of cervical carcinogenesis by high-risk human papillomaviruses: Novel functions of E6 and E7 oncoproteins. Rev. Med. Virol. 2009, 19, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Melar-New, M.; Laimins, L.A. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J. Virol. 2010, 84, 5212–5221. [Google Scholar] [CrossRef] [PubMed]
- Greco, D.; Kivi, N.; Qian, K.; Leivonen, S.K.; Auvinen, P.; Auvinen, E. Human papillomavirus 16 E5 modulates the expression of host microRNAs. PLoS ONE 2011, 6, e21646. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.P.; Degen, M.; Villa, L.L.; Cibas, E.S. Immunomarkers in gynecologic cytology: The search for the ideal ‘biomolecular Papanicolaou test’. Acta Cytol. 2012, 56, 109–121. [Google Scholar] [CrossRef] [PubMed]
- MacCallum, P.; Karimi, L.; Nicholson, L.J. Definition of the transcription factors which bind the differentiation responsive element of the Epstein-Barr virus BZLF1 Z promoter in human epithelial cells. J. Gen. Virol. 1999, 80, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Szekely, L.; Selivanova, G.; Magnusson, K.P.; Klein, G.; Wiman, K.G. EBNA-5, an Epstein-Barr virus-encoded nuclear antigen, binds to the retinoblastoma and p53 proteins. Proc. Natl. Acad. Sci. USA 1993, 90, 5455–5459. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Zhang, F.; Meng, F.; Li, H.; Liu, Q.; Liang, Y.; Dong, Y.; Yang, W.; Han, A. EBV-encoded LMP1 increases nuclear beta-catenin accumulation and its transcriptional activity in nasopharyngeal carcinoma. Tumour Biol. 2011, 32, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.A.; Klingelhutz, A.J.; Raab-Traub, N. Epstein-Barr virus latent membrane protein 2A activates beta-catenin signaling in epithelial cells. J. Virol. 2003, 77, 12276–12284. [Google Scholar] [CrossRef] [PubMed]
- Ruptier, C.; De, G.A.; Ansieau, S.; Granjon, A.; Taniere, P.; Lafosse, I.; Shi, H.; Petitjean, A.; Taranchon-Clermont, E.; Tribollet, V.; et al. TP63 P2 promoter functional analysis identifies beta-catenin as a key regulator of DeltaNp63 expression. Oncogene 2011, 30, 4656–4665. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J.M.; Yip, Y.L.; Lo, K.W.; Deng, W.; To, K.F.; Man, H.P.; Hau, P.M.; Lau, V.M.; Takada, K.; Lui, V.W.; et al. Cyclin D1 overexpression supports stable EBV infection in nasopharyngeal epithelial cells. Proc. Natl. Acad. Sci. USA. 2012, 109, E3473–E3482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikuchi, A.; Yamamoto, H.; Sato, A.; Matsumoto, S. New insights into the mechanism of Wnt signaling pathway activation. Int. Rev. Cell Mol. Biol. 2011, 291, 21–71. [Google Scholar] [PubMed]
- Wiech, T.; Nikolopoulos, E.; Lassman, S.; Heidt, T.; Schopflin, A.; Sarbia, M.; Werner, M.; Shimizu, Y.; Sakka, E.; Ooka, T.; et al. Cyclin D1 expression is induced by viral BARF1 and is overexpressed in EBV-associated gastric cancer. Virchows Arch. 2008, 452, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Byun, S.J.; Kim, Y.A.; Kim, J.E.; Lee, B.L.; Kim, W.H.; Chang, M.S. Cell cycle regulators, APC/beta-catenin, NF-kappaB and Epstein-Barr virus in gastric carcinomas. Pathology 2010, 42, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Buettner, M.; Lang, A.; Tudor, C.S.; Meyer, B.; Cruchley, A.; Barros, M.H.; Farrell, P.J.; Jäck, H.M.; Schuh, W.; Niedobitek, G. Lytic Epstein-Barr virus infection in epithelial cells but not in B-lymphocytes is dependent on Blimp1. J. Gen. Virol. 2012, 93, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
- Reusch, J.A.; Nawandar, D.M.; Wright, K.L.; Kenney, S.C.; Mertz, J.E. Cellular differentiation regulator BLIMP1 induces Epstein-Barr virus lytic reactivation in epithelial and B cells by activating transcription from both the R and Z promoters. J. Virol. 2015, 89, 1731–1743. [Google Scholar] [CrossRef] [PubMed]
- Nawandar, D.M.; Ohashi, M.; Djavadian, R.; Barlow, E.; Makielski, K.; Ali, A.; Lee, D.; Lambert, P.F.; Johannsen, E.; Kenney, S.C. Differentiation-Dependent LMP1 Expression Is Required for Efficient Lytic Epstein-Barr Virus Reactivation in Epithelial Cells. J. Virol. 2017, 91, e02438-16. [Google Scholar] [CrossRef] [PubMed]
- Takacs, M.; Banati, F.; Koroknai, A.; Segesdi, J.; Salamon, D.; Wolf, H.; Niller, H.H.; Minarovits, J. Epigenetic regulation of latent Epstein-Barr virus promoters. Biochim. Biophys. Acta 2010, 1799, 228–235. [Google Scholar] [CrossRef] [PubMed]

| ATG-n | Forward primers |
|---|---|
| 679 | CTGACTAGTCTCATCACGCAACACCCACTGTTT |
| 633 | CTGACTAGTAAGTCAGTCAGGCTGGCCAGG |
| 582 | CTGACTAGTGATCTTGGCATGCCGCCCAGC |
| 468 | CTGACTAGTACCGCAAACACCACTGTGTAGC |
| 410 | CTGACTAGTGGTCGTTGTACACTGCGCGCAG |
| 350 | CTGACTAGTCGATGTCGGCTGTCCTGCAGG |
| 327 | CTGACTAGTAGCTCCGCGTACAGCTTCCTATCC |
| 261 | CTGACTAGTGGCAAAGGCAGGTCTTTCTCATCC |
| 220 | CTGACTAGTCATGGCCCTGAACATGAGGTAGC |
| 156 | CTGACTAGTCACGCCTCGACCGGGGTC |
| 120 | CTGACTAGTGTAGACTTGGCTGGCCTCATGG |
| 103 | CTGACTAGTCATGGTCTCGTCAGGCCAGC |
| 63 | CTGACTAGTTGATAAAATGGGCGTGGCAG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoebe, E.; Wille, C.; Hagemeier, S.; Kenney, S.; Greijer, A.; Middeldorp, J. Epstein–Barr Virus Gene BARF1 Expression is Regulated by the Epithelial Differentiation Factor ΔNp63α in Undifferentiated Nasopharyngeal Carcinoma. Cancers 2018, 10, 76. https://doi.org/10.3390/cancers10030076
Hoebe E, Wille C, Hagemeier S, Kenney S, Greijer A, Middeldorp J. Epstein–Barr Virus Gene BARF1 Expression is Regulated by the Epithelial Differentiation Factor ΔNp63α in Undifferentiated Nasopharyngeal Carcinoma. Cancers. 2018; 10(3):76. https://doi.org/10.3390/cancers10030076
Chicago/Turabian StyleHoebe, Eveline, Coral Wille, Stacy Hagemeier, Shannon Kenney, Astrid Greijer, and Jaap Middeldorp. 2018. "Epstein–Barr Virus Gene BARF1 Expression is Regulated by the Epithelial Differentiation Factor ΔNp63α in Undifferentiated Nasopharyngeal Carcinoma" Cancers 10, no. 3: 76. https://doi.org/10.3390/cancers10030076





