Multi-Combilipases: Co-Immobilizing Lipases with Very Different Stabilities Combining Immobilization via Interfacial Activation and Ion Exchange. The Reuse of the Most Stable Co-Immobilized Enzymes after Inactivation of the Least Stable Ones
Abstract
:1. Introduction
2. Results and Discussion
2.1. Immobilization and Stabilities of the Different Enzymes on Octyl Agarose
2.2. Immobilization and Stability of LEU and RML in a Support Coated with PEI (Octyl-VS-PEI Agarose Beads)
2.3. Covalent Immobilization of CALA, CALB and TLL on Octyl-VS
2.4. Immobilization and Desorption of LEU and RML from Octyl-VS-CALA-PEI
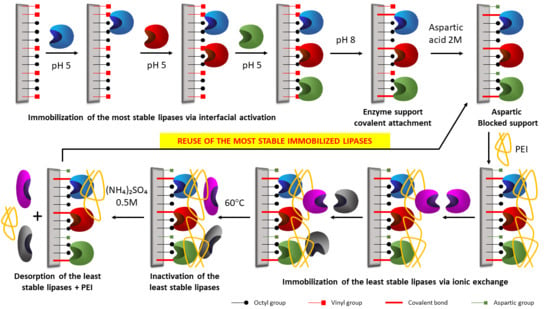
2.5. Building of the Combilipases
2.6. Reuse of the Most Stable Immobilized Enzymes after the Inactivation of the Least Stable Co-Immobilized Enzymes
3. Materials and Methods
3.1. Materials
3.2. Methods
3.2.1. Preparation of Octyl-Vinyl Sulfone Support (Octyl-VS)
3.2.2. Immobilization of Lipases on Octyl Agarose Beads
3.2.3. Immobilization of the Least Stable Enzymes on Octyl-VS-PEI Support
3.2.4. Co-Immobilized Biocatalysts Preparation
3.2.4.1. Immobilization of the Most Stable Lipases on Octyl-VS Support
3.2.4.2. Coating of Immobilized Enzymes with PEI
3.2.4.3. Immobilization of the Least Stable Lipases on Octyl-VS-Enzyme-PEI Biocatalysts
3.2.5. Determination of Enzyme Activity
3.2.5.1. Hydrolysis of p-NPB
3.2.5.2. Hydrolysis of Triacetin
3.2.5.3. Hydrolysis of (R)- or (S)-Methyl Mandelate
3.2.6. Lipase Biocatalysts Thermal Inactivations
3.2.7. Desorption of the Least Stable Lipases from the Supports
3.2.8. Analysis of the Immobilized Enzymes by SDS-PAGE
3.2.9. Titration of Primary Amino Groups in the Biocatalysts
3.2.10. Reuses of the Multi-Combilipases
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hasan, F.; Shah, A.; Javed, S.; Hameed, A. Enzymes used in detergents: Lipases. Afr. J. Biotechnol. 2010, 9, 4836–4844. [Google Scholar]
- Sharma, D.; Sharma, B.; Shukla, A. Biotechnological approach of microbial lipase: A review. Biotechnology 2011, 10, 23–40. [Google Scholar] [CrossRef] [Green Version]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzym. Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Kobayashi, S. Recent developments in lipase-catalyzed synthesis of polyesters. Macromol. Rapid Commun. 2009, 30, 237–266. [Google Scholar] [CrossRef]
- Reetz, M.T. Lipases as practical biocatalysts. Curr. Opin. Chem. Biol. 2002, 6, 145–150. [Google Scholar] [CrossRef]
- Jaeger, K.-E.; Eggert, T. Lipases for biotechnology. Curr. Opin. Biotechnol. 2002, 13, 390–397. [Google Scholar] [CrossRef]
- Stead, D. Microbial lipases: Their characteristics, role in food spoilage and industrial uses. J. Dairy Res. 1986, 53, 481–505. [Google Scholar] [CrossRef] [PubMed]
- Yahya, A.; Anderson, W.; Moo-Young, M. Ester synthesis in lipase-catalyzed reactions. Enzym. Microb. Technol. 1998, 23, 438–450. [Google Scholar] [CrossRef]
- Sharma, S.; Kanwar, S.S. Organic Solvent Tolerant Lipases and Applications. Sci. World J. 2014, 2014, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.P.; Dhar, K.; Kanwar, S.S.; Arora, P.K. Lipase catalysis in organic solvents: Advantages and applications. Biol. Proced. Online 2016, 18, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Priyanka, P.; Tan, Y.; Kinsella, G.K.; Henehan, G.T.; Ryan, B.J. Solvent stable microbial lipases: Current understanding and biotechnological applications. Biotechnol. Lett. 2018, 41, 203–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durand, E.; LeComte, J.; Villeneuve, P. Deep eutectic solvents: Synthesis, application, and focus on lipase-catalyzed reactions. Eur. J. Lipid Sci. Technol. 2013, 115, 379–385. [Google Scholar] [CrossRef]
- Alvarez, E.; Rodriguez, J.; Villa, R.; Gomez, C.; Nieto, S.; Donaire, A.; Lozano, P. Clean enzymatic production of flavor esters in sponge-like ionic liquids. ACS Sustain. Chem. Eng. 2019, 7, 13307–13314. [Google Scholar] [CrossRef]
- Villa, R.; Alvarez, E.; Porcar, R.; García-Verdugo, E.; Luis, S.; Lozano, P. Ionic liquids as an enabling tool to integrate reaction and separation processes. Green Chem. 2019, 21, 6527–6544. [Google Scholar] [CrossRef]
- Lozano, P.; Alvarez, E.; Nieto, S.; Villa, R.; Bernal, J.M.; Donaire, A. Biocatalytic synthesis of panthenyl monoacyl esters in ionic liquids and deep eutectic solvents. Green Chem. 2019, 21, 3353–3361. [Google Scholar] [CrossRef]
- Porcar, R.; Lozano, P.; Burguete, M.I.; Garcia-Verdugo, E.; Luis, S.V. Dimethyl carbonate as a non-innocent benign solvent for the multistep continuous flow synthesis of amino alcohols. React. Chem. Eng. 2018, 3, 572–578. [Google Scholar] [CrossRef]
- De Diego, T.; Lozano, P.; Gmouh, S.; Vaultier, M.; Iborra, J.L. Understanding structure−stability relationships of Candida antartica lipase B in ionic liquids. Biomacromolecules 2005, 6, 1457–1464. [Google Scholar] [CrossRef]
- Lozano, P.; Puente, T.D.D.; Carrié, D.; Vaultier, M.; Iborra, J.L. Continuous green biocatalytic processes using ionic liquids and supercritical carbon dioxide. Chem. Commun. 2002, 692–693. [Google Scholar] [CrossRef]
- Tan, J.-N.; Dou, Y. Deep eutectic solvents for biocatalytic transformations: Focused lipase-catalyzed organic reactions. Appl. Microbiol. Biotechnol. 2020, 104, 1481–1496. [Google Scholar] [CrossRef]
- Dhake, K.P.; Thakare, D.D.; Bhanage, B.M. Lipase: A potential biocatalyst for the synthesis of valuable flavour and fragrance ester compounds. Flavour Fragr. J. 2013, 28, 71–83. [Google Scholar] [CrossRef]
- Ang, X.; Chen, H.; Xiang, J.-Q.; Wei, F.; Quek, S.Y. Preparation and functionality of lipase-catalysed structured phospholipid—A review. Trends Food Sci. Technol. 2019, 88, 373–383. [Google Scholar] [CrossRef]
- Utama, Q.D.; Sitanggang, A.B.; Adawiyah, D.R.; Hariyadi, P. lipase-catalyzed interesterification for the synthesis of medium-long-medium (MLM) structured lipids. Food Technol. Biotechnol. 2019, 57, 305–318. [Google Scholar] [CrossRef]
- Ghanem, A.; Aboul-Enein, H.Y. Lipase-mediated chiral resolution of racemates in organic solvents. Tetrahedron: Asymmetry 2004, 15, 3331–3351. [Google Scholar] [CrossRef]
- Ghanem, A. Trends in lipase-catalyzed asymmetric access to enantiomerically pure/enriched compounds. Tetrahedron 2007, 63, 1721–1754. [Google Scholar] [CrossRef]
- Kapoor, M.; Gupta, M.N. Lipase promiscuity and its biochemical applications. Process Biochem. 2012, 47, 555–569. [Google Scholar] [CrossRef]
- Patel, N.; Rai, D.; Shivam; Shahane, S.; Mishra, U. Lipases: Sources, production, purification, and applications. Recent Patents Biotechnol. 2019, 13, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Filho, D.G.; Silva, A.G.; Guidini, C.Z. Lipases: Sources, immobilization methods, and industrial applications. Appl. Microbiol. Biotechnol. 2019, 103, 7399–7423. [Google Scholar] [CrossRef]
- Melani, N.B.; Tambourgi, E.B.; Silveira, E. Lipases: From production to applications. Sep. Purif. Rev. 2019, 49, 143–158. [Google Scholar] [CrossRef]
- Nawal, G.; Irfan, M.; Ashfaq, S.; Shakir, H.A. An overview of bacterial lipases and their enormous applications. Punjab Univ. J. Zool. 2019, 34, 61–71. [Google Scholar] [CrossRef]
- Verger, R. ‘Interfacial activation’ of lipases: Facts and artifacts. Trends Biotechnol. 1997, 15, 32–38. [Google Scholar] [CrossRef]
- Aloulou, A.; Rodriguez, J.A.; Fernandez, S.; Van Oosterhout, D.; Puccinelli, D.; Carrière, F. Exploring the specific features of interfacial enzymology based on lipase studies. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2006, 1761, 995–1013. [Google Scholar] [CrossRef] [PubMed]
- Reis, P.; Holmberg, K.; Watzke, H.; Leser, M.; Miller, R. Lipases at interfaces: A review. Adv. Colloid Interface Sci. 2009, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Brzozowski, A.M.; Derewenda, U.; Derewenda, Z.S.; Dodson, G.G.; Lawson, D.M.; Turkenburg, J.P.; Bjorkling, F.; Huge-Jensen, B.; Patkar, S.A.; Thim, L. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nat. Cell Biol. 1991, 351, 491–494. [Google Scholar] [CrossRef]
- Van Tilbeurgh, H.; Egloff, M.-P.; Martinez, C.; Rugani, N.; Verger, R.; Cambillau, C. Interfacial activation of the lipase–procolipase complex by mixed micelles revealed by X-ray crystallography. Nat. Cell Biol. 1993, 362, 814–820. [Google Scholar] [CrossRef] [PubMed]
- Derewenda, U.; Brzozowski, A.M.; Lawson, D.M.; Derewenda, Z.S. Catalysis at the interface: The anatomy of a conformational change in a triglyceride lipase. Biochemistry 1992, 31, 1532–1541. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.K.; Song, H.K.; Shin, D.H.; Hwang, K.Y.; Suh, S.W. The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor. Structure 1997, 5, 173–185. [Google Scholar] [CrossRef] [Green Version]
- Derewenda, Z.S.; Derewenda, U.; Dodson, G.G. The crystal and molecular structure of the Rhizomucor miehei triacylglyceride lipase at 1.9 Å resolution. J. Mol. Biol. 1992, 227, 818–839. [Google Scholar] [CrossRef]
- Murty, V.R.; Bhat, J.; Muniswaran, P.K.A. Hydrolysis of oils by using immobilized lipase enzyme: A review. Biotechnol. Bioprocess Eng. 2002, 7, 57–66. [Google Scholar] [CrossRef]
- Mardani, M.; Farmani, J.; Kenari, R. Efficacy of some commercial lipases in hydrolysis of palm olein for production of free fatty acids and diacylglycerol oil. J. Oil Palm Res. 2015, 27, 250–260. [Google Scholar]
- Ferreira, M.M.; De Oliveira, G.F.; Basso, R.C.; Mendes, A.A.; Hirata, D.B. Optimization of free fatty acid production by enzymatic hydrolysis of vegetable oils using a non-commercial lipase from Geotrichum candidum. Bioprocess Biosyst. Eng. 2019, 42, 1647–1659. [Google Scholar] [CrossRef]
- Zhao, X.; Qi, F.; Yuan, C.; Du, W.; Liu, D. Lipase-catalyzed process for biodiesel production: Enzyme immobilization, process simulation and optimization. Renew. Sustain. Energy Rev. 2015, 44, 182–197. [Google Scholar] [CrossRef]
- Zhong, L.; Feng, Y.; Wang, G.; Wang, Z.; Bilal, M.; Lv, H.; Jia, S.; Cui, J. Production and use of immobilized lipases in/on nanomaterials: A review from the waste to biodiesel production. Int. J. Biol. Macromol. 2020, 152, 207–222. [Google Scholar] [CrossRef] [PubMed]
- Poppe, J.K.; Matte, C.R.; Peralba, M.D.C.R.; Fernandez-Lafuente, R.; Rodrigues, R.C.; Ayub, M.A.Z. Optimization of ethyl ester production from olive and palm oils using mixtures of immobilized lipases. Appl. Catal. Gen. 2015, 490, 50–56. [Google Scholar] [CrossRef]
- Alves, J.S.; Vieira, N.S.; Cunha, A.S.; Silva, A.M.; Ayub, M.A.Z.; Fernandez-Lafuente, R.; Rodrigues, R.C. Combi-lipase for heterogeneous substrates: A new approach for hydrolysis of soybean oil using mixtures of biocatalysts. RSC Adv. 2014, 4, 6863–6868. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Lee, N.H.; Lim, J.S.; Um, B.-H.; Park, C.; Kang, S.W.; Kim, S.W. Optimization of the process for biodiesel production using a mixture of immobilized Rhizopus oryzae and Candida rugosa lipases. J. Microbiol. Biotechnol. 2008, 18, 1927–1931. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.H.; Kim, J.M.; Shin, H.Y.; Kang, S.W.; Kim, S.W. Biodiesel production using a mixture of immobilized Rhizopus oryzae and Candida rugosa lipases. Biotechnol. Bioprocess Eng. 2006, 11, 522–525. [Google Scholar] [CrossRef]
- Amoah, J.; Ho, S.-H.; Hama, S.; Yoshida, A.; Nakanishi, A.; Hasunuma, T.; Ogino, C.; Kondo, A. Lipase cocktail for efficient conversion of oils containing phospholipids to biodiesel. Bioresour. Technol. 2016, 211, 224–230. [Google Scholar] [CrossRef] [Green Version]
- Arana-Peña, S.; Carballares, D.; Berenguer-Murcia, Á.; Alcántara, A.R.; Rodrigues, R.C.; Fernández-Lafuente, R. One pot use of combilipases for full modification of oils and fats: Multifunctional and heterogeneous substrates. Catalysts 2020, 10, 605. [Google Scholar] [CrossRef]
- Qiao, H.; Zhang, F.; Guan, W.; Zuo, J.; Feng, D. Optimisation of combi-lipases from Aspergillus niger for the synergistic and efficient hydrolysis of soybean oil. Anim. Sci. J. 2016, 88, 772–780. [Google Scholar] [CrossRef]
- Poppe, J.K.; Matte, C.R.; Fernandez-Lafuente, R.; Rodrigues, R.C.; Ayub, M.A. transesterification of waste frying oil and soybean oil by combi-lipases under ultrasound-assisted reactions. Appl. Biochem. Biotechnol. 2018, 186, 576–589. [Google Scholar] [CrossRef]
- Poppe, J.K.; Matte, C.R.; De Freitas, V.O.; Fernandez-Lafuente, R.; Rodrigues, R.C.; Ayub, M.A.Z. Enzymatic synthesis of ethyl esters from waste oil using mixtures of lipases in a plug-flow packed-bed continuous reactor. Biotechnol. Prog. 2018, 34, 952–959. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.C.; Ayub, M.A. Effects of the combined use of Thermomyces lanuginosus and Rhizomucor miehei lipases for the transesterification and hydrolysis of soybean oil. Process Biochem. 2011, 46, 682–688. [Google Scholar] [CrossRef] [Green Version]
- Araki, C.A.; Marcucci, S.M.P.; Da Silva, L.S.; Maeda, C.H.; Arroyo, P.A.; Zanin, G.M. Effects of a combination of lipases immobilised on desilicated and thiol-modified ZSM-5 for the synthesis of ethyl esters from macauba pulp oil in a solvent-free system. Appl. Catal. A: Gen. 2018, 562, 241–249. [Google Scholar] [CrossRef]
- Pedro, K.; Parreira, J.; Correia, I.; Henriques, C.; Langone, M. Enzymatic biodiesel synthesis from acid oil using a lipase mixture. Química Nova 2017, 41, 284–291. [Google Scholar] [CrossRef]
- Karmee, S.K. Preparation of biodiesel from nonedible oils using a mixture of used lipases. Energy Sources Part Recovery Util. Environ. Eff. 2016, 38, 2727–2733. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, S.B.; Kang, S.W.; Song, Y.S.; Park, C.; Han, S.O.; Kim, S.W. Biodiesel production by a mixture of Candida rugosa and Rhizopus oryzae lipases using a supercritical carbon dioxide process. Bioresour. Technol. 2011, 102, 2105–2108. [Google Scholar] [CrossRef]
- Toro, E.; Rodriguez, D.; Morales, N.; García, L.M.; Godoy, C.A. novel combi-lipase systems for fatty acid ethyl esters production. Catalysts 2019, 9, 546. [Google Scholar] [CrossRef] [Green Version]
- Bin Ibrahim, N.A.; Guo, Z.; Xu, X. Enzymatic interesterification of palm stearin and coconut oil by a dual lipase system. J. Am. Oil Chem. Soc. 2007, 85, 37–45. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, S.B.; Yoo, H.Y.; Lee, J.H.; Park, C.; Han, S.O.; Kim, S.W. Kinetic modeling of biodiesel production by mixed immobilized and co-immobilized lipase systems under two pressure conditions. Korean J. Chem. Eng. 2013, 30, 1272–1276. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, S.B.; Yoo, H.Y.; Suh, Y.J.; Kang, G.B.; Jang, W.I.; Kang, J.; Park, C.; Kim, S.W. Biodiesel production by enzymatic process using Jatropha oil and waste soybean oil. Biotechnol. Bioprocess Eng. 2013, 18, 703–708. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, S.B.; Yoo, H.Y.; Lee, J.H.; Han, S.O.; Park, C.; Kim, S.W. Co-immobilization of Candida rugosa and Rhyzopus oryzae lipases and biodiesel production. Korean J. Chem. Eng. 2013, 30, 1335–1338. [Google Scholar] [CrossRef]
- Shahedi, M.; Yousefi, M.; Habibi, Z.; Mohammadi, M.; As’Habi, M.A. Co-immobilization of Rhizomucor miehei lipase and Candida antarctica lipase B and optimization of biocatalytic biodiesel production from palm oil using response surface methodology. Renew. Energy 2019, 141, 847–857. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, J.H.; Kim, D.S.; Yoo, H.Y.; Park, C.; Kim, S.W. Biodiesel production by lipases co-immobilized on the functionalized activated carbon. Bioresour. Technol. Rep. 2019, 7, 100248. [Google Scholar] [CrossRef]
- Sánchez-Bayo, A.; Morales, V.; Rodríguez, R.; Vicente, G.; Bautista, L.F. Biodiesel production (FAEEs) by heterogeneous combi-lipase biocatalysts using wet extracted lipids from microalgae. Catalysts 2019, 9, 296. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef] [PubMed]
- Arana-Peña, S.; Carballares, D.; Morellon-Sterlling, R.; Berenguer-Murcia, Á.; Alcántara, A.R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Enzyme co-immobilization: Always the biocatalyst designers’ choice…or not? Biotechnol. Adv. in press. [CrossRef]
- López-Serrano, P.; Cao, L.; Van Rantwijk, F.; Sheldon, R. Cross-linked enzyme aggregates with enhanced activity: Application to lipases. Biotechnol. Lett. 2002, 24, 1379–1383. [Google Scholar] [CrossRef]
- Mingarro, I.; Abad, C.; Braco, L. Interfacial activation-based molecular bioimprinting of lipolytic enzymes. Proc. Natl. Acad. Sci. USA 1995, 92, 3308–3312. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Perez, S.; Ghattas, N.; Filice, M.; Guisán, J.M.; Fernández-Lorente, G. Dramatic hyperactivation of lipase of Thermomyces lanuginosa by a cationic surfactant: Fixation of the hyperactivated form by adsorption on sulfopropyl-sepharose. J. Mol. Catal. Enzym. 2015, 122, 199–203. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, W. surfactant imprinting hyperactivated immobilized lipase as efficient biocatalyst for biodiesel production from waste cooking oil. Catalysts 2019, 9, 914. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Lorente, G.; Palomo, J.M.; Mateo, C.; Munilla, R.; Ortiz, C.; Cabrera, Z.; Guisán, A.J.M.; Fernandez-Lafuente, R. Glutaraldehyde cross-linking of lipases adsorbed on aminated supports in the presence of detergents leads to improved performance. Biomacromolecules 2006, 7, 2610–2615. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, J.C.; Garcia-Galan, C.; Rodrigues, R.C.; De Sant’Ana, H.B.; Gonçalves, L.R.B.; Fernandez-Lafuente, R.; Dos Santos, J.C. Stabilizing hyperactivated Lecitase structures through physical treatment with ionic polymers. Process Biochem. 2014, 49, 1511–1515. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Virgen-Ortíz, J.J.; Dos Santos, J.C.; Berenguer-Murcia, Á.; Alcantara, A.R.; Barbosa, O.; Ortiz, C.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports: Immobilization mechanism, advantages, problems, and solutions. Biotechnol. Adv. 2019, 37, 746–770. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos, J.C.; Rueda, N.; Torres, R.T.R.; Barbosa, O.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Evaluation of divinylsulfone activated agarose to immobilize lipases and to tune their catalytic properties. Process Biochem. 2015, 50, 918–927. [Google Scholar] [CrossRef]
- Dos Santos, J.C.S.; Rueda, N.; Sanchez, A.; Villalonga, R.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Versatility of divinylsulfone supports permits the tuning of CALB properties during its immobilization. RSC Adv. 2015, 5, 35801–35810. [Google Scholar] [CrossRef]
- Rueda, N.; Dos Santos, J.C.S.; Torres, R.; López, C.C.O.; Barbosa, O.; Fernandez-Lafuente, R. Improved performance of lipases immobilized on heterofunctional octyl-glyoxyl agarose beads. RSC Adv. 2015, 5, 11212–11222. [Google Scholar] [CrossRef] [Green Version]
- Suescun, A.; Rueda, N.; Dos Santos, J.C.; Castillo, J.J.; López, C.C.O.; Torres, R.; Barbosa, O.; Fernandez-Lafuente, R. Immobilization of lipases on glyoxyl–octyl supports: Improved stability and reactivation strategies. Process Biochem. 2015, 50, 1211–1217. [Google Scholar] [CrossRef]
- Arana-Peña, S.; Mendez-Sanchez, C.; Rios, N.S.; Ortiz, C.; Gonçalves, L.R.; Fernandez-Lafuente, R. New applications of glyoxyl-octyl agarose in lipases co-immobilization: Strategies to reuse the most stable lipase. Int. J. Biol. Macromol. 2019, 131, 989–997. [Google Scholar] [CrossRef]
- Rios, N.S.; Arana-Peña, S.; Mendez-Sanchez, C.; Ortiz, C.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Reuse of lipase from Pseudomonas fluorescens via its step-by-step coimmobilization on glyoxyl-octyl agarose beads with least stable lipases. Catalysts 2019, 9, 487. [Google Scholar] [CrossRef] [Green Version]
- Manoel, E.A.; Dos Santos, J.C.; Freire, D.M.; Rueda, N.; Fernández-Lafuente, R. Immobilization of lipases on hydrophobic supports involves the open form of the enzyme. Enzym. Microb. Technol. 2015, 71, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Peirce, S.; Virgen-Ortíz, J.J.; Tacias-Pascacio, V.G.; Rueda, N.; Bartolome-Cabrero, R.; Fernandez-Lopez, L.; Russo, M.E.; Marzocchella, A.; Fernandez-Lafuente, R. Development of simple protocols to solve the problems of enzyme coimmobilization. Application to coimmobilize a lipase and a β-galactosidase. RSC Adv. 2016, 6, 61707–61715. [Google Scholar] [CrossRef]
- Virgen-Ortíz, J.J.; Dos Santos, J.C.S.; Berenguer-Murcia, Á.; Barbosa, O.; Rodrigues, R.C.; Fernandez-Lafuente, R. Polyethylenimine: A very useful ionic polymer in the design of immobilized enzyme biocatalysts. J. Mater. Chem. B 2017, 5, 7461–7490. [Google Scholar] [CrossRef] [Green Version]
- Abian, O.; Wilson, L.; Mateo, C.; Fernandez-Lorente, G.; Palomo, J.; Fernández-Lafuente, R.; Guisán, J.M.; Re, D.; Tam, A.; Daminatti, M. Preparation of artificial hyper-hydrophilic micro-environments (polymeric salts) surrounding enzyme molecules. J. Mol. Catal. B: Enzym. 2002, 295–303. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Rocha-Martin, J.; Mateo, C.; Cava, F.; Berenguer, J.; Fernandez-Lafuente, R.; Guisan, J.M. Coating of soluble and immobilized enzymes with ionic polymers: Full stabilization of the quaternary structure of multimeric enzymes. Biomacromolecules 2009, 10, 742–747. [Google Scholar] [CrossRef]
- Fernandez-Lopez, L.; Pedrero, S.G.; Lopez-Carrobles, N.; Virgen-Ortíz, J.J.; Gorines, B.C.; Otero, C.; Fernandez-Lafuente, R. Physical crosslinking of lipase from Rhizomucor miehei immobilized on octyl agarose via coating with ionic polymers. Process Biochem. 2017, 54, 81–88. [Google Scholar] [CrossRef]
- Pedroche, J.; Yust, M.D.M.; Mateo, C.; Fernández-Lafuente, R.; Girón-Calle, J.; Alaiz, M.; Vioque, J.; Guisán, J.M.; Millán, F. Effect of the support and experimental conditions in the intensity of the multipoint covalent attachment of proteins on glyoxyl-agarose supports: Correlation between enzyme–support linkages and thermal stability. Enzym. Microb. Technol. 2007, 40, 1160–1166. [Google Scholar] [CrossRef]
- Mateo, C.; Fernandes, B.; Van Rantwijk, F.; Stolz, A.; A Sheldon, R. Stabilisation of oxygen-labile nitrilases via co-aggregation with poly(ethyleneimine). J. Mol. Catal. Enzym. 2006, 38, 154–157. [Google Scholar] [CrossRef]
- Zaak, H.; Fernandez-Lopez, L.; Otero, C.; Sassi, M.; Fernandez-Lafuente, R. Improved stability of immobilized lipases via modification with polyethylenimine and glutaraldehyde. Enzym. Microb. Technol. 2017, 106, 67–74. [Google Scholar] [CrossRef]
- Fernandez-Lopez, L.; Virgen-Ortíz, J.J.; Pedrero, S.G.; Lopez-Carrobles, N.; Gorines, B.C.; Otero, C.; Fernandez-Lafuente, R. Optimization of the coating of octyl-CALB with ionic polymers to improve stability and decrease enzyme leakage. Biocatal. Biotransform. 2017, 36, 47–56. [Google Scholar] [CrossRef]
- Rios, N.S.; Arana-Peña, S.; Mendez-Sanchez, C.; Lokha, Y.; Cortes-Corberan, V.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Increasing the enzyme loading capacity of porous supports by a layer-by-layer immobilization strategy using PEI as glue. Catalysts 2019, 9, 576. [Google Scholar] [CrossRef] [Green Version]
- Arana-Peña, S.; Rios, N.S.; Mendez-Sanchez, C.; Lokha, Y.; Carballares, D.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Coimmobilization of different lipases: Simple layer by layer enzyme spatial ordering. Int. J. Biol. Macromol. 2020, 145, 856–864. [Google Scholar] [CrossRef] [PubMed]
- Arana-Peña, S.; Rios, N.S.; Mendez-Sanchez, C.; Lokha, Y.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Use of polyethylenimine to produce immobilized lipase multilayers biocatalysts with very high volumetric activity using octyl-agarose beads: Avoiding enzyme release during multilayer production. Enzym. Microb. Technol. 2020, 137, 109535. [Google Scholar] [CrossRef]
- Albuquerque, T.L.; Rueda, N.; Dos Santos, J.C.; Barbosa, O.; López, C.C.O.; Binay, B.; Özdemir, E.; Gonçalves, L.R.; Fernandez-Lafuente, R. Easy stabilization of interfacially activated lipases using heterofunctional divinyl sulfone activated-octyl agarose beads. Modulation of the immobilized enzymes by altering their nanoenvironment. Process Biochem. 2016, 51, 865–874. [Google Scholar] [CrossRef]
- Zaak, H.; Kornecki, J.F.; Siar, E.-H.; Fernandez-Lopez, L.; Cortes Corberán, V.; Sassi, M.; Fernandez-Lafuente, R. Coimmobilization of enzymes in bilayers using PEI as a glue to reuse the most stable enzyme: Preventing PEI release during inactivated enzyme desorption. Process Biochem. 2017, 61, 95–101. [Google Scholar] [CrossRef]
- Anderson, E.M.; Larsson, K.M.; Kirk, O. One Biocatalyst–Many Applications: The use of Candida antarctica B-lipase in organic synthesis. Biocatal. Biotransform. 1998, 16, 181–204. [Google Scholar] [CrossRef]
- Monteiro, R.R.; Virgen-Ortiz, J.J.; Berenguer-Murcia, Á.; Da Rocha, T.N.; Dos Santos, J.C.; Alcántara, A.R.; Fernandez-Lafuente, R. Biotechnological relevance of the lipase A from Candida antarctica. Catal. Today. in press. [CrossRef]
- Gotor-Fernández, V.; Busto, E.; Gotor, V. Candida antarctica lipase B: An ideal biocatalyst for the preparation of nitrogenated organic compounds. Adv. Synth. Catal. 2006, 348, 797–812. [Google Scholar] [CrossRef]
- De María, P.D.; Carboni-Oerlemans, C.; Tuin, B.; Bargeman, G.; Van Der Meer, A.; Van Gemert, R. Biotechnological applications of Candida antarctica lipase A: State-of-the-art. J. Mol. Catal. B: Enzym. 2005, 37, 36–46. [Google Scholar] [CrossRef]
- Fernandez-Lafuente, R. Lipase from Thermomyces lanuginosus: Uses and prospects as an industrial biocatalyst. J. Mol. Catal. B: Enzym. 2010, 62, 197–212. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Fernandez-Lafuente, R. Lipase from Rhizomucor miehei as an industrial biocatalyst in chemical process. J. Mol. Catal. B: Enzym. 2010, 64, 1–22. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Fernandez-Lafuente, R. Lipase from Rhizomucor miehei as a biocatalyst in fats and oils modification. J. Mol. Catal. B: Enzym. 2010, 66, 15–32. [Google Scholar] [CrossRef]
- Virgen-Ortíz, J.J.; Dos Santos, J.C.; Ortiz, C.; Berenguer-Murcia, Á.; Barbosa, O.; Rodrigues, R.C.; Fernandez-Lafuente, R. Lecitase ultra: A phospholipase with great potential in biocatalysis. Mol. Catal. 2019, 473, 110405. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos, J.C.; Garcia-Galan, C.; Rodrigues, R.C.; Ana, H.B.D.S.; Gonçalves, L.R.B.; Fernandez-Lafuente, R.; Dos Santos, J.C. Improving the catalytic properties of immobilized Lecitase via physical coating with ionic polymers. Enzym. Microb. Technol. 2014, 60, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lopez, L.; Pedrero, S.G.; Lopez-Carrobles, N.; Gorines, B.C.; Virgen-Ortíz, J.J.; Fernandez-Lafuente, R. Effect of protein load on stability of immobilized enzymes. Enzym. Microb. Technol. 2017, 98, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Zaak, H.; Siar, E.-H.; Kornecki, J.F.; Fernandez-Lopez, L.; Pedrero, S.G.; Virgen-Ortíz, J.J.; Fernandez-Lafuente, R. Effect of immobilization rate and enzyme crowding on enzyme stability under different conditions. The case of lipase from Thermomyces lanuginosus immobilized on octyl agarose beads. Process Biochem. 2017, 56, 117–123. [Google Scholar] [CrossRef]
- Shen, L.; Chen, Z. Critical review of the impact of tortuosity on diffusion. Chem. Eng. Sci. 2007, 62, 3748–3755. [Google Scholar] [CrossRef]
- Pronk, S.; Lindahl, E.; Kasson, P.M. Dynamic heterogeneity controls diffusion and viscosity near biological interfaces. Nat. Commun. 2014, 5, 3034. [Google Scholar] [CrossRef] [Green Version]
- Regan, D.L.; Lilly, M.D.; Dunnill, P. Influence of intraparticle diffusional limitation on the observed kinetics of immobilized enzymes and on catalyst design. Biotechnol. Bioeng. 1974, 16, 1081–1093. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Consolati, T.; Mayr, T.; Nidetzky, B. Shine a light on immobilized enzymes: Real-time sensing in solid supported biocatalysts. Trends Biotechnol. 2013, 31, 194–203. [Google Scholar] [CrossRef]
- Boniello, C.; Mayr, T.; Klimant, I.; Koenig, B.; Riethorst, W.; Nidetzky, B. Intraparticle concentration gradients for substrate and acidic product in immobilized cephalosporin C amidase and their dependencies on carrier characteristics and reaction parameters. Biotechnol. Bioeng. 2010, 106, 528–540. [Google Scholar] [CrossRef]
- Rocha-Martín, J.; Rivas, B.D.L.; Muñoz, R.; Guisán, J.M.; López-Gallego, F. rational co-immobilization of bi-enzyme cascades on porous supports and their applications in bio-redox reactions with in situ recycling of soluble cofactors. ChemCatChem 2012, 4, 1279–1288. [Google Scholar] [CrossRef]
- Palomo, J.M.; Fernandez-Lorente, G.; Mateo, C.; Ortiz, C.; Fernandez-Lafuente, R.; Guisán, J.M. Modulation of the enantioselectivity of lipases via controlled immobilization and medium engineering: Hydrolytic resolution of mandelic acid esters. Enzym. Microb. Technol. 2002, 31, 775–783. [Google Scholar] [CrossRef]
- Chaubey, A.; Parshad, R.; Koul, S.; Taneja, S.C.; Qazi, G.N. Enantioselectivity modulation through immobilization of Arthrobacter sp. lipase: Kinetic resolution of fluoxetine intermediate. J. Mol. Catal. B: Enzym. 2006, 42, 39–44. [Google Scholar] [CrossRef]
- Tacias-Pascacio, V.G.; Peirce, S.; Torrestiana-Sanchez, B.; Yates, M.; Rosales-Quintero, A.; Virgen-Ortíz, J.J.; Fernandez-Lafuente, R. Evaluation of different commercial hydrophobic supports for the immobilization of lipases: Tuning their stability, activity and specificity. RSC Adv. 2016, 6, 100281–100294. [Google Scholar] [CrossRef]
- Ashjari, M.; Mohammadi, M.; Badri, R. Chemical amination of Rhizopus oryzae lipase for multipoint covalent immobilization on epoxy-functionalized supports: Modulation of stability and selectivity. J. Mol. Catal. B: Enzym. 2015, 115, 128–134. [Google Scholar] [CrossRef]
- Lokha, Y.; Arana-Peña, S.; Rios, N.S.; Mendez-Sanchez, C.; Gonçalves, L.R.B.; López-Gallego, F.; Fernandez-Lafuente, R. Modulating the properties of the lipase from Thermomyces lanuginosus immobilized on octyl agarose beads by altering the immobilization conditions. Enzym. Microb. Technol. 2020, 133, 109461. [Google Scholar] [CrossRef]
- Silveira, E.A.; Moreno-Perez, S.; Basso, A.; Serban, S.; Pestana-Mamede, R.; Tardioli, P.W.; Farinas, C.S.; Castejon, N.; Fernandez-Lorente, G.; Rocha-Martin, J.; et al. Biocatalyst engineering of Thermomyces lanuginosus lipase adsorbed on hydrophobic supports: Modulation of enzyme properties for ethanolysis of oil in solvent-free systems. J. Biotechnol. 2019, 289, 126–134. [Google Scholar] [CrossRef]
- Martínez-Sanchez, J.A.; Arana-Peña, S.; Carballares, D.; Yates, M.; Otero, C.; Fernandez-Lafuente, R. Immobilized biocatalysts of Eversa® Transform 2.0 and lipase from Thermomyces lanuginosus: Comparison of some properties and performance in biodiesel production. Catalysts 2020, 10, 738. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bastida, A.; Sabuquillo, P.; Armisen, P.; Fernández-Lafuente, R.; Huguet, J.; Guisán, J.M. A single step purification, immobilization, and hyperactivation of lipases via interfacial adsorption on strongly hydrophobic supports. Biotechnol. Bioeng. 1998, 58, 486–493. [Google Scholar] [CrossRef]
- Boudrant, J.; Woodley, J.M.; Fernandez-Lafuente, R. Parameters necessary to define an immobilized enzyme preparation. Process Biochem. 2020, 90, 66–80. [Google Scholar] [CrossRef]
- Lombardo, D.; Guy, O. Effect of alcohols on the hydrolysis catalyzed by human pancreatic carboxylic-ester hydrolase. Biochim. Biophys. Acta (BBA)-Enzym. 1981, 657, 425–437. [Google Scholar] [CrossRef]
- Hernandez, K.; Porcar, R.; Porcar, R.; Fernandez-Lafuente, R. Hydrolysis of triacetin catalyzed by immobilized lipases: Effect of the immobilization protocol and experimental conditions on diacetin yield. Enzym. Microb. Technol. 2011, 48, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Kornecki, J.F.; Carballares, D.; Morellon-Sterling, R.; Siar, E.H.; Kashefi, S.; Chafiaa, M.; Arana-Peña, S.; Rios, N.S.; Gonçalves, L.R.; Fernandez-Lafuente, R. Influence of phosphate anions on the stability of immobilized enzymes. Effect of enzyme nature, immobilization protocol and inactivation conditions. Process Biochem. 2020, 95, 288–296. [Google Scholar] [CrossRef]
- Zaak, H.; Fernandez-Lopez, L.; Velasco-Lozano, S.; Alcaraz-Fructuoso, M.T.; Sassi, M.; Lopez-Gallego, F.; Fernandez-Lafuente, R. Effect of high salt concentrations on the stability of immobilized lipases: Dramatic deleterious effects of phosphate anions. Process Biochem. 2017, 62, 128–134. [Google Scholar] [CrossRef]
- Laemmli, U.K. cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nat. Cell Biol. 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Trobo-Maseda, L.; Orrego, A.H.; Guisán, J.M.; Rocha-Martin, J. Coimmobilization and colocalization of a glycosyltransferase and a sucrose synthase greatly improves the recycling of UDP-glucose: Glycosylation of resveratrol 3-O-β-D-glucoside. Int. J. Biol. Macromol. 2020, 157, 510–521. [Google Scholar] [CrossRef]










| Ammonium Sulfate, (M) | PEI on the Support, (%) |
|---|---|
| 0.1 | 98 |
| 0.25 | 86 |
| 0.5 | 83 |
| 1 | 84 |
| 2 | 79 |
| 4 | 78 |
| Substrate | ||||
|---|---|---|---|---|
| Biocatalyst | p-NPB | Triacetin | (R)-Methyl Mandelate | (S)-Methyl Mandelate |
| Octyl-CALA | 102.67 ± 5.14 | 0.90 ± 0.04 | 0.255 ± 0.013 | 0.251 ± 0.013 |
| Octyl-VS-CALA | 106.00 ± 5.35 | 1.29 ± 0.07 | 0.092 ± 0.005 | 0.064 ± 0.003 |
| Octyl-CALB | 20.57 ± 1.03 | 8.60 ± 0.43 | 35.797 ± 1.790 | 4.680 ± 0.234 |
| Octyl-VS-CALB | 22.79 ± 1.15 | 8.16 ± 0.41 | 34.708 ± 1.738 | 4.215 ± 0.211 |
| Octyl-TLL | 148.31 ± 7.55 | 34.27 ± 1.72 | 0.041 ± 0.002 | 0.045 ± 0.002 |
| Octyl-VS-TLL | 114.16 ± 5.72 | 32.13 ± 1.71 | 0.016 ± 0.001 | 0.025 ± 0.001 |
| Octyl-LEU | 97.22 ± 4.82 | 6.47 ± 0.31 | 0.026 ± 0.001 | 0.041 ± 0.020 |
| Octyl-VS-PEI-LEU | 114.67 ± 5.75 | 20.97 ± 1.08 | 0.126 ± 0.006 | 0.135 ± 0.006 |
| Octyl-RML | 69.36 ± 3.52 | 21.49 ± 1.20 | 0.038 ± 0.002 | 0.038 ± 0.002 |
| Octyl-VS-PEI-RML | 65.68 ± 3.28 | 27.94 ± 1.35 | 0.292 ± 0.015 | 0.261 ± 0.013 |
| Substrate | ||||
|---|---|---|---|---|
| Biocatalyst | p-NPB | Triacetin | (R)-Methyl Mandelate | (S)-Methyl Mandelate |
| Octyl-VS-CALA | 108.82 ± 5.10 | 1.21 ± 0.06 | 0.094 ± 0.004 | 0.061 ± 0.003 |
| Octyl-VS-CALA-TLL | 136.37 ± 6.82 | 37.21 ± 1.86 | 0.109 ± 0.005 | 0.099 ± 0.005 |
| Octyl-VS-CALA-TLL-CALB | 134.49 ± 5.53 | 40.22 ± 1.90 | 31.447 ± 1.472 | 4.080 ± 0.215 |
| Octyl-VS-CALA-TLL-CALB-PEI | 138.36 ± 6.81 | 40.92 ± 2.12 | 39.901 ± 2.015 | 4.253 ± 0.223 |
| Octyl-VS-CALA-TLL-CALB-PEI-LEU | 153.64 ± 7.70 | 47.54 ± 2.23 | 36.643 ± 1.865 | 4.320 ± 0.224 |
| Octyl-VS-CALA-TLL-CALB-PEI-LEU-RML | 176.52 ± 8.83 | 60.74 ± 3.10 | 37.078 ± 1.855 | 4.602 ± 0.235 |
| Substrate | ||||
|---|---|---|---|---|
| Biocatalyst | p-NPB | Triacetin | (R)-Methyl Mandelate | (S)-Methyl Mandelate |
| Octyl-VS-CALB | 21.26 ± 1.08 | 7.88 ± 0.45 | 34.708 ± 1.738 | 4.104 ± 0.224 |
| Octyl-VS-CALB-TLL | 93.83 ± 4.72 | 34.36 ± 1.79 | 35.605 ± 1.753 | 3.928 ± 0.195 |
| Octyl-VS-CALB-TLL-CALA | 115.45 ± 5.83 | 34.09 ± 1.35 | 34.942 ± 1.497 | 3.909 ± 0.190 |
| Octyl-VS-CALB-TLL-CALA-PEI | 118.75 ± 5.95 | 30.75 ± 1.40 | 35.331 ± 1.698 | 4.144 ± 0.210 |
| Octyl-VS-CALB-TLL-CALA-PEI-RML | 126.23 ± 6.32 | 39.54 ± 2.89 | 33.978 ± 1.707 | 4.131 ± 0.205 |
| Octyl-VS-CALB-TLL-CALA-PEI-RML-LEU | 166.73 ± 8.50 | 56.46 ± 3.05 | 34.790 ± 2.005 | 4.310 ± 0.221 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arana-Peña, S.; Carballares, D.; Cortés Corberan, V.; Fernandez-Lafuente, R. Multi-Combilipases: Co-Immobilizing Lipases with Very Different Stabilities Combining Immobilization via Interfacial Activation and Ion Exchange. The Reuse of the Most Stable Co-Immobilized Enzymes after Inactivation of the Least Stable Ones. Catalysts 2020, 10, 1207. https://doi.org/10.3390/catal10101207
Arana-Peña S, Carballares D, Cortés Corberan V, Fernandez-Lafuente R. Multi-Combilipases: Co-Immobilizing Lipases with Very Different Stabilities Combining Immobilization via Interfacial Activation and Ion Exchange. The Reuse of the Most Stable Co-Immobilized Enzymes after Inactivation of the Least Stable Ones. Catalysts. 2020; 10(10):1207. https://doi.org/10.3390/catal10101207
Chicago/Turabian StyleArana-Peña, Sara, Diego Carballares, Vicente Cortés Corberan, and Roberto Fernandez-Lafuente. 2020. "Multi-Combilipases: Co-Immobilizing Lipases with Very Different Stabilities Combining Immobilization via Interfacial Activation and Ion Exchange. The Reuse of the Most Stable Co-Immobilized Enzymes after Inactivation of the Least Stable Ones" Catalysts 10, no. 10: 1207. https://doi.org/10.3390/catal10101207