A Review of Computer-Aided Expert Systems for Breast Cancer Diagnosis
Abstract
:Simple Summary
Abstract
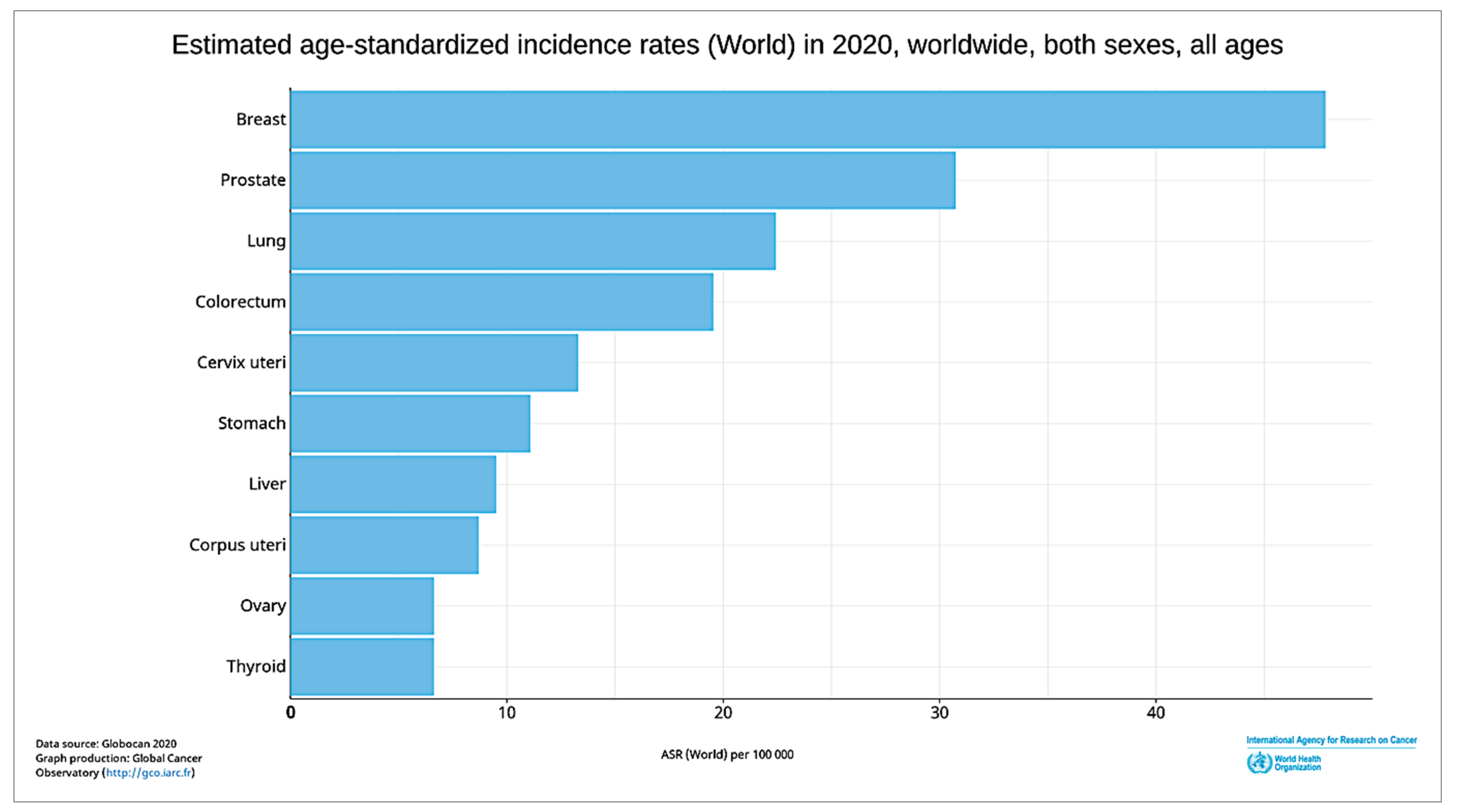
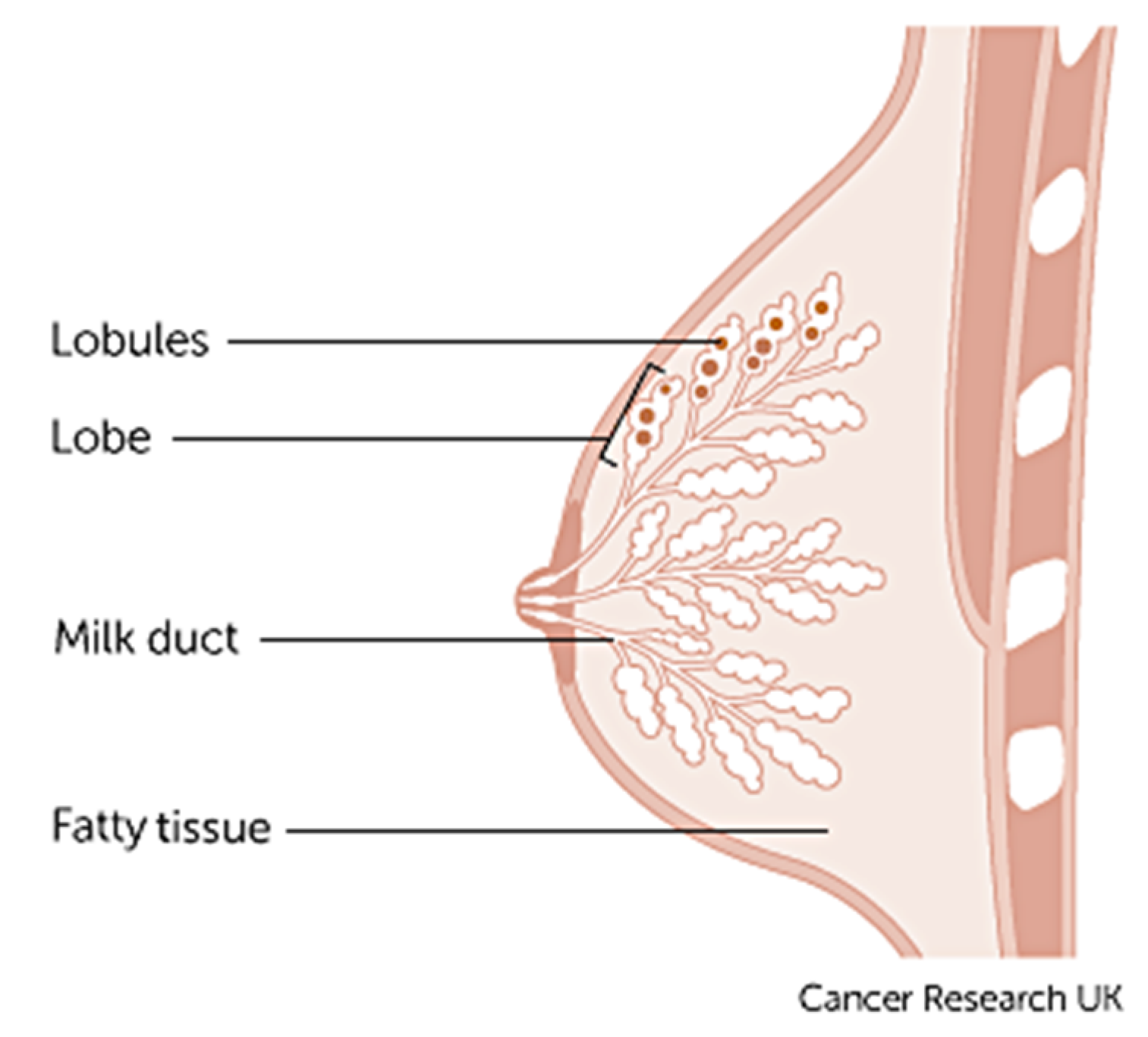
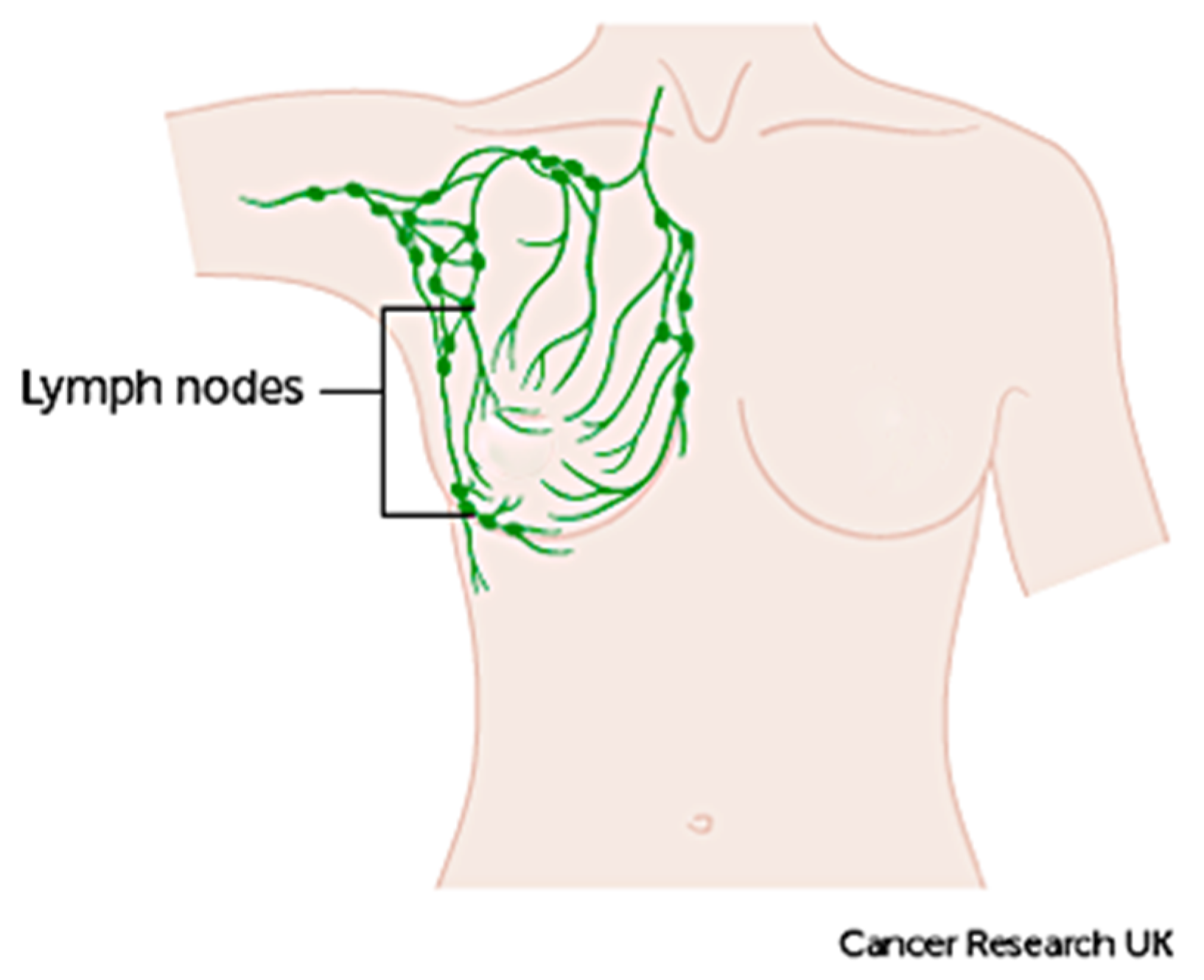
1. Introduction
2. Datasets for Breast Cancer Classification
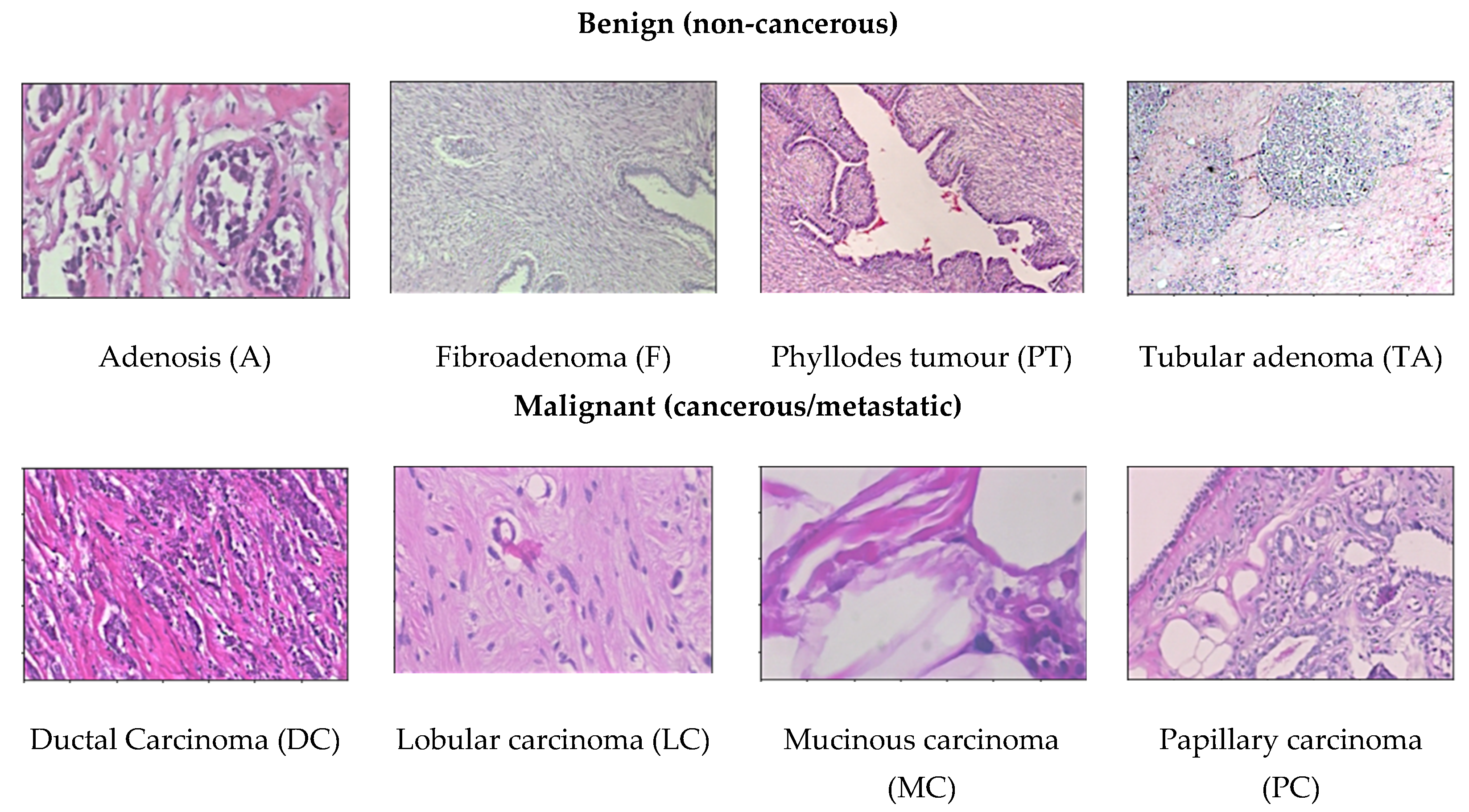
- BreaKHis dataset [15]: This dataset provides 4 different magnification levels of 40×, 100×, 200×, and 400× histology images of size 752 × 582 pixels. It consists of a total number of 7909 images acquired from a clinical study from January 2014 to December 2014 in P&D Laboratory, Brazil by 82 patients. For binary classification, there are two categories of benign and malignant to determine cancer or non-cancerous. There are 1995 images (652 benign and 1370 malignant) in 40× magnification level; 2081 images (644 benign and 1437 malignant) in 100× magnification level; 2013 images (623 benign and 1390 malignant) in 200× magnification level; and 1820 images (588 benign and 1232 malignant) in 400× magnification level in the dataset. To further perform multiclassification, the dataset contains four distinct types for each breast tumours. The category benign type of breast tumour consists of adenosis (A), fibroadenoma (F), phyllodes tumour (PT), and tubular adenoma (TA). The malignant type of breast tumour consists of ductal carcinoma (DC), lobular carcinoma (LC), mucinous carcinoma (MC), and papillary carcinoma (PC). This dataset is the most used dataset by many researchers for CAD breast cancer in histopathology images [11,17,18,19,20,21,22,23,24,25,26,27,28,29,30]. This dataset can be obtained from https://web.inf.ufpr.br/vri/databases/breast-cancer-histopathological-database-breakhis/ (accessed on 16 March 2021).
- Bioimaging Challenge 2015 dataset [31]: This dataset contains 269 images of haematoxylin and eosin (H&E)-stained breast cancer histology images with image size of 2048 × 1536 pixels. Images are provided in 200× magnification level. For binary classification, there are two categories to determine cancer or non-cancerous. To further classify, the non-cancerous categories can be categorized as normal and benign, while the cancerous ones can be categorized as in situ carcinoma and invasive carcinoma. The training set has a total of 249 images form by 55 normal class, 69 benign class, 63 in situ carcinoma class, and 62 invasive carcinoma class, while the test set has a total of 20 images with 5 images for each class. Additionally, there is an extended test set with more diversity provided with a total of 16 images available. In this extended test set, there are 4 images for each class. This dataset can be obtained from https://rdm.inesctec.pt/dataset/nis-2017-003 (accessed on 16 March 2021).
- BACH (BreAst Cancer Histology) dataset [32]: The ICIAR 2018 challenge resulted in the BreAst Cancer Histology (BACH) image dataset, which is an extended version of the Bioimaging 2015 breast histology classification challenge dataset with similar image sizes and magnification levels [31]. The dataset has a total number of 400 images, respectively classified to a total number of 100 normal class, 100 benign class, 100 in situ carcinoma class, and 100 invasive carcinoma class. The test set has a total of 100 images without any labels. The dataset can be obtained from https://iciar2018-challenge.grand-challenge.org/ (accessed on 16 March 2021).
- CAMELYON dataset [33]: The Cancer Metastases in Lymph Nodes Challenge breast cancer metastasis detection dataset combines two datasets collected from CAMELYON16 and CAMELYON17 challenges, with each image approximately 1 × 105 by 2 × 105 pixels at the highest resolution. The first dataset CAMELYON16 consists of a total 400 whole-slide images (WSIs) of haematoxylin and eosin (H&E)-stained lymph node sections collected from Radboud University Medical Center (Nijmegen, The Netherlands) and the University Medical Center Utrecht (Utrecht, The Netherlands). Each image is annotated with a binary label for classification, showing normal and presence of metastases tissue. There are two sets of training datasets, the first has a total number of 170 images, formed of 100 normal class and 70 metastases class, while the second has a total number of 100 images formed of 60 normal class and 40 metastases class. The test set holds a total number of 130 images. The CAMELYON17 dataset consists of a total of 1399 histology breast images. This version is extended from the CAMELYON16 which include patients testing for breast cancer from the CAMELYON16 challenge with an additional three medical centres from the Netherlands, specifically: slides from 130 lymph node resections from Radboud University Medical Center in Nijmegen (RUMC), 144 from Canisius-Wilhelmina Hospital in Nijmegen (CWZ), 129 from University Medical Center Utrecht (UMCU), 168 from Rijnstate Hospital in Arnhem (RST), and 140 from the Laboratory of Pathology East-Netherlands in Hengelo (LPON) [34]. The dataset can be obtained from https://camelyon17.grand-challenge.org (accessed on 16 March 2021).
- PatchCamelyon (PCam) dataset [35]: Whole slide images (WSI) are computationally expensive and only require the small regions of interest (ROIs) from the entire image, therefore it would need to estimate a significantly substantial number of parameters. Thus, this version of dataset is derived from the CAMELYON dataset with a total number of 327.680 histopathologic scans of lymph node sections images, each in the size of 96 × 96 px pixels. Like the CAMELYON dataset, each image is annotated with binary label for classification, showing normal and presence of metastases tissue. The main difference and advantage of this dataset is that it is bigger than CIFAR10, smaller than ImageNet, additionally it is trainable on a single GPU to able to achieve competitive scores in the CAMELYON16 tasks of cancer detection and WSI diagnosis. PCam contributed by supplying the segmented tissue parts that separated tissue and background from the whole slide images. The dataset can be obtained from https://github.com/basveeling/pcam (accessed on 16 March 2021).
- MITOS-12 dataset [36]: The conference ICPR 2012 supplied the MITOS dataset benchmark that consists of 50 histopathology images of haematoxylin and eosin (H&E)-stained slides of breast cancer images from 5 different breast biopsies at 40× magnification level. However, this dataset is too small to produce an exceptionally reliable performance and the robustness of the diagnosis system is limited. Therefore, an extended version of the dataset (MITOS-ATYPIA-14) was presented at ICPR 2014.
- MITOS-ATYPIA-14 dataset [37]: The grand challenge dataset was presented at the ICPR 2014 conference, extended from the MITOS-12 challenge that provides haematoxylin and eosin (H&E)-stained slides of breast cancer images with the size of 1539 × 1376 pixels at 20× and 40× magnification level. There is a training set with a total number of 1200 images acquired from 16 different biopsies and testing set with a total number of 496 images acquired from 5 different breast biopsies. The dataset consists of a significantly diverse variation of stained images in many conditions to elevate the challenge to achieve a more exceptional performance. The dataset can be obtained from https://mitos-atypia-14.grand-challenge.org/ (accessed on 16 March 2021).
- TUPAC16 dataset [38]: The dataset consists of a total number of 73 breast cancer histopathology images at 40× magnification level from three pathology centres in the Netherlands. The dataset is composed of 23 test images with a size of 2000 × 2000 pixels and 50 training images with a size of 5657 × 5657 pixels collected from two separate pathology centres. The images contained in the training dataset are later cropped randomly to the size of 2000 × 2000 pixels. The dataset can be obtained from http://tupac.tue-image.nl/node/3 (accessed on 16 March 2021).
- UCSB bio segmentation benchmark (UCSB-BB) [39]: This dataset contains 50 haematoxylin and eosin (H&E)-stained histopathology images used in breast cancer cell detection with the size of 896 × 768 pixels and a ground truth table. Each image is annotated with binary label for classification, it contains half malignant class and half benign class. The dataset can be obtained from https://bioimage.ucsb.edu/research/bio-segmentation (accessed on 16 March 2021).
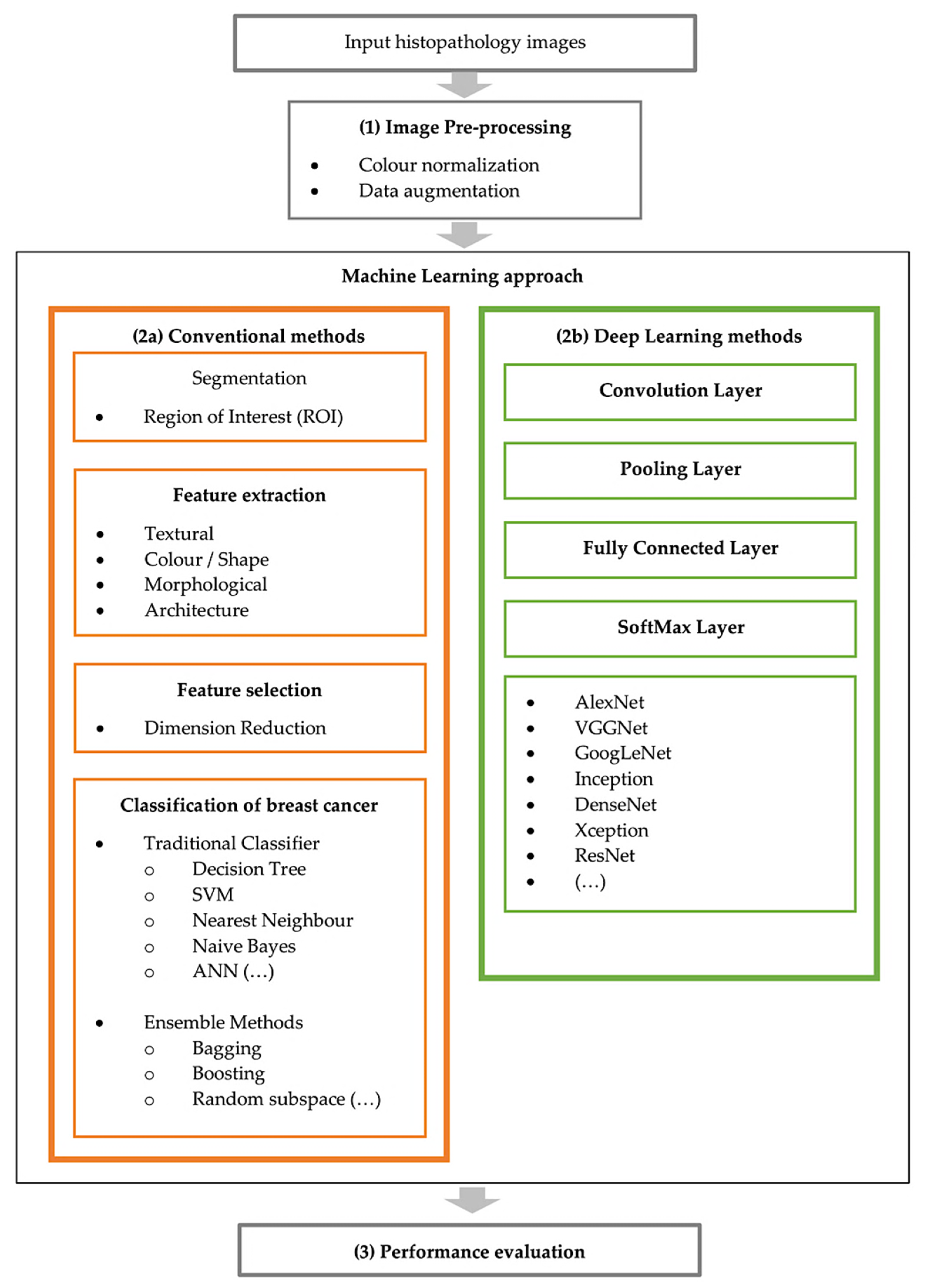
3. Computer-Aided Diagnosis Expert Systems
- Computer-aided detection (CADe) systems, which detect cancer or metastatic tissue.
- Computer-aided diagnosis (CADx) systems, which determine the distinct types of breast cancer.
3.1. Image Pre-Processing
- (1)
- Colour normalisation: The inconsistent various appearances of stained sections is amongst the foremost challenges to analyse histopathological images [40]. This is because the samples are collected under various inconsistent conditions of tissue slices, preparation or image acquisition, noise arising, lightning conditions, and protocols of staining while capturing the digital image [40]. Therefore, these variations could produce samples with different colour intensities [41]. Research studies [18,42] have shown the significant effect of stain normalisation that enhances the performance of breast cancer classification. Here, a few colour normalisation techniques will be investigated by categorizing them into three types of method which are global colour normalisation, the supervised method, and the unsupervised method for stain separation.
- Global colour normalisation: This method is suitable for histology images due to comprehensible values of autocorrelation coefficient or spatial dependency of pixel (intensity). This method separates colour and intensity information using principal component analysis (PCA) [43]. Reinhard et al.’s method was one of the first techniques, which uses a simple statistical analysis to achieve colour correction by comparing one image’s colour boundaries and choosing it as an appropriate source image as a benchmark, applying it as characteristic to all the other images [43]. It uses an unsupervised method to heuristically estimate the absorbance coefficients for the stains for every image and the staining concentrations for every pixel to recompose the images [43].
- Supervised method for stain separation: In this method, images are converted to optical density (OD) space due to Beer’s law [44] that suggests colour stains act linearly in OD space, given in Equation (1).
- Unsupervised method for stain separation: Training is not required because it is expected to learn itself [47]. Macenko et al. first proposed a method to use singular value decomposition method (SVD) to obtain optical density of images to perform quantitative analysis-based colour normalisation [48]. Kothari et al. then proposed a method based on histogram specification using the quantile normalisation based on distinct colour channels obtain from images to match each image to the target image histogram colour channels [49]. Bejnordi et al. later proposed an improved version which relies solely on colour features; their algorithm makes use of spatial information to achieve robustness against severe colour and intensity variations [50]. The comparison of colour normalisation methods is provided in Table 1.Table 1. Comparison of colour normalisation methods.
Ref Proposed Approach Method Advantages Disadvantages [43] Colour transfer algorithm - Convert the colour space of an image from RGB to lαβ [51].
- Transform the background colour of images based on the target colour space.
- Convert images back to RGB colour space.
- All images will have the same consistent range of contrast.
- Structure of images remains.
- Stains in images are not separated properly due to the type of colour space conversion (lαβ).
[48] Fringe search algorithm - Convert the colour space of an image from RGB to lαβ [51].
- Create plane based on calculated two largest SVD.
- Estimate data onto that plane.
- Search for corresponding DOF angles.
- Robust predictions of minimum and maximum are calculated by the αth and (100−α)th percentile.
- Convert these obtain DOF angles values back to OD space.
- Negative coefficient is not found in colour appearance matrix.
- Absence of ambiguity.
- Not ideal for automated tumour detection algorithm because the DOF angle values are estimated observationally.
- Original images are not preserved.
[49] Automated colour segmentation algorithm - Apply pre segmentation by extracting the unique colours in the image to obtain colour map.
- Include knowledges from pre segmented reference images to normalise.
- Apply voting scheme to evaluate on preliminary segmentation labels.
- Apply segmentation to new images with the multiple reference images and combine labels from previous step.
- High accuracy.
- Robustness.
- Makes use of expert domain knowledge.
- Retains the morphology of images.
- Colour map histogram distortion due to chromatic aberration.
- Restricted to segmentation problems with four stain colours.
[46] Nonlinear mapping approach - Map both target image and source images to a representation, where each channel relates to a separate chemical stain.
- Calculate the statistics of each corresponding channel by learning a supervised classification method (RVM).
- Apply a nonlinear correction (mapping) to normalise each separate channel based on previous calculation.
- Reconstruct the normalised source image using the normalised stain channels.
- Satisfactory performance overall for separating stains.
- Performs at pixel level to achieve superior performance.
- High computation complexity.
- Using nonlinear correction (mapping) functions might destroy the original image structure i.e., colour histogram.
- Impossible to convert back to original form of an image after mapping.
[50] Whole-slide image colour standardiser (WSICS) algorithm - Apply hue-saturation-density (HSD) colour transformation [52] to obtain two chromatic components and a density component
- Gather distribution of the transformation of haematoxylin and eosin (H&E).
- Calculate weight contribution of stain in every pixel.
- Convert HSD back to RGB.
- Robustness.
- Remain spatial information of images.
- It is an unsupervised method capable of detecting all stain components correctly.
- Losing the original background colour during the process.
- High processing time.
- Not all information of images is preserved.
- (2)
- Data augmentation: A data-space solution to the problem of limited data by enhancing the size of training datasets to generate a better learning model [56]. Tellex et al. showed that to obtain a particularly reliable performance of CAD system on histopathology images, colour normalisation should be used along with data augmentation [57]. This procedure will imply data wrapping and oversampling over the dataset to increase the sample size of the training dataset as a limited dataset and overfitting is a common challenge [56]. These processed include various image transformations to modify the image morphology [57,58]. If we were to look at one image from a single perspective and make a determination, it is more likely to be prone to error compared to if we were to look at it from several perspectives to make the final determination. Taking this into breast cancer analysis, checking the image with several more perspectives provides a more confident and accurate answer to which class it belongs to. Thus, this procedure provides a broader interpretation to the original image. The comparison of data augmentation techniques applied by several research studies is provided in Table 3.
3.2. Conventional CAD Methods
3.2.1. Segmentation
- Region-based segmentation: There are two main techniques which are (1) region growing and (2) region splitting and merging. Rouhi et al. proposed the application of an automated region growing for segmentation on breast tumour histology images by using an artificial neural network (ANN) to obtain a threshold [62]. Rundo et al. used split and merging algorithms based on the seed selection by an adaptive region growing procedure [63]. Lu et al. applied a multi-scale Laplacian of Gaussian (LoG) [64] to detect the seed points and feed the filtered image to a mean-shift algorithm for segmentation, followed by some morphological operations [65].
- Threshold-based segmentation: To produce a less complex image, the main concept is to transform every pixel based on a threshold value; any pixels with intensity less than a threshold value/limit esteem (constant value) will be replaced with black pixels (0), otherwise replaced with white pixels (1). The input image g (x, y) transformed to a threshold image f (x, y) can be represented mathematically as shown in Equation (3).
- Cluster-based segmentation: This can be described in two clustering methods, hierarchal and partitioning [80]. Hierarchal clustering performs recursively to explore nested clusters in agglomerative (bottom to up) or divisive (top to down) ways [80], whereas partitioning clustering iteratively divides into hard clustering and fuzzy clustering [81]. Kowal et al. applied a cluster approach algorithm for nuclei segmentation from biopsy microscopic images, and achieved a high classification accuracy [82]. Kumar et al. used a k-means clustering based segmentation algorithm and mentioned that this method performs better in comparison to other commonly used segmentation methods [83]. A two-step k-means was applied for segmentation by Shi et al. to consider local correlation of pixels; they first generate a poorly segmented cytoplasm, then in a second step the segmentation does not take into account the nuclei identified during the first clustering; finally, a watershed transform was applied to complete the segmentation [84]. Maqlin et al. suggested a segmentation method based on k-means clustering algorithm to recover the missing edge boundaries based on a convex grouping algorithm, which was suitable for open vesicular and patchy types of nuclei that are commonly obtained in high-risk breast cancers [85].
- Energy-based optimization: This technique defines a cost function, and the process will minimize/maximize the function based on the object of interest (ROI) in the images. A study by Belsare et al. used a spatio-colour-texture graph cut segmentation algorithm to perform segmentation as epithelial lining surrounding the lumen [86]. Wan et al. used a combination of boundary and region information to perform a hybrid active contour method to achieve an automated segmentation of the nuclear region [87], where the energy function was defined as Equation (4).
- Feature-based segmentation: Automatic segmentation based on feature learning has been commonly used for analysing medical images [90]. Song et al. used a multi-scale convolutional network to accurately apply segmentation of cervical cytoplasm and nuclei [91]. Xu and Huang applied a distributed deep neural network architecture to detect cells [92]. Rouhi et al. also proposed a cellular neural network (CNN) to perform segmentation by using genetic algorithm (GA) to determine the parameters [62]. Graham et al. proposed a deep learning method called the HoVer-Net which is a network that targets simultaneous segmentation and classification of nuclei based on the horizontal and vertical distance maps to separate clustered nuclei [93]. Zarella et al. trained an SVM model to learn the features to distinguish between stained pixels and unstained pixels using HSV colour space to identify regions of interest [94]. A summary of different segmentation approaches by several researchers is provided in Table 5.
3.2.2. Feature Extraction
- Morphological Features: Describes the details of the image regarding information in geometric aspects such as the size (radii, perimeter, and area) and shape (smoothness, compactness, symmetry, roundness, and concavity) of a cell [97].
- Textural Features: Collects information of various intensity of every pixel value from histology images by applying several methods to obtain a number of properties such as smoothness, coarseness, and regularity [97].
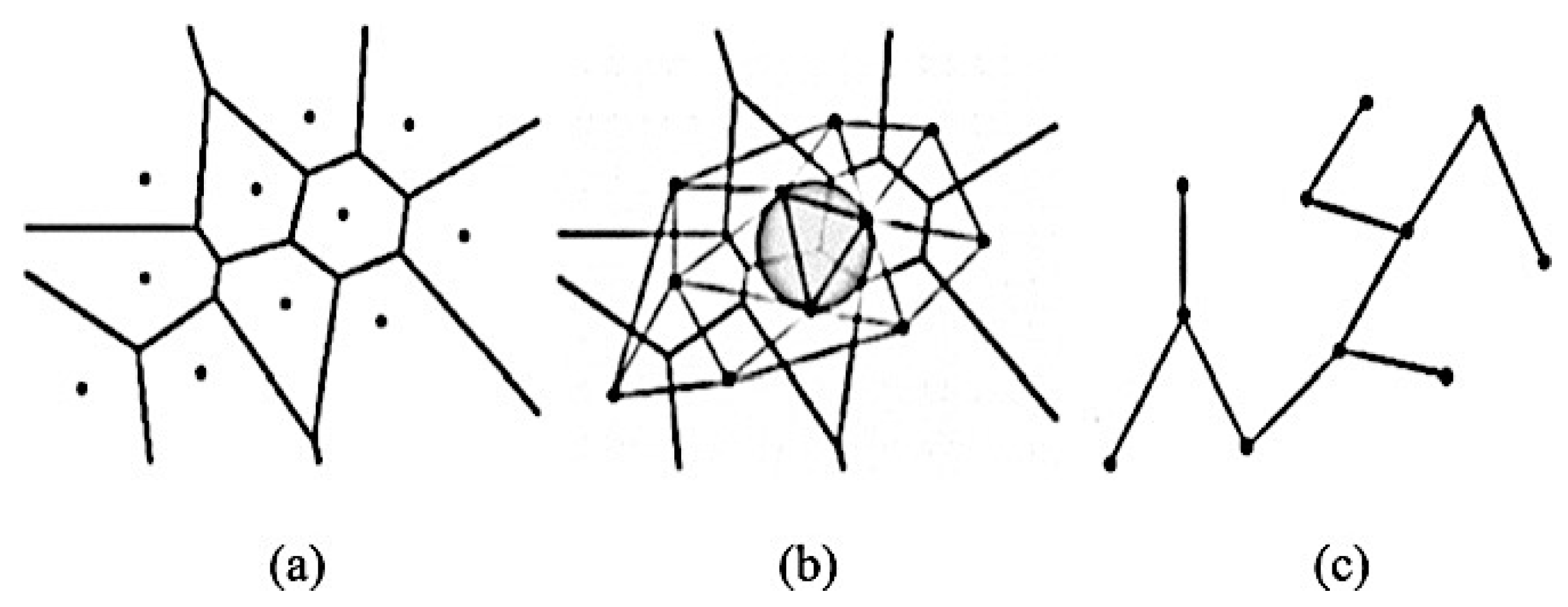
- Graph-Based Topological Features (architectural features): Describes the structure and spatial arrangement of nuclei in a tumour tissue [97]. When dealing with histopathological images, the arrangement and shape of nuclei is connected to the cancer development, therefore this architecture may be calculated using graph-based techniques [98,99]. There are many different topology-based features including the count of number of nodes, edges, edge length, and roundness factor to detect the tissues [100,101]. There are three types of common graph features: Voronoi diagram, Delaunay triangulation, and minimum spanning tree, as shown in Figure 10.
3.2.3. Feature Selection (Dimension Reduction)
3.2.4. Classification
- Nearest Neighbour: A non-parametric approach which falls under supervised learning widely used for both pattern recognition and classification applications [120]. The algorithm predicts each new point being input to the closest distance frame arrival point in the data; the calculation for distance varies but Euclidean distance is a common approach [121]. Let p and q be two datapoints of n-dimensions, then distance between x and y can be expressed by Euclidean distance shown in Equation (5).
- Support Vector Machine (SVM): Vapnik et al. proposed this method which works by mapping input information (feature vectors) to a higher dimensional space to obtain a hyperplane that can separate the labels/classes [122]. An optimal hyperplane can be obtained by maximizing the distances between support vectors (the data points closest to the boundary of the class) of two classes [123,124,125]. Recently, several research studies on breast cancer using histopathology images were performed by applying SVM classifiers [15,30,31,54]. Korkmaz and Poyraz proposed a classification framework focusing on minimum redundancy, maximum relevance feature selection, and least square SVM (LSSVM); their results claimed to be 100% accurate with only four false negatives for benign tumours in a three-class problem; however, no further evaluation was performed [126]. Chan and Tuszynski applied SVM classifier on their fractal features; their results achieved 97.9% F-score for magnification level 40× on the BreaKHis dataset [23]. Bardou et al. have also applied an approach of SVM to classify the images using handcrafted features [26].
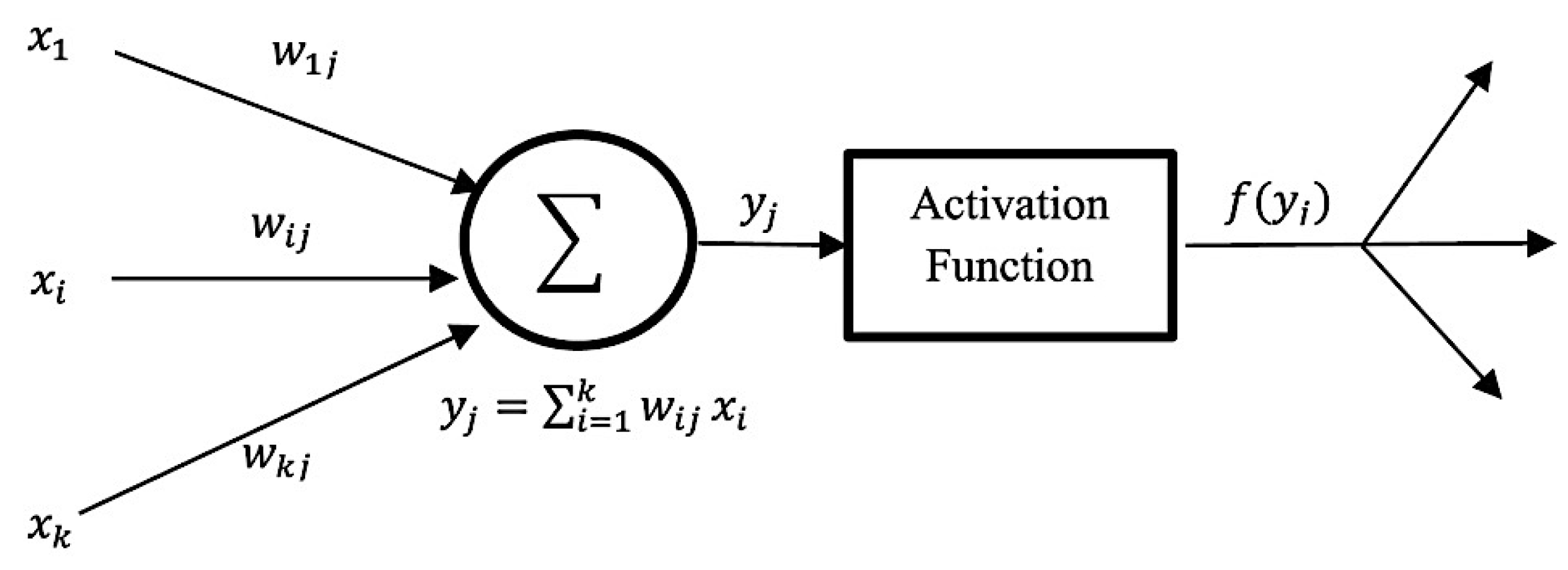
- Artificial Neural Network (ANN): ANN is inspired by human perception that can models complex nonlinear functions. The basic architecture of ANN starts by receiving input data , calculating each of the pieces of input information by multiplying to its corresponding weight , and obtaining a weighted output f (xj), with the support of a defined activation function until reaching the output layer. Figure 11 below demonstrates the basic structure of a single neuron in a feed-forward ANN [127]. Kassani et al. applied a multi-layer perceptron classifier on four different benchmark datasets and achieved the highest accuracy of 98.13% [19].
- Decision Tree: A decision tree algorithm is a supervised learning method for classification derived from the concept of ‘divide and conquer’ methodology. A complete decision tree is built based on feature space and labels; every new prediction will traverse from the root to the leaf node to produce an output. Asri et al. applied classification by using the C4.5 algorithm, an approach with a total of 11 features, and obtained 91.13% accuracy [128]. The extreme gradient boosting (XGBoost) is a new tree-based algorithm that has been increasing in popularity for data classification recently, and has proved to be a highly effective method for data classification [129]. Vo et al. have also applied gradient boosted trees as their breast cancer detection classifier [18].
- Bayesian Network: Bayesian network (BN) calculates probabilistic statistics to form a representation of relationships among a set of features space using an acrylic graph as shown in Figure 12, along with the value of conditional probabilities for each feature [130]. This type of classifier is commonly used for calculating probability estimations rather than predictions [116].
- Ensemble Classifier: This approach simply combines a few classifier methods instead of using a single classifier to produce a more accurate result. Commonly used methods to build an ensemble classifier are bagging, boosting, and random subspace method [131]. T.K. Ho proposed a random subspace classifier, in which a random feature subset is picked up from the original dataset for training each classifier; a voting scheme is then applied to produce a unique output from the from all the outputs in the combined classifiers [132]. Alkassar et al. applied an ensemble classifier that chooses the maximum score of prediction that includes a combination of decision tree, linear and quadratic discriminant, logistic regression, naive Bayes, SVM, and KNN [22].
3.3. Deep Learning CAD Methods
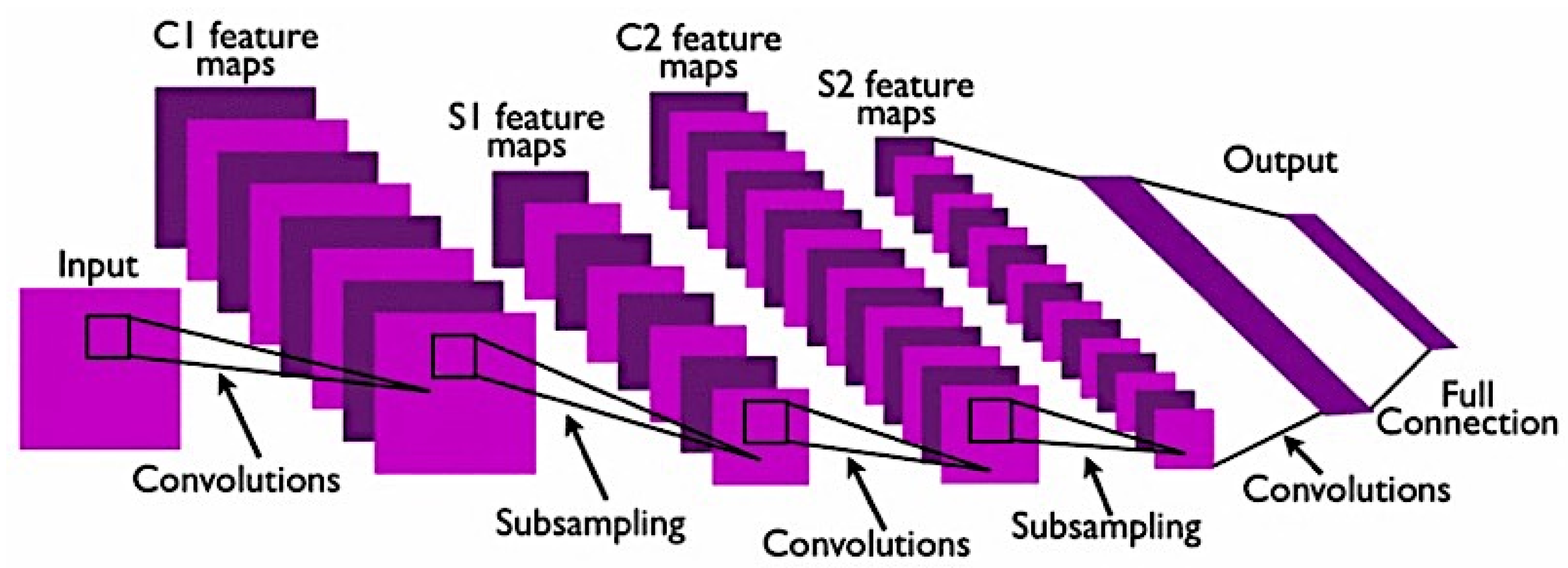
- Training from scratch: This method requires a large amount of input on histopathology images of breast cancer to train the CNN model. It requires more effort and skills to achieve a reliable performance CNN model when it comes to selecting hyperparameters such as learning rate, number of layers, convolutional filters and more, which can be a challenging task. This implementation also requires a high GPU processing power to perform training as CNN training can be time consuming because of the complex architecture [142].
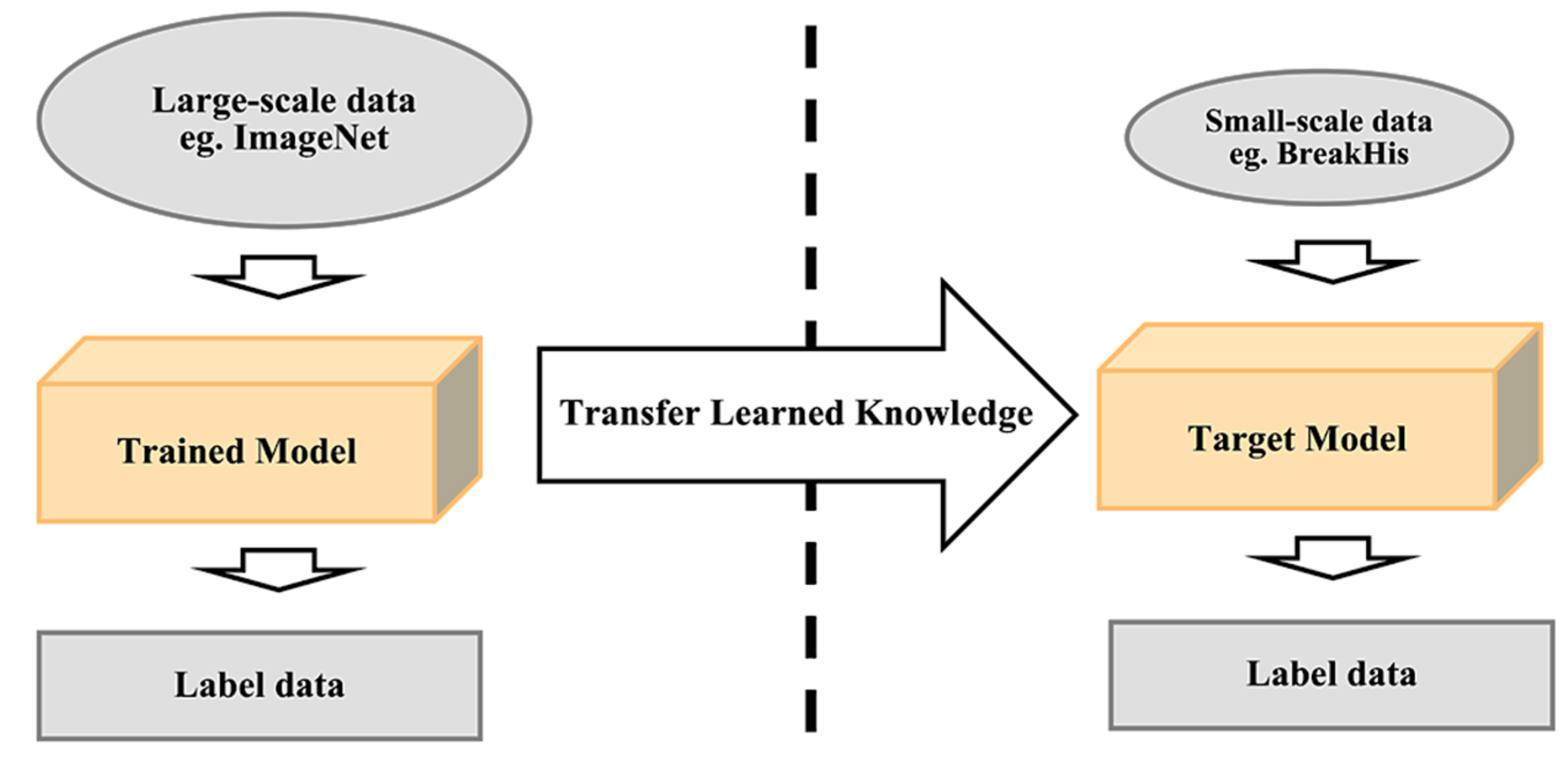
- Transfer learning: Most publicly available datasets for breast histology images are considered as small datasets for training a deep learning model, which can be highly prone to overfitting due to the inferior performance of generalizability. The transfer learning method provides a solution to this by performing transfer knowledge tasks on the model based on a source domain that provides a large amount of sample data to the target domain. Pre-trained models can sufficiently prepare the small-scale histology dataset in a deep learning model. It can be used to: (1) perform as a baseline model, which uses the architecture of the pre-trained network and builds the model from scratch by random initialization of weights [143]; (2) perform as a feature extractor, which extracts key features and the outputs which go into the convolutional base are fed directly to the classifier without modifying any weights or convolutional parameters [143]; and (3) perform fine tuning where weights will be passed into the designed network from the pre-trained network by fine tuning the layer or performing partial training of the network [143]. Figure 14 illustrates the transfer learning approach.
3.4. Performance Evaluation
- Sensitivity represents the percentage of positive numbers of samples classified correctly. The formula to calculate this is shown in Equation (6).
- Specificity represents the percentage of negative numbers of samples classified correctly. The formula to calculate this is shown in Equation (7).
- Accuracy represents the percentage of correct classification rate. The formula to calculate this is shown in Equation (8).
- Precision also known as PPV (Positive Predictive Value) represents the statistical variability measurement (total number of positive results). The formula to calculate this is shown in Equation (9).
- Recall represents the proportion of negative numbers of samples classified correctly. The formula to calculate this is shown in Equation (10).
- F1-measure represents the weighted mean of precision and recall. The formula to calculate this is shown in Equation (11).
4. Discussion and Conclusions
- Image enhancement: Original histology images may contain visuals like noise, colour variation, intensity variation, low pixelation or more because of the staining processing during image acquisition. It is challenging to focus on the target area; therefore, image processing plays a role in standardising and improving the quality of histology images.
- Detecting the cells or nuclei: Segmentation procedure assists in locating and identifying every cell in the image. This plays a role in obtaining the accurate region of interest to further measure the existence of cancer in the cell.
- Learning the features: This process of feature extraction provides the geometrical information of the detected cell which will be later considered as knowledge to determine the possibility of cancer. The CNN approach on this matter provides a robust solution with automated learning.
- Justification on diagnosis results: There always exists a situation where pathologists might examine an incorrect result due to several factors such as lack of experience, heavy workload, human error, or miscalculation. Thus, a CAD system can provide a second perspective on or verification of the diagnosis results by pathologists under the microscope assessment.
- Fast diagnosis results: As discussed in this paper, one of the benefits of a CAD system is to help breast cancer patients in early diagnosis to treat it before it progresses to more advanced stages. Pathologists often face challenges while diagnosing breast cancer because it requires an extensive amount time, effort, and process to perform microscope examination on histology images, therefore a CAD system can efficiently provide a faster solution.
- Improve productivity: The advancement of machine learning techniques produces higher productivity in a pathologist’s microscope examination and possible reduction of the number of false negatives associated with morphologic detection of tumour cells for deep learning techniques [55].
- Data limitations: Working with complex and large amounts of medical data can be challenging as they require high processing power and huge memory storage. Machine learning, especially deep learning, requires a large amount of data to train the model to produce a reliable and correct result. Some of the research papers acquired small datasets from private institutions, which are more likely to perform differently when being used in the real-world hospital environment. For publicly available datasets, most of them are considered as small datasets which are also most likely not applicable when it comes to performing in a real-world environment. Looking at the largest public dataset, for example the BreaKHis dataset, it does not satisfy the condition of a dataset with enough patient samples. Therefore, existing CAD systems do not have sufficient knowledge learned that is ready to be applicable in the real-world environment.
- Bias and imbalance class: This problem among datasets can lead to undesired classification for the diagnosis result. When a CAD system is built upon a dataset with imbalanced classes, the results will be more likely to be biased and therefore produce wrong diagnosis. When a trained model is biased to a specific class due to the imbalanced dataset it destroys the reliability of a CAD system because it will increase the rate of wrong classification. There are solutions to deal with problems like these by applying oversampling, undersampling, and algorithm-level methods [152]. Therefore, there are insufficient investigations performed on solutions that show significant improvement for imbalanced data to be able to practically use it in hospitals.
5. Future Directions
- Recently, the investigation and proposed CNN models have been increasing to provide an efficient solution to solve task-specific problems. In the future, there is always space for a new and more powerful CNN model that combines and utilises all the existing CNN’s good characteristics to be discovered. For breast cancer classification, specifically a model that performs segmentation on cancer and non-cancerous regions.
- Most research studies focus on the indicators of accuracy and performance metrics while developing a diagnosis system. However, when it comes to the applicability in real-world hospital environments the performance is undefined. Problems like class imbalance and large-scale diagnosis systems require extensive investigation in unpredictable real-world environments to obtain reliable CAD systems. Therefore, further investigation needs to be performed and will require many years of clinical practice of a CAD system in the real-world environment to constantly adapt and improve to be able gain credibility for clinical adoption in the future.
- Currently, the development of pre-trained CNNs on histopathology breast cancer image datasets does not exist. Most of the current research studies apply feature extraction that uses pre-trained CNNs on the general ImageNet data. Therefore, future researchers can explore building a large-scale pre-trained CNN focusing on breast cancer histopathology images that is task specific to assist breast cancer diagnosis.
- In recent research studies, the authors in [26] have applied feature descriptors of scale invariant feature transform (DSIFT) features and speeded-up robust features (SURF). However, oriented fast and rotated brief (ORB) features have outperformed both SIFT and SURF [151]. In the future, further analysis can investigate the ORB features on a breast cancer classification task.
- It will be important to investigate a reliable-performance CAD system over a longer period with various settings to understand the strengths and weaknesses to ensure the confidence and reliability of the system to be integrated in practical healthcare in the future of medical diagnosis.
- Recently, new algorithms like eXtreme Gradient Boosting (XGBoost) [153] have shown increased popularity because of their reliable performance and can be experimented with and integrated in CAD systems.
- Developing a mobile-based compatible expert system for breast cancer diagnosis to provide further convenience for more users to access, especially those with limited access to computer-based systems.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. Available online: https://tinyurl.com/ugemlbs (accessed on 16 March 2021).
- Cancer Research UK Breast Cancer Statistics|Cancer Research UK. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/breast-cancer#heading-Zero (accessed on 16 March 2021).
- Sizilio, G.R.M.A.; Leite, C.R.M.; Guerreiro, A.M.G.; Neto, A.D.D. Fuzzy Method for Pre-Diagnosis of Breast Cancer from the Fine Needle Aspirate Analysis. Biomed. Eng. Online 2012, 11. [Google Scholar] [CrossRef] [Green Version]
- Robertson, S.; Azizpour, H.; Smith, K.; Hartman, J. Digital Image Analysis in Breast Pathology—From Image Processing Techniques to Artificial Intelligence. Transl. Res. 2018, 194, 19–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krawczyk, B.; Schaefer, G.; Woźniak, M. A Hybrid Cost-Sensitive Ensemble for Imbalanced Breast Thermogram Classification. Artif. Intell. Med. 2015, 65. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Tiwari, A. Breast Cancer Diagnosis Using Genetically Optimized Neural Network Model. Expert Syst. Appl. 2015, 42. [Google Scholar] [CrossRef]
- Chen, H.L.; Yang, B.; Liu, J.; Liu, D.Y. A Support Vector Machine Classifier with Rough Set-Based Feature Selection for Breast Cancer Diagnosis. Expert Syst. Appl. 2011, 38. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- What Is Breast Cancer?|Cancer Research UK. Available online: https://www.cancerresearchuk.org/about-cancer/breast-cancer/about (accessed on 16 March 2021).
- Breast Cancer Organization. What Is Breast Cancer?|Breastcancer.Org. 2016. pp. 1–19. Available online: https://www.breastcancer.org/symptoms/understand_bc/what_is_bc (accessed on 16 March 2021).
- Alom, M.Z.; Yakopcic, C.; Nasrin, M.S.; Taha, T.M.; Asari, V.K. Breast Cancer Classification from Histopathological Images with Inception Recurrent Residual Convolutional Neural Network. J. Digit. Imaging 2019, 32. [Google Scholar] [CrossRef] [Green Version]
- Akram, M.; Iqbal, M.; Daniyal, M.; Khan, A.U. Awareness and Current Knowledge of Breast Cancer. Biol. Res. 2017, 50, 33. [Google Scholar] [CrossRef] [Green Version]
- Pantanowitz, L.; Evans, A.; Pfeifer, J.; Collins, L.; Valenstein, P.; Kaplan, K.; Wilbur, D.; Colgan, T. Review of the Current State of Whole Slide Imaging in Pathology. J. Pathol. Inform. 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.U.; Islam, N.; Jan, Z.; Ud Din, I.; Rodrigues, J.J.P.C. A Novel Deep Learning Based Framework for the Detection and Classification of Breast Cancer Using Transfer Learning. Pattern Recognit. Lett. 2019, 125. [Google Scholar] [CrossRef]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A Dataset for Breast Cancer Histopathological Image Classification. IEEE Trans. Biomed. Eng. 2016, 63. [Google Scholar] [CrossRef]
- Gurcan, M.N.; Boucheron, L.E.; Can, A.; Madabhushi, A.; Rajpoot, N.M.; Yener, B. Histopathological Image Analysis: A Review. IEEE Rev. Biomed. Eng. 2009, 2. [Google Scholar] [CrossRef] [Green Version]
- Bayramoglu, N.; Kannala, J.; Heikkila, J. Deep Learning for Magnification Independent Breast Cancer Histopathology Image Classification. In Proceedings of the International Conference on Pattern Recognition, Cancun, Mexico, 4–8 December 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 2440–2445. [Google Scholar] [CrossRef] [Green Version]
- Vo, D.M.; Nguyen, N.Q.; Lee, S.W. Classification of Breast Cancer Histology Images Using Incremental Boosting Convolution Networks. Inf. Sci. 2019, 482. [Google Scholar] [CrossRef]
- Kassani, S.H.; Kassani, P.H.; Wesolowski, M.J.; Schneider, K.A.; Deters, R. Classification of Histopathological Biopsy Images Using Ensemble of Deep Learning Networks. arXiv 2019, arXiv:1909.11870. [Google Scholar]
- Murtaza, G.; Shuib, L.; Mujtaba, G.; Raza, G. Breast Cancer Multi-Classification through Deep Neural Network and Hierarchical Classification Approach. Multimed. Tools Appl. 2020, 79. [Google Scholar] [CrossRef]
- Toğaçar, M.; Özkurt, K.B.; Ergen, B.; Cömert, Z. BreastNet: A Novel Convolutional Neural Network Model through Histopathological Images for the Diagnosis of Breast Cancer. Phys. A Stat. Mech. Its Appl. 2020, 545. [Google Scholar] [CrossRef]
- Alkassar, S.; Jebur, B.A.; Abdullah, M.A.M.; Al-Khalidy, J.H.; Chambers, J.A. Going Deeper: Magnification-Invariant Approach for Breast Cancer Classification Using Histopathological Images. IET Comput. Vis. 2021, 15, 151–164. [Google Scholar] [CrossRef]
- Chan, A.; Tuszynski, J.A. Automatic Prediction of Tumour Malignancy in Breast Cancer with Fractal Dimension. R. Soc. Open Sci. 2016, 3. [Google Scholar] [CrossRef] [Green Version]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. Breast Cancer Histopathological Image Classification Using Convolutional Neural Networks. In Proceedings of the International Joint Conference on Neural Networks, Vancouver, BC, Canada, 24–29 July 2016; IEEE: Piscataway, NJ, USA, 2016. [Google Scholar]
- Han, Z.; Wei, B.; Zheng, Y.; Yin, Y.; Li, K.; Li, S. Breast Cancer Multi-Classification from Histopathological Images with Structured Deep Learning Model. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Bardou, D.; Zhang, K.; Ahmad, S.M. Classification of Breast Cancer Based on Histology Images Using Convolutional Neural Networks. IEEE Access 2018, 6. [Google Scholar] [CrossRef]
- Gandomkar, Z.; Brennan, P.C.; Mello-Thoms, C. MuDeRN: Multi-Category Classification of Breast Histopathological Image Using Deep Residual Networks. Artif. Intell. Med. 2018, 88. [Google Scholar] [CrossRef]
- Budak, Ü.; Cömert, Z.; Rashid, Z.N.; Şengür, A.; Çıbuk, M. Computer-Aided Diagnosis System Combining FCN and Bi-LSTM Model for Efficient Breast Cancer Detection from Histopathological Images. Appl. Soft Comput. J. 2019, 85. [Google Scholar] [CrossRef]
- George, K.; Faziludeen, S.; Sankaran, P.; Paul, J.K. Deep Learned Nucleus Features for Breast Cancer Histopathological Image Analysis Based on Belief Theoretical Classifier Fusion. In Proceedings of the IEEE Region 10 Annual International Conference, Proceedings/TENCON, Kochi, India, 17–20 October 2019; IEEE: Piscataway, NJ, USA, 2019. [Google Scholar]
- Sudharshan, P.J.; Petitjean, C.; Spanhol, F.; Oliveira, L.E.; Heutte, L.; Honeine, P. Multiple Instance Learning for Histopathological Breast Cancer Image Classification. Expert Syst. Appl. 2019, 117. [Google Scholar] [CrossRef]
- Araujo, T.; Aresta, G.; Castro, E.; Rouco, J.; Aguiar, P.; Eloy, C.; Polonia, A.; Campilho, A. Classification of Breast Cancer Histology Images Using Convolutional Neural Networks. PLoS ONE 2017, 12. [Google Scholar] [CrossRef]
- Aresta, G.; Araújo, T.; Kwok, S.; Chennamsetty, S.S.; Safwan, M.; Alex, V.; Marami, B.; Prastawa, M.; Chan, M.; Donovan, M.; et al. BACH: Grand Challenge on Breast Cancer Histology Images. Med. Image Anal. 2019, 56. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Bandi, P.; Bejnordi, B.E.; Geessink, O.; Balkenhol, M.; Bult, P.; Halilovic, A.; Hermsen, M.; van de Loo, R.; Vogels, R.; et al. 1399 H&E-Stained Sentinel Lymph Node Sections of Breast Cancer Patients: The CAMELYON Dataset. Gigascience 2018, 7, giy065. [Google Scholar]
- Bándi, P.; Geessink, O.; Manson, Q.; Van Dijk, M.; Balkenhol, M.; Hermsen, M.; Ehteshami Bejnordi, B.; Lee, B.; Paeng, K.; Zhong, A.; et al. From Detection of Individual Metastases to Classification of Lymph Node Status at the Patient Level: The CAMELYON17 Challenge. IEEE Trans. Med. Imaging 2019, 38. [Google Scholar] [CrossRef] [Green Version]
- Veeling, B.S.; Linmans, J.; Winkens, J.; Cohen, T.; Welling, M. Rotation Equivariant CNNs for Digital Pathology. In Proceedings of the 21st International Conference, Granada, Spain, 16–20 September 2018; Spinger: Cham, Switzerland, 2018. Lecture Notes in Computer Science. Volume 11071. [Google Scholar]
- Roux, L.; Racoceanu, D.; Loménie, N.; Kulikova, M.; Irshad, H.; Klossa, J.; Capron, F.; Genestie, C.; Naour, G.; Gurcan, M. Mitosis Detection in Breast Cancer Histological Images an ICPR 2012 Contest. J. Pathol. Inform. 2013, 4. [Google Scholar] [CrossRef]
- MITOS-ATYPIA-14 Grand Challenge. Available online: https://mitos-atypia-14.grand-challenge.org/ (accessed on 17 March 2021).
- Veta, M.; Heng, Y.J.; Stathonikos, N.; Bejnordi, B.E.; Beca, F.; Wollmann, T.; Rohr, K.; Shah, M.A.; Wang, D.; Rousson, M.; et al. Predicting Breast Tumor Proliferation from Whole-Slide Images: The TUPAC16 Challenge. Med. Image Anal. 2019, 54. [Google Scholar] [CrossRef] [Green Version]
- Drelie Gelasca, E.; Obara, B.; Fedorov, D.; Kvilekval, K.; Manjunath, B.S. A Biosegmentation Benchmark for Evaluation of Bioimage Analysis Methods. BMC Bioinform. 2009, 10. [Google Scholar] [CrossRef] [Green Version]
- Kaushal, C.; Bhat, S.; Koundal, D.; Singla, A. Recent Trends in Computer Assisted Diagnosis (CAD) System for Breast Cancer Diagnosis Using Histopathological Images. IRBM 2019, 40, 211–227. [Google Scholar] [CrossRef]
- De Matos, J.; De Souza Britto, A., Jr.; Oliveira, L.E.S.; Koerich, A.L. Histopathologic Image Processing: A Review. arXiv 2019, arXiv:1904.07900. [Google Scholar]
- Ciompi, F.; Geessink, O.; Bejnordi, B.E.; De Souza, G.S.; Baidoshvili, A.; Litjens, G.; Van Ginneken, B.; Nagtegaal, I.; Van Der Laak, J. The Importance of Stain Normalization in Colorectal Tissue Classification with Convolutional Networks. In Proceedings of the International Symposium on Biomedical Imaging, Melbourne, Australia, 18–21 April 2017; IEEE: Piscataway, NJ, USA, 2017. [Google Scholar]
- Reinhard, E.; Ashikhmin, M.; Gooch, B.; Shirley, P. Color Transfer between Images. IEEE Comput. Graph. Appl. 2001, 21. [Google Scholar] [CrossRef]
- Helmenstine, A.M. Beer’s Law Defintion and Equation; ThoughtCo.: New York, NY, USA, 2019. [Google Scholar]
- Ruifrok, A.C.; Johnston, D.A. Quantification of Histochemical Staining by Color Deconvolution. Anal. Quant. Cytol. Histol. 2001, 23, 291–299. [Google Scholar]
- Khan, A.M.; Rajpoot, N.; Treanor, D.; Magee, D. A Nonlinear Mapping Approach to Stain Normalization in Digital Histopathology Images Using Image-Specific Color Deconvolution. IEEE Trans. Biomed. Eng. 2014, 61. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; kumar Jain, A.; Lal, S.; Kini, J. A Study about Color Normalization Methods for Histopathology Images. Micron 2018, 114, 42–61. [Google Scholar] [CrossRef] [PubMed]
- Macenko, M.; Niethammer, M.; Marron, J.S.; Borland, D.; Woosley, J.T.; Guan, X.; Schmitt, C.; Thomas, N.E. A Method for Normalizing Histology Slides for Quantitative Analysis. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, ISBI 2009, Boston, MA, USA, 28 June–1 July 2009; IEEE: Piscataway, NJ, USA, 2009; pp. 1107–1110. [Google Scholar]
- Kothari, S.; Phan, J.H.; Moffitt, R.A.; Stokes, T.H.; Hassberger, S.E.; Chaudry, Q.; Young, A.N.; Wang, M.D. Automatic Batch-Invariant Color Segmentation of Histological Cancer Images. In Proceedings of the International Symposium on Biomedical Imaging, Chicago, IL, USA, 30 March–2 April 2011; IEEE: Piscataway, NJ, USA, 2011. [Google Scholar]
- Bejnordi, B.E.; Litjens, G.; Timofeeva, N.; Otte-Höller, I.; Homeyer, A.; Karssemeijer, N.; Van Der Laak, J.A.W.M. Stain Specific Standardization of Whole-Slide Histopathological Images. IEEE Trans. Med. Imaging 2016, 35. [Google Scholar] [CrossRef]
- Sandid, F.; Douik, A. Texture Descriptor Based on Local Combination Adaptive Ternary Pattern. IET Image Process. 2015, 9. [Google Scholar] [CrossRef] [Green Version]
- Van Der Laak, J.A.W.M.; Pahlplatz, M.M.M.; Hanselaar, A.G.J.M.; De Wilde, P.C.M. Hue-Saturation-Density (HSD) Model for Stain Recognition in Digital Images from Transmitted Light Microscopy. Cytometry 2000, 39. [Google Scholar] [CrossRef]
- Rakhlin, A.; Shvets, A.; Iglovikov, V.; Kalinin, A.A. Deep Convolutional Neural Networks for Breast Cancer Histology Image Analysis. In Proceedings of the 15th International Conference, ICIAR 2018, Póvoa de Varzim, Portugal, 27–29 June 2018; IEEE: Piscataway, NJ, USA, 2018; Volume 10882 LNCS. [Google Scholar]
- Li, Y.; Wu, J.; Wu, Q. Classification of Breast Cancer Histology Images Using Multi-Size and Discriminative Patches Based on Deep Learning. IEEE Access 2019, 7. [Google Scholar] [CrossRef]
- Liu, Y.; Kohlberger, T.; Norouzi, M.; Dahl, G.E.; Smith, J.L.; Mohtashamian, A.; Olson, N.; Peng, L.H.; Hipp, J.D.; Stumpe, M.C. Artificial Intelligence–Based Breast Cancer Nodal Metastasis Detection Insights into the Black Box for Pathologists. Arch. Pathol. Lab. Med. 2019, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shorten, C.; Khoshgoftaar, T.M. A Survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6. [Google Scholar] [CrossRef]
- Tellez, D.; Litjens, G.; Bándi, P.; Bulten, W.; Bokhorst, J.M.; Ciompi, F.; van der Laak, J. Quantifying the Effects of Data Augmentation and Stain Color Normalization in Convolutional Neural Networks for Computational Pathology. Med. Image Anal. 2019, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxena, S.; Gyanchandani, M. Machine Learning Methods for Computer-Aided Breast Cancer Diagnosis Using Histopathology: A Narrative Review. J. Med. Imaging Radiat. Sci. 2020, 51, 42–61. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Arsalan, M.; Owais, M.; Lee, M.B.; Park, K.R. Artificial Intelligence-Based Mitosis Detection in Breast Cancer Histopathology Images Using Faster R-CNN and Deep CNNs. J. Clin. Med. 2020, 9, 749. [Google Scholar] [CrossRef] [Green Version]
- Mehra, R. Breast Cancer Histology Images Classification: Training from Scratch or Transfer Learning? ICT Express 2018, 4. [Google Scholar] [CrossRef]
- Cheng, H.D.; Jiang, X.H.; Sun, Y.; Wang, J. Color Image Segmentation: Advances and Prospects. Pattern Recognit. 2001, 34. [Google Scholar] [CrossRef]
- Rouhi, R.; Jafari, M.; Kasaei, S.; Keshavarzian, P. Benign and Malignant Breast Tumors Classification Based on Region Growing and CNN Segmentation. Expert Syst. Appl. 2015, 42. [Google Scholar] [CrossRef]
- Rundo, L.; Militello, C.; Vitabile, S.; Casarino, C.; Russo, G.; Midiri, M.; Gilardi, M.C. Combining Split-and-Merge and Multi-Seed Region Growing Algorithms for Uterine Fibroid Segmentation in MRgFUS Treatments. Med. Biol. Eng. Comput. 2016, 54. [Google Scholar] [CrossRef] [PubMed]
- Marr, D.; Hildreth, E. Theory of Edge Detection. Proc. R. Soc. Lond. Biol. Sci. 1980, 207. [Google Scholar] [CrossRef]
- Lu, C.; Ji, M.; Ma, Z.; Mandal, M. Automated Image Analysis of Nuclear Atypia in High-Power Field Histopathological Image. J. Microsc. 2015, 258. [Google Scholar] [CrossRef]
- Vincent, O.; Folorunso, O. A Descriptive Algorithm for Sobel Image Edge Detection. In Proceedings of the 2009 InSITE Conference, Macon, GA, USA, 12–15 June 2009; Informing Science Institute California: Santa Rosa, CA, USA, 2009. [Google Scholar]
- Salman, N. Image Segmentation Based on Watershed and Edge Detection Techniques. Int. Arab. J. Inf. Technol. 2006, 3, 104–110. [Google Scholar] [CrossRef]
- Prewitt, J. Object enhancement and extraction. In Picture Processing and Psychopictorics; Elsevier: Amsterdam, The Netherlands, 1970; Volume 10. [Google Scholar]
- Stehfest, H. Algorithm 368: Numerical Inversion of Laplace Transforms [D5]. Commun. ACM 1970, 13. [Google Scholar] [CrossRef]
- Canny, J. A Computational Approach to Edge Detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, 8. [Google Scholar] [CrossRef]
- George, Y.M.; Zayed, H.H.; Roushdy, M.I.; Elbagoury, B.M. Remote Computer-Aided Breast Cancer Detection and Diagnosis System Based on Cytological Images. IEEE Syst. J. 2014, 8. [Google Scholar] [CrossRef]
- Gonzalez, R.C.; Woods, R.E.; Masters, B.R. Digital Image Processing, Third Edition. J. Biomed. Opt. 2009, 14. [Google Scholar] [CrossRef]
- Faridi, P.; Danyali, H.; Helfroush, M.S.; Jahromi, M.A. An Automatic System for Cell Nuclei Pleomorphism Segmentation in Histopathological Images of Breast Cancer. In Proceedings of the 2016 IEEE Signal Processing in Medicine and Biology Symposium, SPMB 2016, Philadelphia, PA, USA, 3 December 2016; IEEE: Piscataway, NJ, USA, 2017. [Google Scholar]
- Otsu, N. Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst Man Cybern 1979, 9. [Google Scholar] [CrossRef] [Green Version]
- Zarella, M.D.; Garcia, F.U.; Breen, D.E. A Template Matching Model for Nuclear Segmentation in Digital Images of H&E Stained Slides. In Proceedings of the 9th International Conference on Bioinformatics and Biomedical Technology, Lisbon, Portugal, 14–16 May 2017. [Google Scholar]
- Saha, M.; Agarwal, S.; Arun, I.; Ahmed, R.; Chatterjee, S.; Mitra, P.; Chakraborty, C. Histogram Based Thresholding for Automated Nucleus Segmentation Using Breast Imprint Cytology. In Advancements of Medical Electronics; Springer: New Delhi, India, 2015. [Google Scholar]
- Moncayo, R.; Romo-Bucheli, D.; Romero, E. A Grading Strategy for Nuclear Pleomorphism in Histopathological Breast Cancer Images Using a Bag of Features (BOF). In Proceedings of the 20th Iberoamerican Congress, CIARP 2015, Montevideo, Uruguay, 9–12 November 2015; Volume 9423. [Google Scholar]
- Khairuzzaman, A.K.M.; Chaudhury, S. Multilevel Thresholding Using Grey Wolf Optimizer for Image Segmentation. Expert Syst. Appl. 2017, 86. [Google Scholar] [CrossRef]
- Sirinukunwattana, K.; Khan, A.M.; Rajpoot, N.M. Cell Words: Modelling the Visual Appearance of Cells in Histopathology Images. Comput. Med. Imaging Graph. 2015, 42. [Google Scholar] [CrossRef]
- Rokach, L.; Maimon, O. Clustering Methods. In Data Mining and Knowledge Discovery Handbook; Maimon, O., Rokach, L., Eds.; Springer US: Boston, MA, USA, 2005; pp. 321–352. ISBN 978-0-387-25465-4. [Google Scholar]
- De Carvalho, F.D.A.T.; Lechevallier, Y.; De Melo, F.M. Partitioning Hard Clustering Algorithms Based on Multiple Dissimilarity Matrices. Pattern Recognit. 2012, 45. [Google Scholar] [CrossRef]
- Kowal, M.; Filipczuk, P.; Obuchowicz, A.; Korbicz, J.; Monczak, R. Computer-Aided Diagnosis of Breast Cancer Based on Fine Needle Biopsy Microscopic Images. Comput. Biol. Med. 2013, 43. [Google Scholar] [CrossRef]
- Kumar, R.; Srivastava, R.; Srivastava, S. Detection and Classification of Cancer from Microscopic Biopsy Images Using Clinically Significant and Biologically Interpretable Features. J. Med. Eng. 2015. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Zhong, J.; Huang, R.; Lin, J. Automated Quantitative Image Analysis of Hematoxylin-Eosin Staining Slides in Lymphoma Based on Hierarchical Kmeans Clustering. In Proceedings of the 2016 8th International Conference on Information Technology in Medicine and Education, ITME 2016, Fuzhou, China, 23–25 December 2016; IEEE: Piscataway, NJ, USA, 2017. [Google Scholar]
- Maqlin, P.; Thamburaj, R.; Mammen, J.J.; Manipadam, M.T. Automated Nuclear Pleomorphism Scoring in Breast Cancer Histopathology Images Using Deep Neural Networks. In Proceedings of the Third International Conference, MIKE 2015, Hyderabad, India, 9–11 December 2015; Volume 9468. [Google Scholar]
- Belsare, A.D.; Mushrif, M.M.; Pangarkar, M.A.; Meshram, N. Classification of Breast Cancer Histopathology Images Using Texture Feature Analysis. In Proceedings of the IEEE Region 10 Annual International Conference, Proceedings/TENCON; IEEE: Piscataway, NJ, USA, 2016. [Google Scholar]
- Wan, T.; Cao, J.; Chen, J.; Qin, Z. Automated Grading of Breast Cancer Histopathology Using Cascaded Ensemble with Combination of Multi-Level Image Features. Neurocomputing 2017, 229. [Google Scholar] [CrossRef]
- Zhang, W.; Li, H. Automated Segmentation of Overlapped Nuclei Using Concave Point Detection and Segment Grouping. Pattern Recognit. 2017, 71. [Google Scholar] [CrossRef]
- Jia, D.; Zhang, C.; Wu, N.; Guo, Z.; Ge, H. Multi-Layer Segmentation Framework for Cell Nuclei Using Improved GVF Snake Model, Watershed, and Ellipse Fitting. Biomed. Signal. Process. Control 2021, 67, 102516. [Google Scholar] [CrossRef]
- Janowczyk, A.; Madabhushi, A. Deep Learning for Digital Pathology Image Analysis: A Comprehensive Tutorial with Selected Use Cases. J. Pathol. Inform. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, L.; Chen, S.; Ni, D.; Lei, B.; Wang, T. Accurate Segmentation of Cervical Cytoplasm and Nuclei Based on Multiscale Convolutional Network and Graph Partitioning. IEEE Trans. Biomed. Eng. 2015, 62. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Huang, J. Detecting 10,000 Cells in One Second. In Proceedings of the 19th International Conference, Athens, Greece, 17–21 October 2016; Volume 9901 LNCS. [Google Scholar]
- Graham, S.; Vu, Q.D.; Raza, S.E.A.; Azam, A.; Tsang, Y.W.; Kwak, J.T.; Rajpoot, N. Hover-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images. Med. Image Anal. 2019, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarella, M.D.; Breen, D.E.; Reza, M.A.; Milutinovic, A.; Garcia, F.U. Lymph Node Metastasis Status in Breast Carcinoma Can Be Predicted via Image Analysis of Tumor Histology. Anal. Quant. Cytol. Histol. 2015, 37, 273–285. [Google Scholar]
- Belsare, A.D.; Mushrif, M.M.; Pangarkar, M.A.; Meshram, N. Breast Histopathology Image Segmentation Using Spatio-Colour-Texture Based Graph Partition Method. J. Microsc. 2016, 262. [Google Scholar] [CrossRef]
- Krig, S. Computer Vision Metrics: Survey, Taxonomy, and Analysis; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Das, A.; Nair, M.S.; Peter, S.D. Computer-Aided Histopathological Image Analysis Techniques for Automated Nuclear Atypia Scoring of Breast Cancer: A Review. J. Digit. Imaging 2020, 33, 1091–1121. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Jiang, Z.; Xie, F.; Zhang, H.; Ma, Y.; Shi, H.; Zhao, Y. Feature Extraction from Histopathological Images Based on Nucleus-Guided Convolutional Neural Network for Breast Lesion Classification. Pattern Recognition. 2017, 71. [Google Scholar] [CrossRef]
- Sharma, H.; Zerbe, N.; Lohmann, S.; Kayser, K.; Hellwich, O.; Hufnagl, P. A Review of Graph-Based Methods for Image Analysis in Digital Histopathology. Diagn. Pathol. 2015, 1. [Google Scholar] [CrossRef]
- Shi, J.; Wu, J.; Li, Y.; Zhang, Q.; Ying, S. Histopathological Image Classification with Color Pattern Random Binary Hashing-Based PCANet and Matrix-Form Classifier. IEEE J. Biomed. Health Inform. 2017, 21. [Google Scholar] [CrossRef] [PubMed]
- Ehteshami Bejnordi, B.; Lin, J.; Glass, B.; Mullooly, M.; Gierach, G.L.; Sherman, M.E.; Karssemeijer, N.; Van Der Laak, J.; Beck, A.H. Deep Learning-Based Assessment of Tumor-Associated Stroma for Diagnosing Breast Cancer in Histopathology Images. In Proceedings of the Proceedings International Symposium on Biomedical Imaging, Melbourne, Australia, 18–21 April 2017; IEEE: Piscataway, NJ, USA, 2017. [Google Scholar]
- Zwanenburg, A.; Leger, S.; Vallières, M.; Löck, S. Image biomarker standardisation initiative-feature definitions. arXiv 2016, arXiv:1612.07003. [Google Scholar]
- Balazsi, M.; Blanco, P.; Zoroquiain, P.; Levine, M.D.; Burnier, M.N. Invasive Ductal Breast Carcinoma Detector That Is Robust to Image Magnification in Whole Digital Slides. J. Med. Imaging 2016, 3. [Google Scholar] [CrossRef] [Green Version]
- Gupta, V.; Bhavsar, A. Breast Cancer Histopathological Image Classification: Is Magnification Important? In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Rezaeilouyeh, H.; Mollahosseini, A.; Mahoor, M.H. Microscopic Medical Image Classification Framework via Deep Learning and Shearlet Transform. J. Med. Imaging 2016, 3. [Google Scholar] [CrossRef]
- Shukla, K.K.; Tiwari, A.; Sharma, S. Classification of Histopathological Images of Breast Cancerous and Non Cancerous Cells Based on Morphological Features. Biomed. Pharmacol. J. 2017, 10. [Google Scholar] [CrossRef]
- Tambasco Bruno, D.O.; Do Nascimento, M.Z.; Ramos, R.P.; Batista, V.R.; Neves, L.A.; Martins, A.S. LBP Operators on Curvelet Coefficients as an Algorithm to Describe Texture in Breast Cancer Tissues. Expert Syst. Appl. 2016, 55. [Google Scholar] [CrossRef] [Green Version]
- Wan, T.; Zhang, W.; Zhu, M.; Chen, J.; Achim, A.; Qin, Z. Automated Mitosis Detection in Histopathology Based on Non-Gaussian Modeling of Complex Wavelet Coefficients. Neurocomputing 2017, 237. [Google Scholar] [CrossRef] [Green Version]
- Gandomkar, Z.; Brennan, P.C.; Mello-Thoms, C. Computer-Assisted Nuclear Atypia Scoring of Breast Cancer: A Preliminary Study. J. Digit. Imaging 2019, 32. [Google Scholar] [CrossRef]
- Khan, A.M.; Sirinukunwattana, K.; Rajpoot, N. A Global Covariance Descriptor for Nuclear Atypia Scoring in Breast Histopathology Images. IEEE J. Biomed. Health Inform. 2015, 19. [Google Scholar] [CrossRef] [PubMed]
- Maroof, N.; Khan, A.; Qureshi, S.A.; Rehman, A.U.; Khalil, R.K.; Shim, S.O. Mitosis Detection in Breast Cancer Histopathology Images Using Hybrid Feature Space. Photodiagn. Photodyn. Ther. 2020, 31. [Google Scholar] [CrossRef]
- Tashk, A.; Helfroush, M.S.; Danyali, H.; Akbarzadeh-jahromi, M. Automatic Detection of Breast Cancer Mitotic Cells Based on the Combination of Textural, Statistical and Innovative Mathematical Features. Appl. Math. Model. 2015, 39. [Google Scholar] [CrossRef]
- Burges, C.J.C. Dimension Reduction: A Guided Tour. In Foundation and Trends in Machine Learning; now Publishers Inc.: Hanover, MA, USA, 2009; Volume 2. [Google Scholar] [CrossRef]
- Clarke, R.; Ressom, H.W.; Wang, A.; Xuan, J.; Liu, M.C.; Gehan, E.A.; Wang, Y. The Properties of High-Dimensional Data Spaces: Implications for Exploring Gene and Protein Expression Data. Nat. Rev. Cancer 2008, 8, 37–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster Analysis and Display of Genome-Wide Expression Patterns. Proc. Natl. Acad. Sci. USA 1998, 95. [Google Scholar] [CrossRef] [Green Version]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine Learning Applications in Cancer Prognosis and Prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef] [Green Version]
- Sakri, S.B.; Abdul Rashid, N.B.; Muhammad Zain, Z. Particle Swarm Optimization Feature Selection for Breast Cancer Recurrence Prediction. IEEE Access 2018, 6. [Google Scholar] [CrossRef]
- Cruz-Roa, A.; Gilmore, H.; Basavanhally, A.; Feldman, M.; Ganesan, S.; Shih, N.; Tomaszewski, J.; Madabhushi, A.; González, F. High-Throughput Adaptive Sampling for Whole-Slide Histopathology Image Analysis (HASHI) via Convolutional Neural Networks: Application to Invasive Breast Cancer Detection. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Dhahri, H.; Al Maghayreh, E.; Mahmood, A.; Elkilani, W.; Faisal Nagi, M. Automated Breast Cancer Diagnosis Based on Machine Learning Algorithms. J. Healthc. Eng. 2019. [Google Scholar] [CrossRef]
- Rajaguru, H.; Sannasi Chakravarthy, S.R. Analysis of Decision Tree and K-Nearest Neighbor Algorithm in the Classification of Breast Cancer. Asian Pac. J. Cancer Prev. 2019, 20. [Google Scholar] [CrossRef] [Green Version]
- Wadkar, K.; Pathak, P.; Wagh, N. Breast Cancer Detection Using ANN Network and Performance Analysis With SVM. Int. J. Comput. Eng. Technol. 2019, 10. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20. [Google Scholar] [CrossRef] [Green Version]
- Introduction to Support Vector Machines—OpenCV 2.4.13.7 Documentation. Available online: https://docs.opencv.org/2.4/doc/tutorials/ml/introduction_to_svm/introduction_to_svm.html (accessed on 17 March 2021).
- Ayat, N.E.; Cheriet, M.; Suen, C.Y. Automatic Model Selection for the Optimization of SVM Kernels. Pattern Recognit. 2005, 38. [Google Scholar] [CrossRef]
- Wang, H.; Zheng, B.; Yoon, S.W.; Ko, H.S. A Support Vector Machine-Based Ensemble Algorithm for Breast Cancer Diagnosis. Eur. J. Oper. Res. 2018, 267. [Google Scholar] [CrossRef]
- Korkmaz, S.A.; Poyraz, M. Least Square Support Vector Machine and Minumum Redundacy Maximum Relavance for Diagnosis of Breast Cancer from Breast Microscopic Images. Procedia Soc. Behav. Sci. 2015, 174. [Google Scholar] [CrossRef] [Green Version]
- Jha, G.K. Artificial Neural Networks and Its Applications; Indian Agricultural Statistics Research Institute (I.C.A.R.): New Delhi, India, 2007. [Google Scholar]
- Asri, H.; Mousannif, H.; Al Moatassime, H.; Noel, T. Using Machine Learning Algorithms for Breast Cancer Risk Prediction and Diagnosis. Procedia Comput. Sci. 2016, 83, 1064–1069. [Google Scholar]
- Parashar, J.; Rai, M. Breast Cancer Images Classification by Clustering of ROI and Mapping of Features by CNN with XGBOOST Learning. Mater. Today Proc. 2020. [Google Scholar] [CrossRef]
- Kotsiantis, S.B. Supervised Machine Learning: A Review of Classification Techniques. Informatica 2007, 31, 249–268. [Google Scholar]
- Min, S.H. A Genetic Algorithm-Based Heterogeneous Random Subspace Ensemble Model for Bankruptcy Prediction. Int. J. Appl. Eng. Res. 2016, 11, 2927–2931. [Google Scholar]
- Ho, T.K. The Random Subspace Method for Constructing Decision Forests. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20. [Google Scholar] [CrossRef] [Green Version]
- Wahab, N.; Khan, A.; Lee, Y.S. Two-Phase Deep Convolutional Neural Network for Reducing Class Skewness in Histopathological Images Based Breast Cancer Detection. Comput. Biol. Med. 2017, 85. [Google Scholar] [CrossRef]
- LeCun, Y.; Kavukcuoglu, K.; Farabet, C. Convolutional Networks and Applications in Vision. In Proceedings of the ISCAS 2010—2010 IEEE International Symposium on Circuits and Systems: Nano-Bio Circuit Fabrics and Systems, Paris, France, 2 June–30 May 2010. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Li, F. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; IEEE: Piscataway, NJ, USA, 2015. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on Machine Learning, ICML 2015, Lille, France, 7–9 July 2015. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; IEEE: Piscataway, NJ, USA, 2016. [Google Scholar]
- Bevilacqua, V.; Brunetti, A.; Guerriero, A.; Trotta, G.F.; Telegrafo, M.; Moschetta, M. A Performance Comparison between Shallow and Deeper Neural Networks Supervised Classification of Tomosynthesis Breast Lesions Images. Cogn. Syst. Res. 2019, 53, 3–19. [Google Scholar] [CrossRef]
- Sharma, S.; Mehra, R. Conventional Machine Learning and Deep Learning Approach for Multi-Classification of Breast Cancer Histopathology Images—A Comparative Insight. J. Digit. Imaging 2020, 33, 632–654. [Google Scholar] [CrossRef]
- Komura, D.; Ishikawa, S. Machine Learning Methods for Histopathological Image Analysis. Comput. Struct. Biotechnol. J. 2018, 16, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Sun, X.; Zhou, N.; Han, X.; Yao, J. Efficient Mitosis Detection in Breast Cancer Histology Images by RCNN. In Proceedings of the International Symposium on Biomedical Imaging, Venice, Italy, 8–11 April 2019. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Z.; Sun, L.; Huo, Q. An Anchor-Free Region Proposal Network for Faster R-CNN-Based Text Detection Approaches. IJDAR 2019, 22, 315–327. [Google Scholar] [CrossRef] [Green Version]
- Donahue, J.; Jia, Y.; Vinyals, O.; Hoffman, J.; Zhang, N.; Tzeng, E.; Darrell, T. DeCAF: A Deep Convolutional Activation Feature for Generic Visual Recognition. In Proceedings of the 31st International Conference on Machine Learning, ICML 2014, Beijing, China, 21–26 June 2014. [Google Scholar]
- Spanhol, F.A.; Cavalin, P.R.; Oliveira, L.S.; Petitjean, C.; Heutte, L. Deep Features for Breast Cancer Histopathological Image Classification. In Proceedings of the 2017 IEEE International Conference on Systems, Man, and Cybernetics, SMC 2017, Banff, AB, Canada, 5–8 October 2017. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9. [Google Scholar] [CrossRef]
- Rublee, E.; Rabaud, V.; Konolige, K.; Bradski, G. ORB: An Efficient Alternative to SIFT or SURF. In Proceedings of the IEEE International Conference on Computer Vision 2011, Barcelona, Spain, 6–13 November 2011; IEEE: Piscataway, NJ, USA, 2011. [Google Scholar]
- He, H.; Garcia, E.A. Learning from Imbalanced Data. IEEE Trans. Knowl. Data Eng. 2009, 21. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; Association for Computing Machinery: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]




| Ref | Year | Adapted Colour Normalisation Method |
|---|---|---|
| [42] | 2017 | Bejnordi et al. and Macenko et al. methodology |
| [31] | 2017 | Macenko et al. methodology |
| [27] | 2018 | Reinhard et al. and Kothari et al. methodology |
| [53] | 2018 | Macenko et al. methodology |
| [18] | 2019 | Macenko et al. methodology |
| [19] | 2019 | Macenko et al. methodology |
| [54] | 2019 | Reinhard et al. methodology |
| [29] | 2019 | Macenko et al. methodology |
| [55] | 2019 | Simplified version of Bejnordi et al. methodology |
| [22] | 2021 | Khan et al. methodology |
| Ref | Flipping | Cropping/Shearing | Rotation | Translation | Shifting | Scaling | Zooming | Contrast | Fill Mode | Brightness |
|---|---|---|---|---|---|---|---|---|---|---|
| [11] | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| [27] | ✓ | ✓ | ✓ | |||||||
| [18] | ✓ | ✓ | ✓ | ✓ | ||||||
| [53] | ✓ | ✓ | ||||||||
| [19] | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| [17] | ✓ | ✓ | ✓ | |||||||
| [21] | ✓ | ✓ | ✓ | ✓ | ||||||
| [26] | ✓ | ✓ | ||||||||
| [59] | ✓ | ✓ | ✓ | ✓ | ||||||
| [60] | ✓ | |||||||||
| [20] | ✓ | ✓ | ✓ | ✓ | ✓ |
| Approach | Segmentation Technique | Definition | Advantages | Limitations |
|---|---|---|---|---|
| Image processing | Region-based | Each pixel will be separated into groups (regions) in a homogeneous way based on a seed point. |
|
|
| Edge-based | Edges are defined based on the sharp discontinuity (i.e., intensity) in the image. |
|
| |
| Thresholding-based | Transform every pixel based on a threshold value obtained from a histogram of image that corresponds to regions. |
|
| |
| Machine learning | Cluster-based | Objects in image will be categorised into specific regions (groups) based on their similarity in pixels. |
|
|
| Energy-based optimization | Contour object of interest by minimizing/maximizing a predefined cos function. |
|
| |
| Feature-based | Uses a model to train and learn the features to determine which pixels are ROI. |
|
|
| Type of Technique(s) Employed | Ref | Year | Approach(es) | Remarks |
|---|---|---|---|---|
| Cluster-based | [82] | 2013 | Cluster algorithm |
|
| [83] | 2015 | K-means clustering algorithm |
| |
| [84] | 2017 | Two step k-means clustering, and watershed transform |
| |
| [85] | 2015 | Segmentation: k-means clustering algorithm. Recover edges: Convex grouping algorithm. |
| |
| Edge-based | [71] | 2014 | Watershed |
|
| [73] | 2016 | Distance regularized level set evolution (DRLSE) algorithm |
| |
| Energy-based optimization | [86] | 2016 | Graph cut: Spatio-colour-texture graph segmentation algorithm [95]. |
|
| [87] | 2017 | Hybrid active contour method |
| |
| [88] | 2017 | Three-phase level set method to set contour |
| |
| [89] | 2021 | Watershed and improved gradient vector flow (GVF) snake model |
| |
| Feature-based | [94] | 2015 | Support vector machine (SVM) |
|
| [91] | 2015 | Multi-scale convolutional network |
| |
| [92] | 2016 | Distributed deep neural network |
| |
| [62] | 2016 | Cellular neural network (CNN) trained on genetic algorithm (GA) parameters |
| |
| [93] | 2019 | Deep learning using HoVer-Net |
| |
| [59] | 2020 | Faster R-CNN |
| |
| Region-based | [62] | 2015 | Automated region growing using ANN to obtain threshold |
|
| [65] | 2015 | Mean-shift algorithm |
| |
| [63] | 2016 | Split and merging algorithm based on adaptive region growing |
| |
| Threshold-based | [77] | 2015 | Maximally Stable Extreme Regions (MSER) |
|
| [78] | 2017 | Multilevel thresholding based on Grey Wolf Optimizer (GWO) algorithm using Kapur’s entropy and Otsu’s between class variance functions. |
| |
| [76] | 2015 | Histogram-based thresholding |
| |
| [79] | 2015 | Dictionary, thresholding |
| |
| [75] | 2017 | Otsu thresholding |
|
| Approach on CAD Method | Ref | Year | Dataset | Classification Type | Methods | Results |
|---|---|---|---|---|---|---|
| Conventional | [83] | 2015 | 2828 histology images | Binary | KNN | Accuracy: 92.2% Specificity: 94.02% Sensitivity: 82% F1-measure: 75.94% |
| [126] | 2015 | Firat University Medicine Faculty Pathology Laboratory | Multi-class | SVM (Least Square Support Vector Machine) | Accuracy: 100%; four FN for benign tumours in a three-class problem | |
| [15] | 2016 | BreaKHis | Multi-class | SVM, Random Forest, QDA (Quadratic Discriminant Analysis), Nearest Neighbour | Accuracy: 80% to 85% | |
| [23] | 2016 | BreaKHis | Binary and Multi-class | SVM | Highest F1-score: 97.9% | |
| [128] | 2016 | Wisconsin Breast Cancer dataset | Binary | Decision tree: C4.5 algorithm | Accuracy: 91.13% | |
| [128] | 2016 | Wisconsin Breast Cancer dataset | Binary | SVM | Accuracy: 97.13% | |
| [30] | 2019 | BreaKHis | Binary | SVM | Prr: 92.1% | |
| [22] | 2021 | BreaKHis | Binary and Multi-class | Ensemble Classifier | Highest accuracy: 99% | |
| Deep Learning | [26] | 2018 | BreaKHis | Binary and Multi-class | SVM and CNN | Accuracy: 96.15–98.33% (binary); 83.31–88.23% (multi-class) |
| [17] | 2016 | BreaKHis | Binary and Multi-class | Single-task CNN (malignancy); Multi-task CNN (magnification level) | Prr: 83.13%. Prr: 80.10% | |
| [24] | 2016 | BreaKHis | Multi-class | AlexNet CNN | Prr: 90% | |
| [30] | 2019 | BreaKHis | Multi-class | MIL-CNN (Multiple Instance Learning-CNN) | Prr: 92.1% | |
| [53] | 2018 | BACH | Binary and Multi-class | ResNet-50, InceptionV3, VGG-16 and Gradient boosted trees | Accuracy: 87.2% (for binary) and 93.8% (for multi) AUC: 97.3% Sensitivity: 96.5 Specificity: 88.0% | |
| [60] | 2018 | BreaKHis | Binary | VGG16, VGG19, ResNet5 and Logistic regression | Accuracy: 92.60% AUC: 95.65% Precision: 95.95% | |
| [149] | 2017 | BreaKHis | Multi-class | Modified AlexNet and DeCAF (Deep Convolutional Activation Feature) | Accuracy: 81.5–86.3%F1-score: 86.7%-90.3% | |
| [31] | 2017 | Bioimaging Challenge 2015 | Binary and Multi-class | CNN and SVM | Accuracy: 83.3%(binary); 77.8% (multi-class) Sensitivity: 95.6%. | |
| [25] | 2017 | BreaKHis | Multi-class | Custom CSDCNN (Class Structure-based Deep Convolutional Neural Network) based on GoogLeNet | Accuracy: 93.2% | |
| [27] | 2018 | BreaKHis | Binary and Multi-class | ResNet CNN | Accuracy: 98.77% (Binary) Prr: 96.25% (Multi class) | |
| [145] | 2019 | MITOS-ATYPIA-14, TUPAC-16 | Binary | Modified faster-RCNN | Precision: 76% Recall: 72% F1 score: 73.6% | |
| [18] | 2019 | BreaKHisBioimaging Challenge 2015 | Multi-class | Inception and ResNet CNN (IRRCNN) and Gradient boosting trees | Accuracy: 99.5% (binary); 96.4% (multi-class) | |
| [29] | 2019 | BreaKHis | Multi-class | Pre-trained CNN (AlexNet, ResNet-18 and ResNet-50) and SVM | Accuracy: 96.88% Sensitivity: 97.30% Specificity: 95.97% AUC: 0.9942 | |
| [54] | 2019 | Bioimaging Challenge 2015 | Multi-class | Pre-trained ResNet50 with SVM classifier | Accuracy: 95% Recall: 89% | |
| [28] | 2019 | BreaKHis | Binary | FCN (Fully Convolutional Network) based on AlexNet and Bi-LSTM (Bidirectional Long Short-Term Memory) | Accuracy: 91.90% Sensitivity: 96.8% Specificity: 91% | |
| [11] | 2019 | BreaKHis andBioimaging Challenge 2015 | Binary and Multi-class | Inception Recurrent Residual CNN (IRRCNN) | Accuracy: 99.05% (for binary) and 98.59% (for multi) | |
| [55] | 2019 | Camelyon16 | Binary | LYNA algorithm based on Inception-v3 | AUC: 99% Sensitivity: 91% | |
| [19] | 2020 | BACH, BreaKHis, PatchCamelyon, and Bioimaging 2015 | Binary | Pre-trained VGG19, MobileNet, and DenseNet with MLP (Multi-Layer Perceptron) | Accuracy: 92.71% Precision: 95.74% Recall: 89.80% F-score: 92.43% | |
| [21] | 2020 | BreaKHis | Multi-class | CNN features with MLP (Multi-Layer Perceptron) | Accuracy: 98.80% | |
| [59] | 2020 | MITOS-12MITOS-ATYPIA-14 | Multi-class | Faster-RCNN and a score-level fusion of Resnet-50 and Densenet-201 CNNs | Precision: 87.6% Recall: 84.1% F1-measure: 85.8% | |
| [20] | 2020 | BreaKHis | Multi-class | BMIC_Net: Pre-trained AlexNet and KNN | Accuracy: 95.48% | |
| [22] | 2021 | BreaKHis | Binary and Multi-class | Xception and DenseNet CNNs | Accuracy: 99% (binary); 92% (multi-class) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liew, X.Y.; Hameed, N.; Clos, J. A Review of Computer-Aided Expert Systems for Breast Cancer Diagnosis. Cancers 2021, 13, 2764. https://doi.org/10.3390/cancers13112764
Liew XY, Hameed N, Clos J. A Review of Computer-Aided Expert Systems for Breast Cancer Diagnosis. Cancers. 2021; 13(11):2764. https://doi.org/10.3390/cancers13112764
Chicago/Turabian StyleLiew, Xin Yu, Nazia Hameed, and Jeremie Clos. 2021. "A Review of Computer-Aided Expert Systems for Breast Cancer Diagnosis" Cancers 13, no. 11: 2764. https://doi.org/10.3390/cancers13112764






