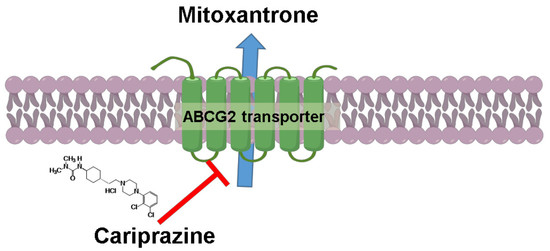
Cariprazine, A Dopamine D2/D3 Receptor Partial Agonist, Modulates ABCG2-Mediated Multidrug Resistance in Cancer
Abstract
:1. Introduction
2. Results
2.1. Cariprazine Significantly Increases the Efficacy of Mitoxantrone in Cancer Cell Lines Overexpressing the ABCG2 Transporter
2.2. Cariprazine Synergistically Increases the Efficacy of MX
2.3. Cariprazine Decreases the Protein Expression Levels of the ABCG2 Transporter Protein
2.4. Cariprazine Inhibits the Efflux Function of the ABCG2 Transporter
2.5. Molecular Docking Analysis of the Interaction of Cariprazine with A Human ABCG2.
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Lines and Cell Culture
4.3. Cell Cytotoxicity through Colororimeteric MTT Assay
4.4. Cell Morphological Analysis
4.5. Protein Estimation: Cell Lysate Preparation and Bicinchoninic Acid (BCA) Analysis
4.6. Western Blot Analysis
4.7. Rhodamine 123 Accumulation Assay
4.8. The Effect of Cariprazine on the Efficacy of MX
4.9. Molecular Docking Studies
4.9.1. Ligand Structure Preparation
4.9.2. Protein Structure Preparation
4.9.3. Docking Protocol
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ejendal, K.F.; Hrycyna, C.A. Multidrug resistance and cancer: The role of the human ABC transporter ABCG2. Curr. Protein Pept. Sci. 2002, 3, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Gillet, J.P.; Gottesman, M.M. Mechanisms of multidrug resistance in Cancer. In Multi-Drug Resistance in Cancer; Humana Press: New York, NY USA, 2010; pp. 47–76. [Google Scholar]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of ATP–dependent transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Higgins, C.F. Multiple molecular mechanisms for multidrug resistance transporters. Nature 2007, 446, 749–757. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.; Tomlinson, B. Targeting the ABCG2-overexpressing multidrug resistant (MDR) cancer cells by PPARγ agonists. Br. J. Pharmacol. 2013, 170, 1137–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dean, M.; Allikmets, R. Complete characterization of the human ABC gene family. J. Bioenerg. Biomembr. 2001, 33, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H.; Yu, A.M. ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Curr. Pharm. Des. 2014, 20, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.L.; Huang, F.; Lu, H.Z. Drug-resistant proteins in breast cancer: Recent progress in multidrug resistance. Ai zheng Aizheng Chin. J. Cancer 2003, 22, 441–444. [Google Scholar]
- Litman, T.; Jensen, U.; Hansen, A.; Covitz, K.M.; Zhan, Z.; Fetsch, P.; Abati, A.; Hansen, P.R.; Horn, T.; Skovsgaard, T.; et al. Use of peptide antibodies to probe for the mitoxantrone resistance-associated protein MXR/BCRP/ABCP/ABCG2. Acta Biochim. Biophys. Sin. 2002, 1565, 6–16. [Google Scholar] [CrossRef]
- Miyake, K.; Mickley, L.; Litman, T.; Zhan, Z.; Robey, R.; Cristensen, B.; Brangi, M.; Greenberger, L.; Dean, M.; Fojo, T. Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone-resistant cells. Cancer Res. 1999, 59, 8–13. [Google Scholar] [PubMed]
- Ambudkar, S.V.; Dey, S.; Hrycyna, C.A.; Ramachandra, M.; Pastan, I.; Gottesman, M.M. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu. Rev. Pharmacol. Toxicol. 1999, 39, 361–398. [Google Scholar] [CrossRef] [PubMed]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Rabindran, S.K.; Ross, D.D.; Doyle, L.A.; Yang, W.; Greenberger, L.M.; Fumitremorgin, C. Reverses multidrug resistance in cells transfected with the breast cancer resistance protein. Cancer Res. 2000, 60, 47–50. [Google Scholar] [PubMed]
- Allen, J.D.; van Loevezijn, A.; Lakhai, J.M.; van der Valk, M.; van Tellingen, O.; Reid, G.; Schellens, J.H.; Koomen, G.J.; Schinkel, A.H. Potent and specific inhibition of the breast cancer resistance protein multidrug transporter in vitro and in mouse intestine by a novel analogue of fumitremorgin C. Mol. Cancer Ther. 2002, 1, 417–425. [Google Scholar] [PubMed]
- Li, X.Q.; Wang, L.; Lei, Y.; Hu, T.; Zhang, F.L.; Cho, C.H.; To, K.K. Reversal of P-gp and BCRP-mediated MDR by tariquidar derivatives. Eur. J. Med. Chem. 2015, 101, 560–572. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.K.; Sodani, K.; Dai, C.L.; Abuznait, A.H.; Singh, S.; Xiao, Z.J.; Patel, A.; Talele, T.T.; Fu, L.; Kaddoumi, A.; et al. Nilotinib potentiates anticancer drug sensitivity in murine ABCB1-, ABCG2-, and ABCC10-multidrug resistance xenograft models. Cancer Lett. 2013, 328, 307–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, A.; Li, T.W.; Anreddy, N.; Wang, D.S.; Sodani, K.; Gadhia, S.; Kathawala, R.; Yang, D.H.; Cheng, C.; Chen, Z.S. Suppression of ABCG2 mediated MDR in vitro and in vivo by a novel inhibitor of ABCG2 drug transport. Pharmacol. Res. 2017, 121, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Matsson, P.; Englund, G.; Ahlin, G.; Bergström, C.A.S.; Norinder, U.; Artursson, P. A global drug inhibition pattern for the human ATP-binding cassette transporter breast cancer resistance protein (abcg2). J. Pharmacol. Exp. Ther. 2007, 323, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Hussein, N.; Amawi, H.; Karthikeyan, C.; Hall, F.S.; Mittal, R.; Trivedi, P.; Ashby, C.R.; Tiwari, A.K. The dopamine D 3 receptor antagonists PG01037, NGB2904, SB277011A, and U99194 reverse ABCG2 transporter-mediated drug resistance in cancer cell lines. Cancer Lett. 2017, 396, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Ágai-Csongor, É.; Domány, G.; Nógrádi, K.; Galambos, J.; Vágó, I.; Keserű, G.M.; Greiner, I.; Laszlovszky, I.; Gere, A.; Schmidt, É.; et al. Discovery of cariprazine (RGH-188): A novel antipsychotic acting on dopamine D3/D2 receptors. Bioorg. Med. Chem. Lett. 2012, 22, 3437–3440. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.K.; Sodani, K.; Wang, S.R.; Kuang, Y.H.; Ashby Jr, C.R.; Chen, X.; Chen, Z.S. Nilotinib (AMN107, Tasigna®) reverses multidrug resistance by inhibiting the activity of the ABCB1/Pgp and ABCG2/BCRP/MXR transporters. Biochem. Pharmacol. 2009, 78, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.M.; Manolaridis, I.; Kowal, J.; Zechner, M.; Taylor, N.M.I.; Bause, M.; Bauer, S.; Bartholomaeus, R.; Bernhardt, G.; Koenig, B.; et al. Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat. Struct. Mol. Biol. 2018, 25, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, C.; Malla, R.; Ashby, C.R., Jr.; Amawi, H.; Abbott, K.L.; Moore, J.; Chen, J.; Balch, C.; Lee, C.; Flannery, P.C.; et al. Pyrimido(1″,2″:1,5) pyrazolo(3,4-b)quinolines: Novel compounds that reverse ABCG2-mediated resistance in cancer cells. Cancer Lett. 2016, 376, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Baumert, C.; Hilgeroth, A. Recent advances in the development of P-gp inhibitors. Anticancer Agents Med. Chem. 2009, 9, 415–436. [Google Scholar] [CrossRef] [PubMed]
- Pena-Solorzano, D.; Stark, S.A.; Konig, B.; Sierra, C.A.; Ochoa-Puentes, C. ABCG2/BCRP: Specific and Nonspecific Modulators. Med. Res. Rev. 2017, 37, 987–1050. [Google Scholar] [CrossRef] [PubMed]
- McCormack, P.L. Cariprazine: First Global Approval. Drugs 2015, 75, 2035–2043. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Chen, Z.; McCarty, N. Synergistic anticancer effects of arsenic trioxide with bortezomib in mantle cell lymphoma. Am. J. Hematol. 2012, 87, 1057–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.K.; James, J.; Annunziata, C.M. Topotecan synergizes with CHEK1 (CHK1) inhibitor to induce apoptosis in ovarian cancer cells. BMC Cancer 2015, 15, 196. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.; Hwang, D.J.; Barrett, C.M.; Yang, J.; Duke, C.B.; Miller, D.D.; Dalton, J.T. A novel bis-indole destabilizes microtubules and displays potent in vitro and in vivo antitumor activity in prostate cancer. Cancer Chemother. Pharmacol. 2011, 67, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Cuestas, M.L.; Castillo, A.I.; Sosnik, A.; Mathet, V.L. Downregulation of mdr1 and abcg2 genes is a mechanism of inhibition of efflux pumps mediated by polymeric amphiphiles. Bioorg. Med. Chem. Lett. 2012, 22, 6577–6579. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, H.; Wei, L.; Li, G.; Yu, J.; Gao, Y.; Gao, P.; Zhang, X.; Wei, F.; Yin, D. Transcriptional modulation of BCRP gene to reverse multidrug resistance by toremifene in breast adenocarcinoma cells. Breast Cancer Res. Treat. 2010, 123, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, R.; Nishiyama, Y.; Furuta, T.; Hatano, H.; Igarashi, Y.; Asakawa, N.; Kodaira, H.; Takahashi, H.; Aiyama, R.; Matsuzaki, T. Novel acrylonitrile derivatives, YHO-13177 and YHO-13351, reverse BCRP/ABCG2-mediated drug resistance in vitro and in vivo. Mol. Cancer Ther. 2011, 10, 1252–1263. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Yao, Q.; Zhang, A.M.; Lin, S.; Wang, X.X.; Wu, L.; Sun, J.G.; Chen, Z.T. The effects of artesunate on the expression of EGFR and ABCG2 in A549 human lung cancer cells and a xenograft model. Molecules 2011, 16, 10556–10569. [Google Scholar] [CrossRef] [PubMed]
- Amawi, H.; Hussein, N.A.; Karthikeyan, C.; Manivannan, E.; Wisner, A.; Williams, F.E.; Samuel, T.; Trivedi, P.; Ashby, C.R., Jr.; Tiwari, A.K. HM015k, a Novel Silybin Derivative, Multi-Targets Metastatic Ovarian Cancer Cells and Is Safe in Zebrafish Toxicity Studies. Front. Pharmacol. 2017, 8, 498. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Stiegler, A.L.; Boggon, T.J.; Kobayashi, S.; Halmos, B. EGFR-mutated lung cancer: A paradigm of molecular oncology. Oncotarget 2010, 1, 497–514. [Google Scholar] [PubMed]
- Antczak, C.; Wee, B.; Radu, C.; Bhinder, B.; Holland, E.C.; Djaballah, H. A high-content assay strategy for the identification and profiling of ABCG2 modulators in live cells. Assay Drug Dev. Technol. 2014, 12, 28–42. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, T.; Ross, D.D. Breast cancer resistance protein (BCRP/ABCG2): Its role in multidrug resistance and regulation of its gene expression. Chin. J. Cancer 2012, 31, 73–99. [Google Scholar] [CrossRef] [PubMed]
- Van Loevezijn, A.; Allen, J.D.; Schinkel, A.H.; Koomen, G.J. Inhibition of BCRP-mediated drug efflux by fumitremorgin-type indolyl diketopiperazines. Bioorg. Med. Chem. Lett. 2001, 11, 29–32. [Google Scholar] [CrossRef]
- Shiozawa, K.; Oka, M.; Soda, H.; Yoshikawa, M.; Ikegami, Y.; Tsurutani, J.; Nakatomi, K.; Nakamura, Y.; Kitazaki, T.; Mizuta, Y. Reversal of breast cancer resistance protein (BCRP/ABCG2)-mediated drug resistance by novobiocin, a coumermycin antibiotic. Int. J. Cancer 2004, 108, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Houghton, P.J.; Germain, G.S.; Harwood, F.C.; Schuetz, J.D.; Stewart, C.F.; Buchdunger, E.; Traxler, P. Imatinib mesylate is a potent inhibitor of the ABCG2 (BCRP) transporter and reverses resistance to topotecan and SN-38 in vitro. Cancer Res. 2004, 64, 2333–2337. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, J.; DeGraff, W.G.; Gazdar, A.F.; Minna, J.D.; Mitchell, J.B. Evaluation of a tetrazolium-based semiautomated colorimetric assay: Assessment of chemosensitivity testing. Cancer Res. 1987, 47, 936–942. [Google Scholar] [PubMed]
- Shi, Z.; Liang, Y.J.; Chen, Z.S.; Wang, X.W.; Wang, X.H.; Ding, Y.; Chen, L.M.; Yang, X.P.; Fu, L.W. Reversal of MDR1/P-glycoprotein-mediated multidrug resistance by vector-based RNA interference in vitro and in vivo. Cancer Biol. Ther. 2006, 5, 39–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]







| Treatment | IC50 ± SEM (µM) a | |||
|---|---|---|---|---|
| H460 | FR b | H460-MX20 | FR b | |
| Mitoxantrone | 0.22 ± 0.01 | 1 | 6 ± 0.1 | 28 |
| +Cariprazine 1 µM | 0.2 ± 0.09 | 1 | 4.36 ± 0.3 * | 20 |
| +Cariprazine 10 µM | 0.2 ± 0.06 | 1 | 1 ± 0.5 ** | 5 |
| +Cariprazine 20 µM | 0.2 ± 0.02 | 1 | 0.2 ± 0.04 ** | 1 |
| +Nilotinib 5 µM | 0.2 ± 0.04 | 1 | 0.4 ± 0.01 ** | 2 |
| Treatment | IC50 ± SEM (µM) a | |||
|---|---|---|---|---|
| S1 | FR b | S1M1-80 | FR b | |
| Mitoxantrone | 0.5 ± 0.01 | 1 | 46 ± 0.57 | 93 |
| +Cariprazine 1 µM | 0.5 ± 0.02 | 0.9 | 26.4 ± 0.12 *** | 53 |
| +Cariprazine 10 µM | 0.43 ± 0.01 | 0.9 | 10 ± 0.09 *** | 20 |
| +Cariprazine 20 µM | 0.6 ± 0.01 | 1 | 0.66 ± 0.007 *** | 1.332 |
| +Nilotinib 5 µM | 0.44 ± 0.02 | 0.9 | 0.44 ± 0.01 *** | 0.888 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussein, N.; Ashby, C.R., Jr.; Amawi, H.; Nyinawabera, A.; Vij, A.; Khare, V.M.; Karthikeyan, C.; Tiwari, A.K. Cariprazine, A Dopamine D2/D3 Receptor Partial Agonist, Modulates ABCG2-Mediated Multidrug Resistance in Cancer. Cancers 2018, 10, 308. https://doi.org/10.3390/cancers10090308
Hussein N, Ashby CR Jr., Amawi H, Nyinawabera A, Vij A, Khare VM, Karthikeyan C, Tiwari AK. Cariprazine, A Dopamine D2/D3 Receptor Partial Agonist, Modulates ABCG2-Mediated Multidrug Resistance in Cancer. Cancers. 2018; 10(9):308. https://doi.org/10.3390/cancers10090308
Chicago/Turabian StyleHussein, Noor, Charles R. Ashby, Jr., Haneen Amawi, Angelique Nyinawabera, Atul Vij, Vishwa M. Khare, Chandrabose Karthikeyan, and Amit K. Tiwari. 2018. "Cariprazine, A Dopamine D2/D3 Receptor Partial Agonist, Modulates ABCG2-Mediated Multidrug Resistance in Cancer" Cancers 10, no. 9: 308. https://doi.org/10.3390/cancers10090308