Enhanced Hsa-miR-181d/p-STAT3 and Hsa-miR-181d/p-STAT5A Ratios Mediate the Anticancer Effect of Garcinol in STAT3/5A-Addicted Glioblastoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Drugs and Chemicals
2.2. Analyses of Cancer RNAseq Dataset
2.3. Cell lines and Primary Culture Cell Culture
2.4. Sulforhodamine B (SRB) Viability Assay
2.5. Western Blot Analysis
2.6. Immunohistochemical (IHC) Staining
2.7. Immunofluorescence Staining and Quantification
2.8. Tumorsphere Formation Assay
2.9. Colony Formation Assay
2.10. Invasion Assay
2.11. Wound-Healing Migration Assay
2.12. Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
2.13. PE-Annexin V/7-AAD Cell Death Assay
2.14. Mir-181 Transfection Assay
2.15. Mice Tumor Xenograft Study
2.16. Statistical Analysis
3. Results
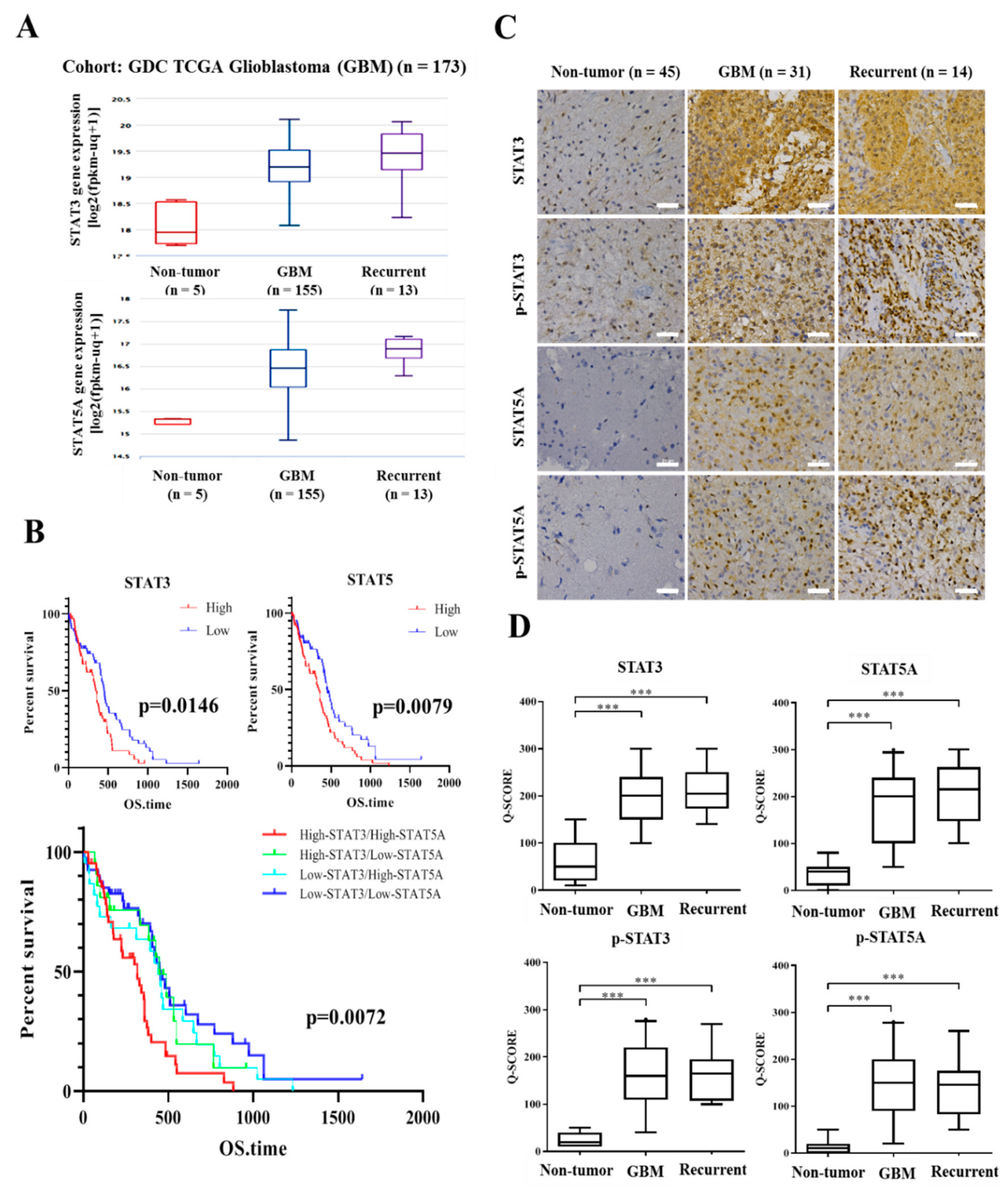
3.1. STAT-3 and STAT-5 Are Highly Expressed in Primary and Recurrent Glioblastoma, and Their Expression Negatively Correlates with Overall Survival Rates
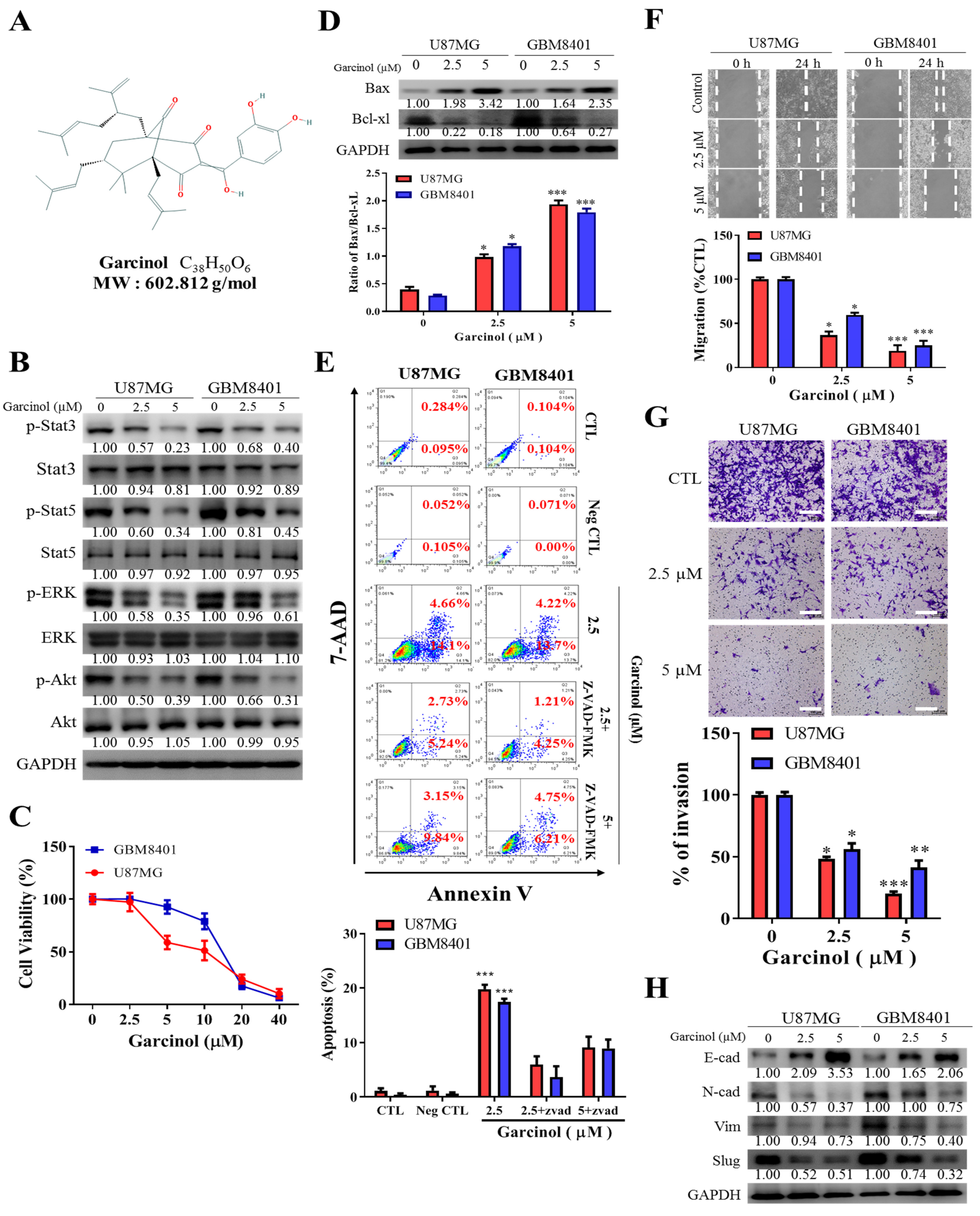
3.2. Garcinol Significantly Inhibits GBM Cell Viability and Oncogenicity through Induction of STAT3/5A Signaling and Enhanced Apoptosis
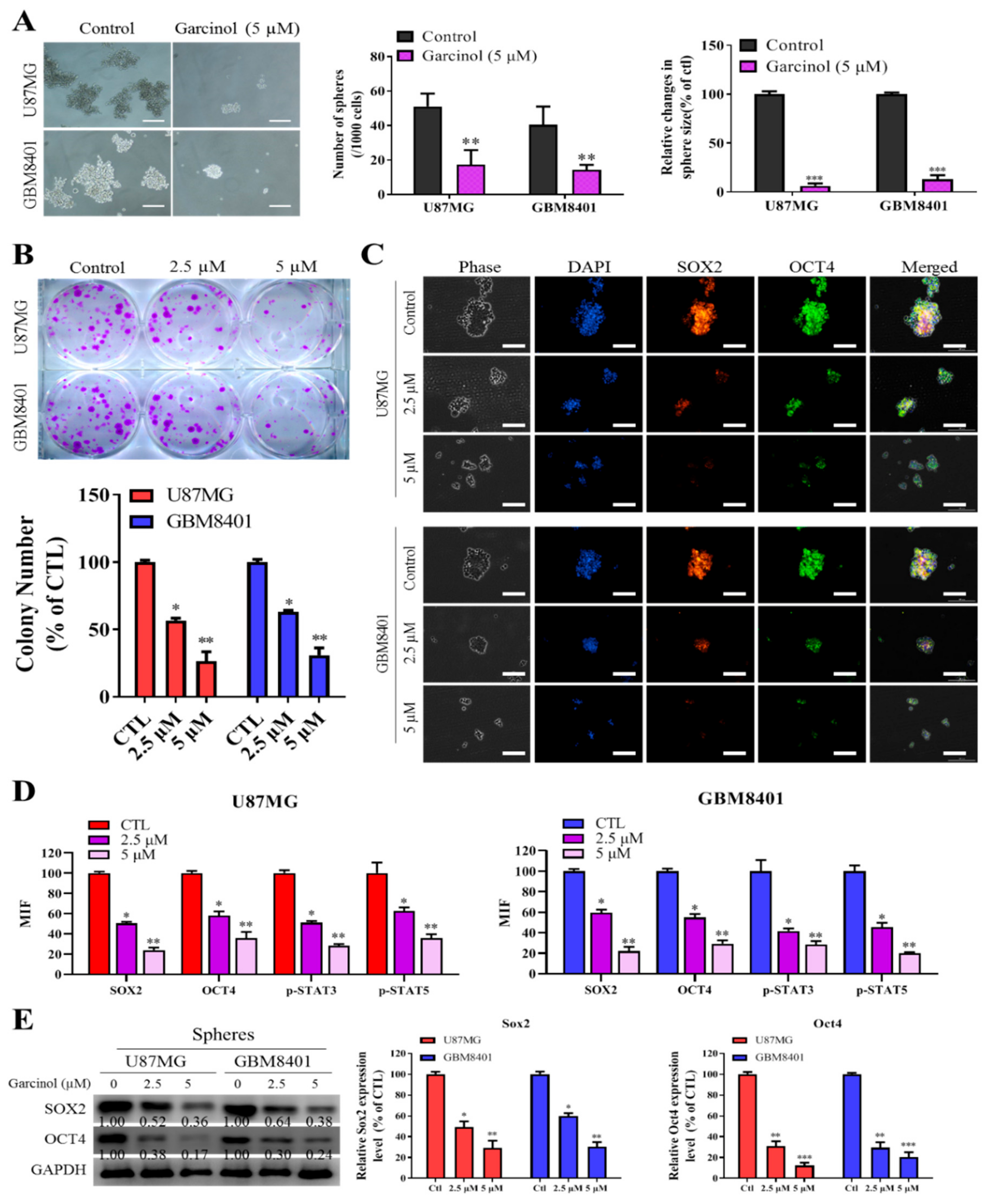
3.3. Garcinol Negatively Impacts GBM Stem Cell-Like Phenotypes
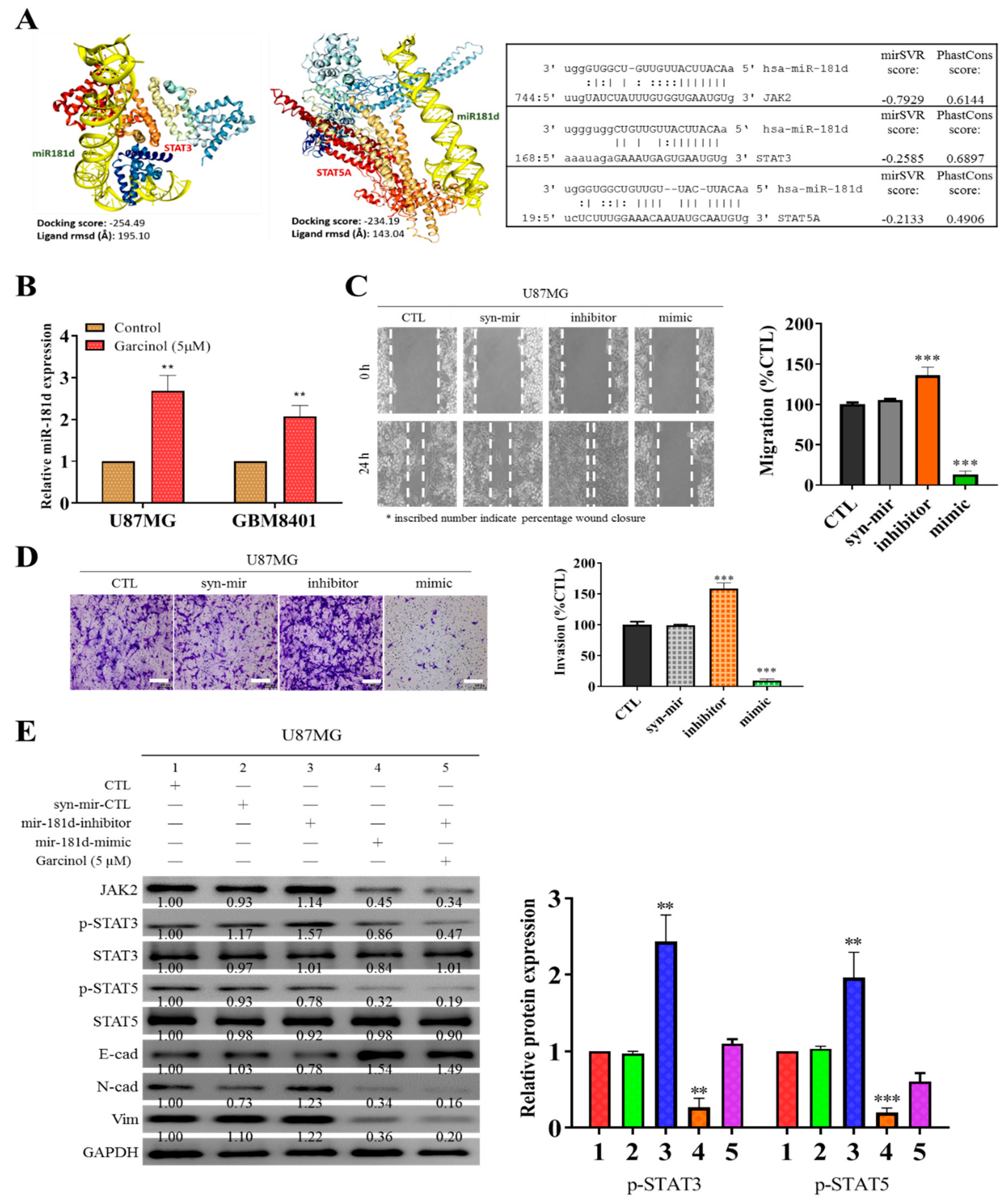
3.4. Garcinol Increases the Expression of Hsa-miR181d, which Has Inhibitory Effects on STAT3 and STAT5 Activation
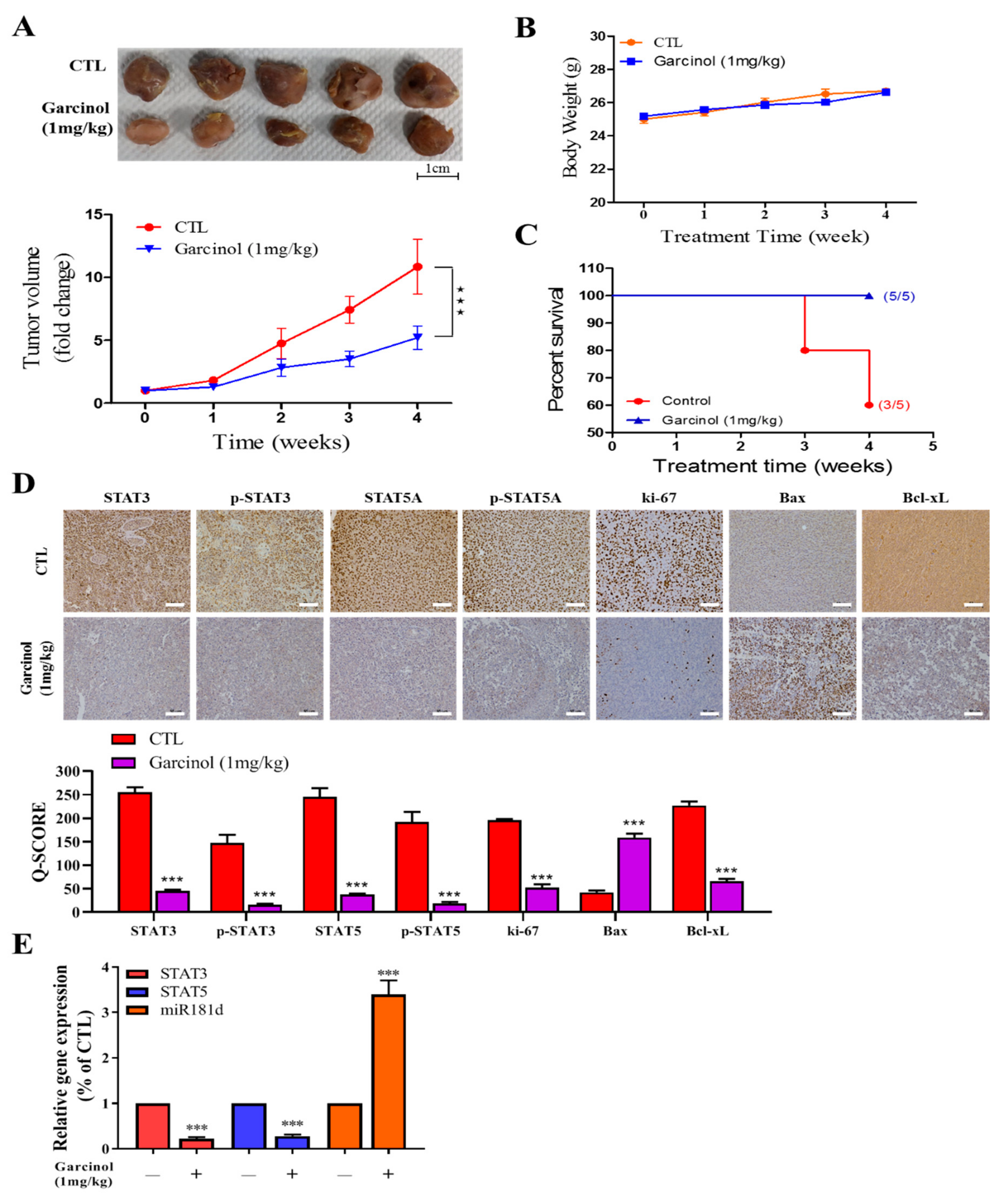
3.5. Garcinol Inhibits Tumor Growth in GBM Mice Models through Inversely Correlated STAT3/5A and hsa-miR-181d Expressions
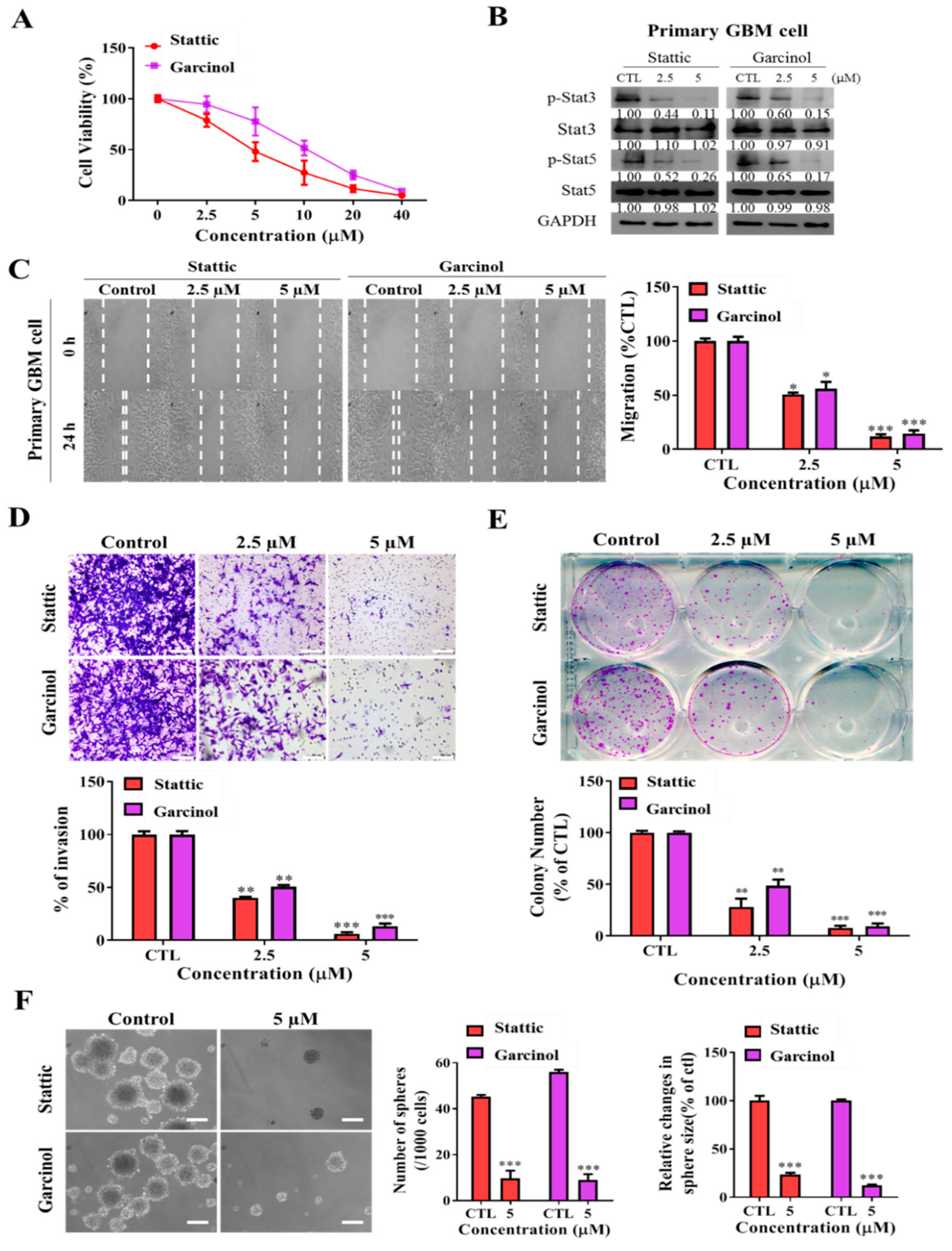
3.6. Garcinol, Akin to Stattic, a Selective Inhibitor of STAT3/5A Activation, Inhibits the Metastatic and Cancer Stem Cell-Like Phenotypes of Primary GBM Culture Cells
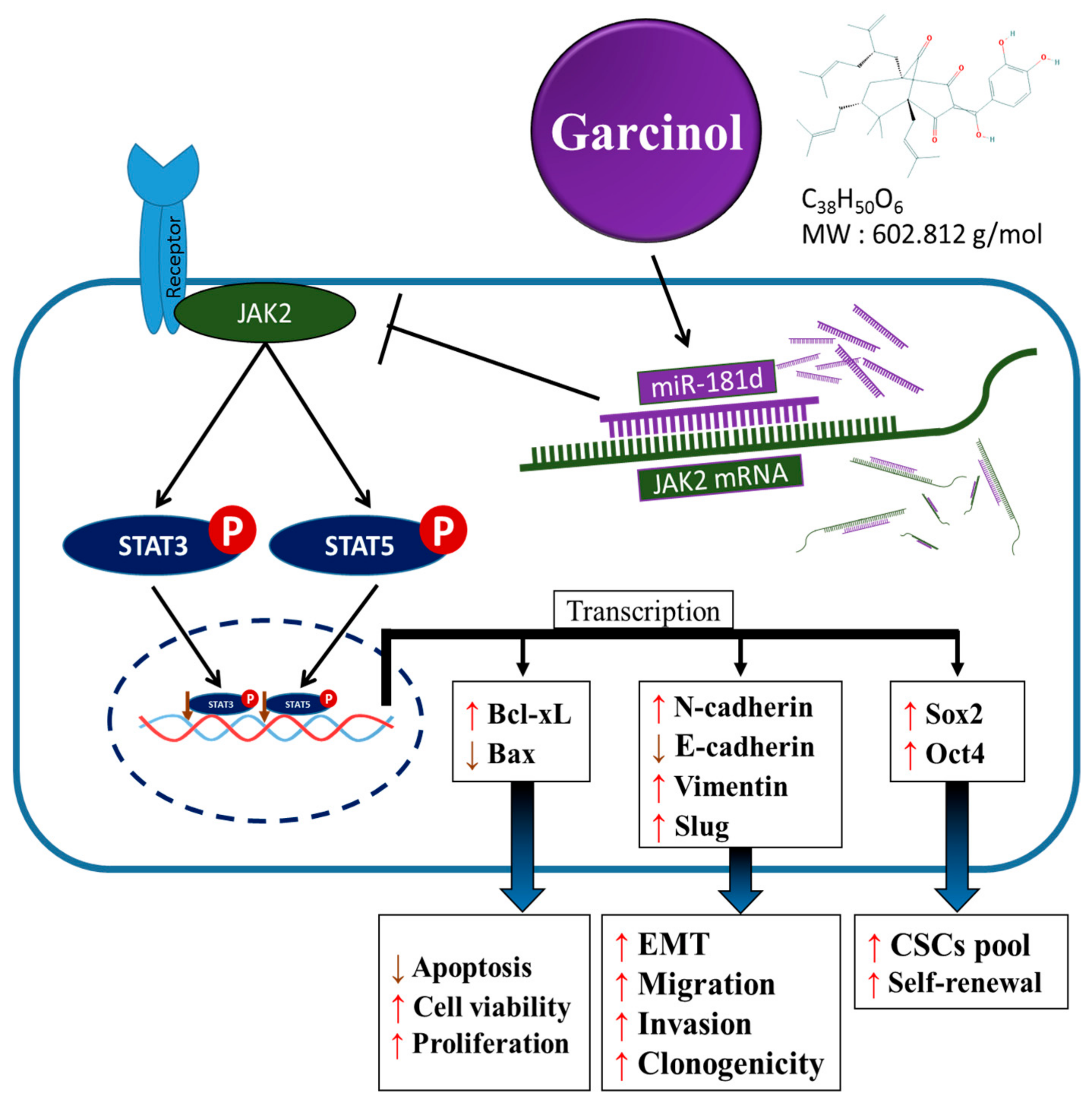
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Adamson, C.; Kanu, O.O.; Mehta, A.I.; Di, C.; Lin, N.; Mattox, A.K.; Bigner, D.D. Glioblastoma multiforme: A review of where we have been and where we are going. Expert Opin. Investig. Drugs 2009, 18, 1061–1083. [Google Scholar] [CrossRef] [PubMed]
- Omuro, A.; DeAngelis, L.M. Glioblastoma and other malignant gliomas: A clinical review. JAMA 2013, 310, 1842–1850. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Kreisl, T.N.; Kim, L.; Moore, K.; Duic, P.; Royce, C.; Stroud, I.; Fine, H.A. Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. J. Clin. Oncol. 2009, 27, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, A.O.; Stupp, R. Epilepsy in brain tumor patients. Curr. Opin. Neurol. 2010, 23, 603–609. [Google Scholar] [CrossRef]
- Colak, S.; Medema, J.P. Cancer stem cells—Important players in tumor therapy resistance. FEBS J. 2014, 281, 4779–4791. [Google Scholar] [CrossRef]
- Frank, N.Y.; Schatton, T.; Frank, M.H. The therapeutic promise of the cancer stem cell concept. J. Clin. Investig. 2010, 120, 41–50. [Google Scholar] [CrossRef]
- Jackson, M.; Hassiotou, F.; Nowak, A. Glioblastoma stem-like cells: At the root of tumor recurrence and a therapeutic target. Carcinogenesis 2015, 36, 177–185. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 2018, 1–16. [Google Scholar] [CrossRef]
- Toledo-Guzmán, M.E.; Bigoni-Ordóñez, G.D.; Hernández, M.I.; Ortiz-Sánchez, E. Cancer stem cell impact on clinical oncology. World J. Stem Cells 2018, 10, 183–195. [Google Scholar] [CrossRef]
- Cheng, C.C.; Shi, L.H.; Wang, X.J.; Wang, S.X.; Wan, X.Q.; Liu, S.R.; Wang, Y.F.; Lu, Z.; Wang, L.H.; Ding, Y. Stat3/Oct-4/c-Myc signal circuit for regulating stemness-mediated doxorubicin resistance of triple-negative breast cancer cells and inhibitory effects of WP1066. Int. J. Oncol. 2018, 53, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Do, D.V.; Ueda, J.; Messerschmidt, D.M.; Lorthongpanich, C.; Zhou, Y.; Feng, B.; Guo, G.; Lin, P.J.; Hossain, M.Z.; Zhang, W.; et al. A genetic and developmental pathway from STAT3 to the OCT4–NANOG circuit is essential for maintenance of ICM lineages in vivo. Genes Dev. 2013, 27, 1378–1390. [Google Scholar] [CrossRef] [PubMed]
- Matthews, J.R.; Sansom, O.J.; Clarke, A.R. Absolute requirement for STAT3 function in small-intestine crypt stem cell survival. Cell Death Differ. 2011, 18, 1934–1943. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.; Turkson, J. Targeting STAT3 in cancer: How successful are we? Expert Opin. Investig. Drugs 2009, 18, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Guryanova, O.A.; Wu, Q.; Cheng, L.; Lathia, J.D.; Huang, Z.; Yang, J.; MacSwords, J.; Eyler, C.E.; McLendon, R.E.; Heddleston, J.M.; et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell 2011, 19, 498–511. [Google Scholar] [CrossRef]
- Carro, M.S.; Lim, W.K.; Alvarez, M.J.; Bollo, R.J.; Zhao, X.D.; Snyder, E.Y.; Sulman, E.P.; Anne, S.L.; Doetsch, F.; Colman, H.; et al. The transcriptional network for mesenchymal transformation of brain tumors. Nature 2010, 463, 318–325. [Google Scholar] [CrossRef]
- Li, G.; Wei, H.; Chen, Z.; Lv, S.; Yin, C.; Wang, D. STAT3 silencing with lentivirus inhibits growth and induces apoptosis and differentiation of U251 cells. J. Neuro-Oncology 2009, 91, 165–174. [Google Scholar] [CrossRef]
- Cao, S.; Wang, C.; Zheng, Q.; Qiao, Y.; Xu, K.; Jiang, T.; Wu, A. STAT5 regulates glioma cell invasion by pathways dependent and independent of STAT5 DNA binding. Neurosci. Lett. 2011, 487, 228–233. [Google Scholar] [CrossRef]
- Latha, K.; Li, M.; Chumbalkar, V.; Gururaj, A.; Hwang, Y.; Dakeng, S.; Furnari, F.B. Nuclear EGFRvIII-STAT5b complex contributes to glioblastoma cell survival by direct activation of the Bcl-XL promoter. Int. J. Cancer 2013, 132, 509–520. [Google Scholar] [CrossRef]
- Roos, A.; Dhruv, H.D.; Peng, S.; Inge, L.J.; Tuncali, S.; Pineda, M.; Millard, N.; Mayo, Z.; Eschbacher, J.M.; Loftus, J.C.; et al. EGFRvIII-Stat5 Signaling Enhances Glioblastoma Cell Migration and Survival. Mol. Cancer Res. 2018, 16, 1185–1195. [Google Scholar] [CrossRef]
- Fan, Q.W.; Cheng, C.K.; Gustafson, W.C.; Charron, E.; Zipper, P.; Wong, R.A.; Chen, J.; Lau, J.; Knobbe-Thomsen, C.; Weller, M.; et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell 2013, 24, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, V.; Nowicki, M.O.; Idriss, M.; Jimenez, M.A.; Lugli, G.; Hayes, J.L.; Mahmoud, A.B.; Zane, R.E.; Passaro, C.; Ligon, K.L.; et al. The functional synergism of microRNA clustering provides therapeutically relevant epigenetic interference in glioblastoma. Nat. Commun. 2019, 10, 442. [Google Scholar] [CrossRef] [PubMed]
- Floyd, D.; Purow, B. Micro-masters of glioblastoma biology and therapy: Increasingly recognized roles for microRNAs. Neuro-Oncology 2014, 16, 622–627. [Google Scholar] [CrossRef]
- Pop-Bica, C.; Pintea, S.; Cojocneanu-Petric, R.; Del Sal, G.; Piazza, S.; Wu, Z.H.; Alencar, A.J.; Lossos, I.S.; Berindan-Neagoe, I.; Calin, G.A. MiR-181 family-specific behavior in different cancers: A meta-analysis view. Cancer Metastasis Rev. 2018, 37, 17–32. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; De Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMTGene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.H.; Zhang, P.; Fisher, B.J.; Macdonald, D.R.; McElroy, J.P.; Lesser, G.J.; Fleming, J.; Chakraborty, A.R.; Liu, Z.; Becker, A.P.; et al. Association of MGMT Promoter Methylation Status With Survival Outcomes in Patients With High-Risk Glioma Treated With Radiotherapy and Temozolomide: An Analysis From the NRG Oncology/RTOG 0424 Trial. JAMA Oncol. 2018, 4, 1405–1409. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, J.; Hoadley, K.; Kushwaha, D.; Ramakrishnan, V.; Li, S.; Kang, C.; You, Y.; Jiang, C.; Song, S.W.; et al. miR-181d: A predictive glioblastoma biomarker that downregulates MGMT expression. Neuro-Oncology 2012, 14, 712–719. [Google Scholar] [CrossRef]
- Yang, F.; Liu, X.; Liu, Y.; Liu, Y.; Zhang, C.; Wang, Z.; Jiang, T.; Wang, Y. miR-181d/MALT1 regulatory axis attenuates mesenchymal phenotype through NF-κB pathways in glioblastoma. Cancer Lett. 2017, 396, 1–9. [Google Scholar] [CrossRef]
- Saadat, N.; Gupta, S.V. Potential Role of Garcinol as an Anticancer Agent. J. Oncol. 2012, 2012, 647206. [Google Scholar] [CrossRef]
- Huang, W.C.; Kuo, K.T.; Adebayo, B.O.; Wang, C.H.; Chen, Y.J.; Jin, K.; Yeh, C.T. Garcinol inhibits cancer stem cell-like phenotype via suppression of the Wnt/β-catenin/STAT3 axis signalling pathway in human non-small cell lung carcinomas. J. Nutr. Biochem. 2018, 54, 140–150. [Google Scholar] [CrossRef]
- Arshad, L.; Haque, M.A.; Bukhari, S.N.A.; Jantan, I. An overview of structure–activity relationship studies of curcumin analogs as antioxidant and anti-inflammatory agents. Futur. Med. Chem. 2017, 9, 605–626. [Google Scholar] [CrossRef]
- Hong, J.; Kwon, S.J.; Sang, S.; Ju, J.; Zhou, J.N.; Ho, C.T.; Huang, M.T.; Yang, C.S. Effects of garcinol and its derivatives on intestinal cell growth: Inhibitory effects and autoxidation-dependent growth-stimulatory effects. Free. Radic. Biol. Med. 2007, 42, 1211–1221. [Google Scholar] [CrossRef]
- Ahmad, A.; Sarkar, S.H.; Aboukameel, A.; Ali, S.; Biersack, B.; Seibt, S.; Li, Y.; Bao, B.; Kong, D.; Banerjee, S.; et al. Anticancer action of garcinol in vitro and in vivo is in part mediated through inhibition of STAT-3 signaling. Carcinogenesis 2012, 33, 2450–2456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, A.; Wang, Z.; Ali, R.; Maitah, M.Y.; Kong, D.; Banerjee, S.; Padhye, S.; Sarkar, F.H. Apoptosis-inducing effect of garcinol is mediated by NF-κB signaling in breast cancer cells. J. Cell. Biochem. 2010, 109, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Su, Y.K.; Lin, C.M.; Chao, T.Y.; Huang, S.P.; Huynh, T.T.; Jan, H.J.; Whang-Peng, J.; Chiou, J.F.; Wu, A.T.; et al. Preclinical investigation of ibrutinib, a Bruton’s kinase tyrosine (Btk) inhibitor, in suppressing glioma tumorigenesis and stem cell phenotypes. Oncotarget 2016, 7, 69961–69975. [Google Scholar] [CrossRef]
- Lathia, J.D.; Mack, S.C.; Mulkearns-Hubert, E.E.; Valentim, C.L.; Rich, J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015, 29, 1203–1217. [Google Scholar] [CrossRef] [Green Version]
- Beier, D.; Schulz, J.B.; Beier, C.P. Chemoresistance of glioblastoma cancer stem cells—Much more complex than expected. Mol. Cancer 2011, 10, 128. [Google Scholar] [CrossRef] [Green Version]
- Mohanty, S.K.; Yagiz, K.; Pradhan, D.; Luthringer, D.J.; Amin, M.B.; Alkan, S.; Cinar, B. STAT3 and STAT5A are potential therapeutic targets in castration-resistant prostate cancer. Oncotarget 2017, 8, 85997–86010. [Google Scholar] [CrossRef] [Green Version]
- Ghanem, S.; Friedbichler, K.; Boudot, C.; Bourgeais, J.; Gouilleux-Gruart, V.; Régnier, A.; Herault, O.; Moriggl, R.; Gouilleux, F. STAT5A/5B-specific expansion and transformation of hematopoietic stem cells. Blood Cancer J. 2017, 7, e514. [Google Scholar] [CrossRef] [Green Version]
- Gleixner, K.V.; Schneeweiss, M.A.; Herrmann, H.; Blatt, K.; Berger, D.; Eisenwort, G.; Greiner, G.; Byrgazov, K.; Hoermann, G.; Konopleva, M.; et al. Combined Targeting of STAT3 and STAT5: A Novel Approach to Overome Drug Resistance in Ph+ Cml. Blood 2016, 128, 4241. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, F.; Niu, R. Multiple regulation pathways and pivotal biological functions of STAT3 in cancer. Sci. Rep. 2015, 5, 17663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, S.J.; Kwon, O.S.; Kim, K.T.; Go, Y.H.; Yu, S.I.; Lee, B.H.; Miyoshi, H.; Oh, E.; Cho, S.J.; Cha, H.J. Role of MEK partner-1 in cancer stemness through MEK/ERK pathway in cancerous neural stem cells, expressing EGFRviii. Mol. Cancer 2017, 16, 140. [Google Scholar] [CrossRef]
- Shi, M.; Ji, H.; Jiang, H.; Li, D.; Chen, G.; Wang, Z. MicroRNA-181d is a tumor suppressor in human esophageal squamous cell carcinoma inversely regulating Derlin-1. Oncol. Rep. 2016, 36, 2041–2048. [Google Scholar]
- Wang, X.F.; Shi, Z.M.; Wang, X.R.; Cao, L.; Wang, Y.Y.; Zhang, J.X.; Yin, Y.; Luo, H.; Kang, C.S.; Liu, N.; et al. MiR-181d acts as a tumor suppressor in glioma by targeting K-ras and Bcl-2. J. Cancer Res. Clin. Oncol. 2012, 138, 573–584. [Google Scholar] [CrossRef]
- Luo, N.; Balko, J.M. Role of JAK-STAT Pathway in Cancer Signaling. In Predictive Biomarkers in Oncology; Badve, S., Kumar, G., Eds.; Springer: Cham, Switzerland, 2019; pp. 311–319. [Google Scholar]
- Zhuang, G.; Wu, X.; Jiang, Z.; Kasman, I.; Yao, J.; Guan, Y.; Oeh, J.; Modrusan, Z.; Bais, C.; Sampath, D.; et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012, 31, 3513–3523. [Google Scholar] [CrossRef]
- Lam, D.; Barré, B.; Guette, C.; Coqueret, O. Circulating miRNAs as new activators of the JAK-STAT3 pathway. JAK STAT 2013, 2, e22996. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Gao, X.; Zhang, W.; Zhu, T.; Bi, W.; Zhang, Y. MicroRNA-204 deregulation in lung adenocarcinoma controls the biological behaviors of endothelial cells potentially by modulating Janus kinase 2-signal transducer and activator of transcription 3 pathway. IUBMB Life 2017, 70, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Ho, K.H.; Chen, P.H.; Hsi, E.; Shih, C.M.; Chang, W.C.; Cheng, C.H.; Lin, C.W.; Chen, K.C. Identification of IGF-1-enhanced cytokine expressions targeted by miR-181d in glioblastomas via an integrative miRNA/mRNA regulatory network analysis. Sci. Rep. 2017, 7, 732. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.E.; Patel, M.; Ruzevick, J.; Jackson, C.M.; Lim, M. STAT3 Activation in Glioblastoma: Biochemical and Therapeutic Implications. Cancers 2014, 6, 376–395. [Google Scholar] [CrossRef] [Green Version]
- Ramalingam, D.; Ziegelbauer, J.M. Viral microRNAs Target a Gene Network, Inhibit STAT Activation, and Suppress Interferon Responses. Sci. Rep. 2017, 7, 40813. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.-W.; Lee, P.M.; Bamodu, O.A.; Su, Y.-K.; Fong, I.-H.; Yeh, C.-T.; Chien, M.-H.; Kan, I.-H.; Lin, C.-M. Enhanced Hsa-miR-181d/p-STAT3 and Hsa-miR-181d/p-STAT5A Ratios Mediate the Anticancer Effect of Garcinol in STAT3/5A-Addicted Glioblastoma. Cancers 2019, 11, 1888. https://doi.org/10.3390/cancers11121888
Liu H-W, Lee PM, Bamodu OA, Su Y-K, Fong I-H, Yeh C-T, Chien M-H, Kan I-H, Lin C-M. Enhanced Hsa-miR-181d/p-STAT3 and Hsa-miR-181d/p-STAT5A Ratios Mediate the Anticancer Effect of Garcinol in STAT3/5A-Addicted Glioblastoma. Cancers. 2019; 11(12):1888. https://doi.org/10.3390/cancers11121888
Chicago/Turabian StyleLiu, Heng-Wei, Peter Mingjui Lee, Oluwaseun Adebayo Bamodu, Yu-Kai Su, Iat-Hang Fong, Chi-Tai Yeh, Ming-Hsien Chien, I-Hung Kan, and Chien-Min Lin. 2019. "Enhanced Hsa-miR-181d/p-STAT3 and Hsa-miR-181d/p-STAT5A Ratios Mediate the Anticancer Effect of Garcinol in STAT3/5A-Addicted Glioblastoma" Cancers 11, no. 12: 1888. https://doi.org/10.3390/cancers11121888






