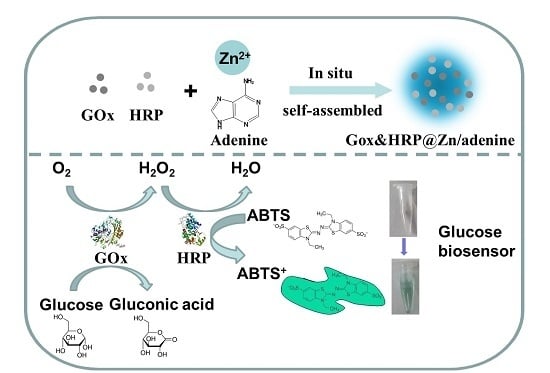
In-Situ Self-Assembly of Zinc/Adenine Hybrid Nanomaterials for Enzyme Immobilization
Abstract
:1. Introduction
2. Results and Discussion
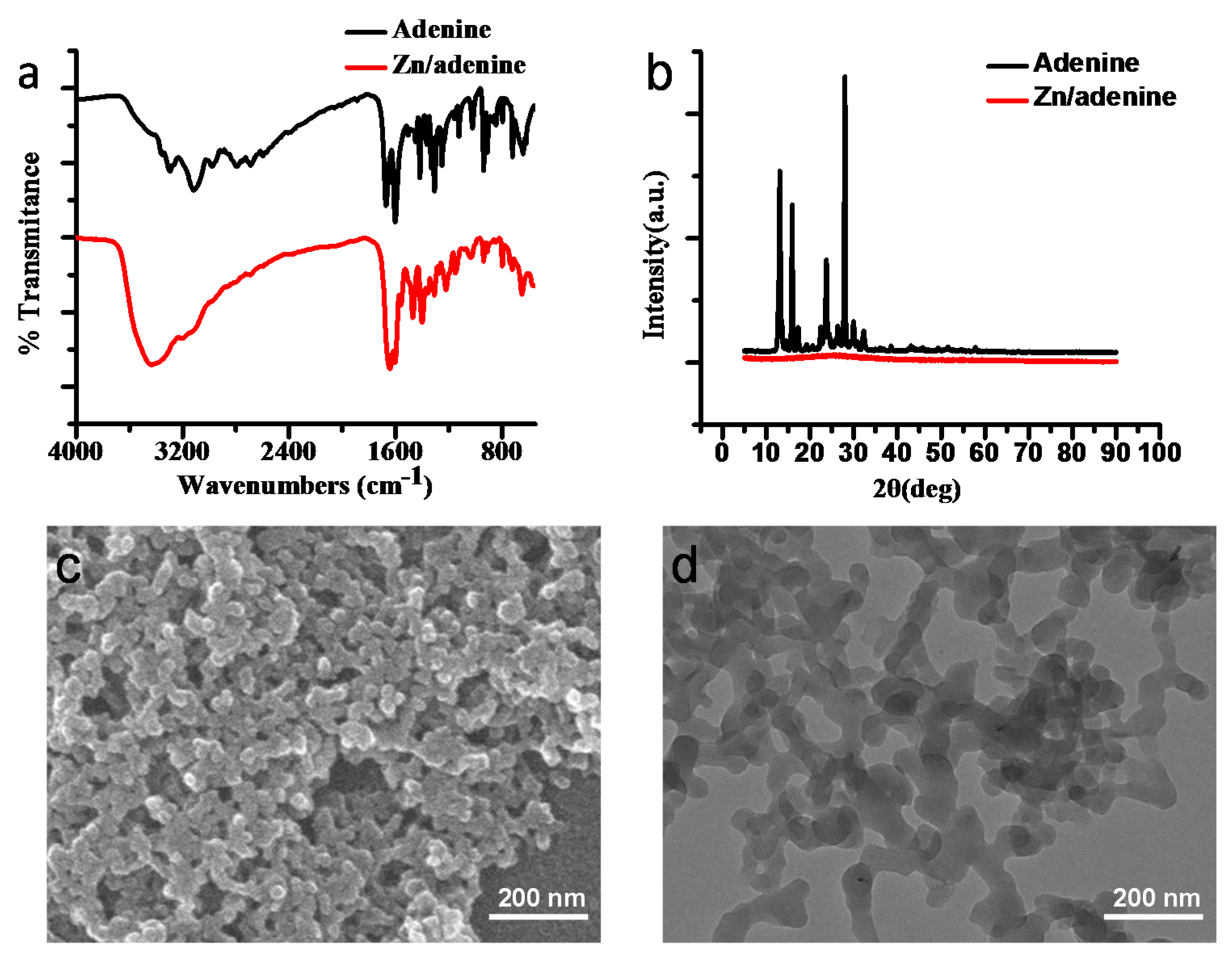
2.1. Preparation and Characterization of Zn/Adenine Complexes
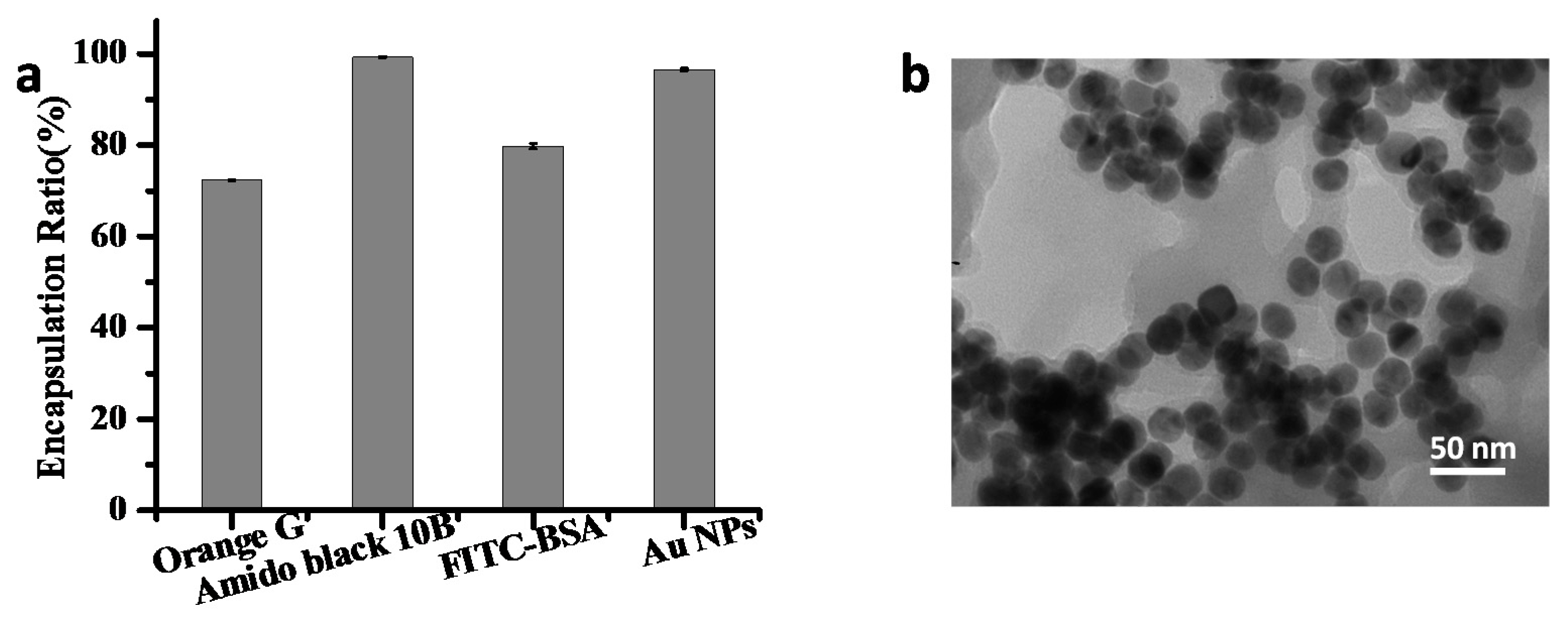
2.2. Encapsulation Property of Zn/Adenine Complexes
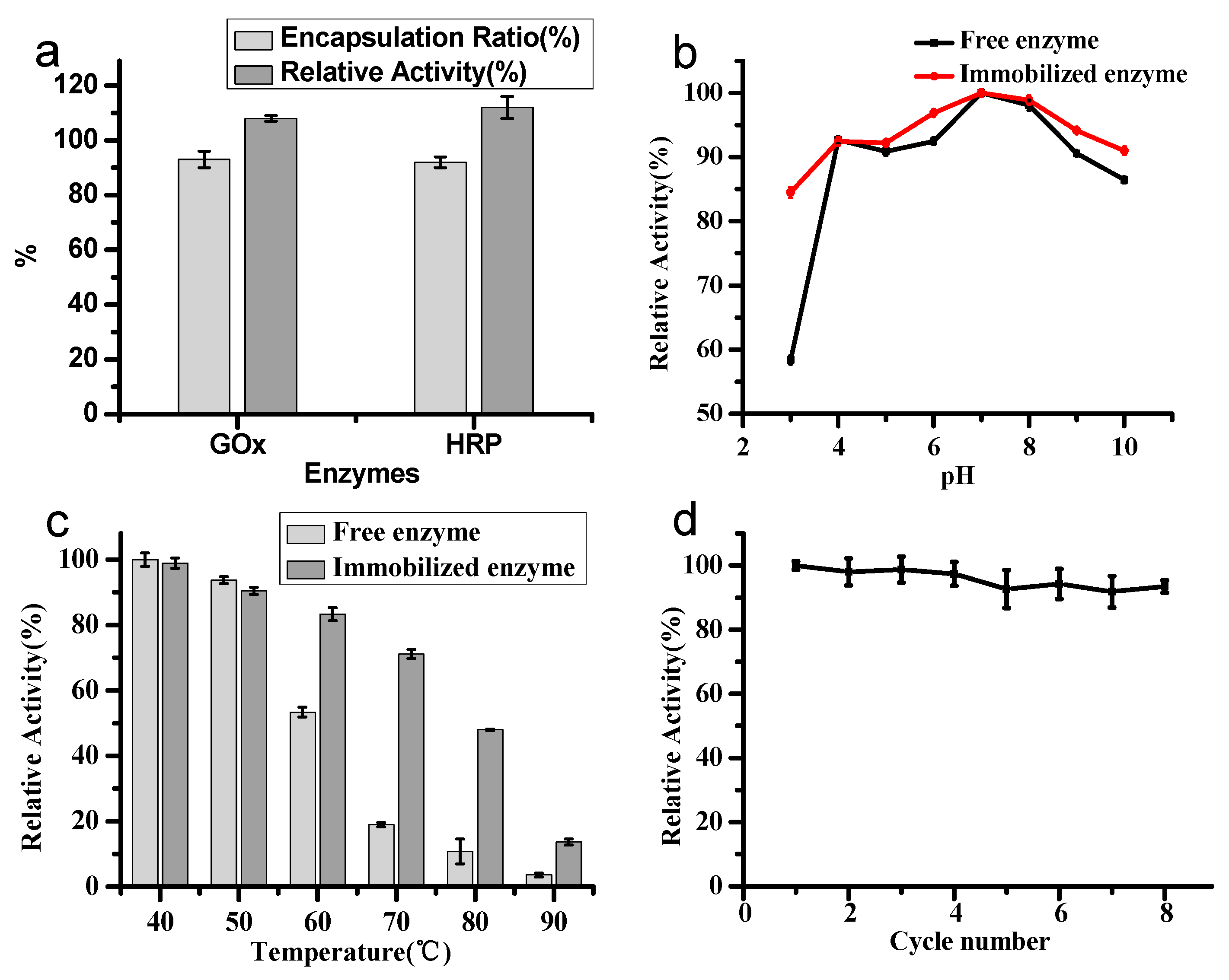
2.3. Immobilization of Single Enzyme
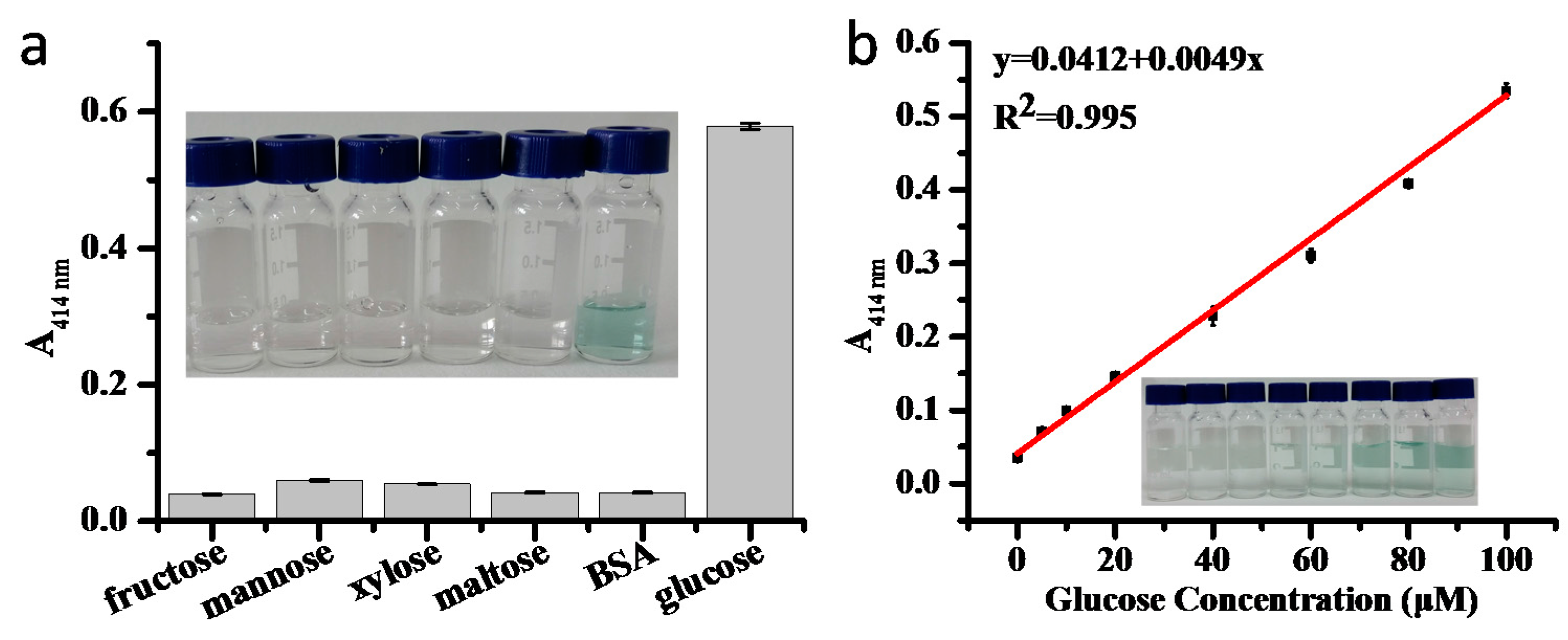
2.4. Co-Immobilization of GOx and HRP
3. Materials and Methods
3.1. Materials
3.2. Preparation of Zn and Adenine Coordinated Complexes
3.3. Study of the Zn/Adenine Complexes at Different Concentrations and pH of Buffer, as Well as Different Ionic Strengths
3.4. Stoichiometry and Structural Characterization of Zn/Adenine Complexes
3.5. Encapsulation Experiment
3.6. Immobilization of Single Enzymes
3.7. Enzyme Stability Test
3.8. Co-Immobilization of GOx and HRP
3.9. Glucose Detection with GOx–HRP–Zn/Adenine Complexes
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bornscheuer, U.T.; Huisman, G.W.; Kazlauskas, R.J.; Lutz, S.; Moore, J.C.; Robins, K. Engineering the third wave of biocatalysis. Nature 2012, 485, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Han, S.S.; Kim, H.S. Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnol. Adv. 2015, 33, 1443–1454. [Google Scholar] [CrossRef] [PubMed]
- Kirk, O.; Borchert, T.V.; Fuglsang, C.C. Industrial enzyme applications [review]. Curr. Opin. Biotechnol. 2002, 13, 345–351. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Tran, D.N.; Balkus, K.J.B., Jr. Perspective of recent progress in immobilization of enzymes. ACS Catal. 2011, 1, 956–968. [Google Scholar] [CrossRef]
- Wu, X.; Hou, M.; Ge, J. Metal-organic frameworks and inorganic nanoflowers: A type of emerging inorganic crystal nanocarrier for enzyme immobilization. Catal. Sci. Technol. 2015, 5, 5077–5085. [Google Scholar] [CrossRef]
- Cao, S.; Xu, P.; Ma, Y.; Yao, X.; Yao, Y.; Zong, M.; Li, X.; Lou, W. Recent advances in immobilized enzymes on nanocarriers. Chin. J. Catal. 2016, 37, 1814–1823. [Google Scholar] [CrossRef]
- Altinkaynak, C.; Tavlasoglu, S.; Ÿzdemir, N.; Ocsoy, I. A new generation approach in enzyme immobilization: Organic-inorganic hybrid nanoflowers with enhanced catalytic activity and stability. Enzyme Microb. Technol. 2016, 93–94, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jiang, S.; Zhao, X.; Liang, H. Co-immobilization of enzymes and magnetic nanoparticles by metal-nucleotide hydrogelnanofibers for improving stability and recycling. Molecules 2017, 22, 179. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Jiang, S.; Yuan, Q.; Li, G.; Wang, F.; Zhang, Z.; Liu, J. Co-immobilization of multiple enzymes by metal coordinated nucleotide hydrogel nanofibers: Improved stability and an enzyme cascade for glucose detection. Nanoscale 2016, 8, 6071–6078. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Liu, B.; Yuan, Q.; Liu, J. Magnetic iron oxide nanoparticle seeded growth of nucleotide coordinated polymers. ACS Appl. Mater. Interfaces 2016, 8, 15615–15622. [Google Scholar] [CrossRef] [PubMed]
- Lyu, F.; Zhang, Y.; Zare, R.N.; Ge, J.; Liu, Z. One-pot synthesis of protein-embedded metal-organic frameworks with enhanced biological activities. Nano Lett. 2014, 14, 5761–5765. [Google Scholar] [CrossRef] [PubMed]
- Brady, D.; Jordaan, J. Advances in enzyme immobilisation. Biotechnol. Lett. 2009, 31, 1639. [Google Scholar] [CrossRef] [PubMed]
- Govardhan, C.P. Crosslinking of enzymes for improved stability and performance. Curr. Opin. Biotechnol. 1999, 10, 331–335. [Google Scholar] [CrossRef]
- Kim, J.; Grate, J.W.; Wang, P. Nanostructures for enzyme stabilization. Chem. Eng. Sci. 2006, 61, 1017–1026. [Google Scholar] [CrossRef]
- Lei, C.; Shin, Y.; Magnuson, J.K.; Fryxell, G.; Lasure, L.L.; Elliott, D.C.; Liu, J.; Ackerman, E.J. Characterization of functionalized nanoporous supports for protein confinement. Nanotechnology 2006, 17, 5531–5538. [Google Scholar] [CrossRef] [PubMed]
- Nadar, S.S.; Rathod, V.K. Magnetic macromolecular cross linked enzyme aggregates (cleas) of glucoamylase. Enzyme Microb. Technol. 2016, 83, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Schoevaart, R.; Wolbers, M.W.; Golubovic, M.; Ottens, M.; Kieboom, A.P.; Van, R.F.; La, V.D.W.; Sheldon, R.A. Preparation, optimization, and structures of cross-linked enzyme aggregates (cleas). Biotechnol. Bioeng. 2004, 87, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Sharma, A.; Gupta, M.N. Preparation of cross-linked enzyme aggregates by using bovine serum albumin as a proteic feeder. Anal. Biochem. 2006, 351, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A. Cross-linked enzyme aggregates (cleas): Stable and recyclable biocatalysts. Biochem. Soc. Trans. 2007, 35, 1583–1587. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A.; Van, P.S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Bao, W.J.; Wang, J.; Wang, K.; Xu, J.J.; Chen, H.Y.; Xia, X.H. A green approach to the synthesis of novel “desert rose stone”-like nanobiocatalytic system with excellent enzyme activity and stability. Sci. Rep. 2014, 4, 6606. [Google Scholar] [CrossRef] [PubMed]
- Cook, T.R.; Zheng, Y.R.; Stang, P.J. Metal-organic frameworks and self-assembled supramolecular coordination complexes: Comparing and contrasting the design, synthesis and functionality of metal-organic materials. Chem. Rev. 2013, 113, 734–777. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cohen Stuart, M.A.; Marcelis, A.T.M.; Colomb-Delsuc, M.; Otto, S.; van der Gucht, J. Stable polymer micelles formed by metal coordination. Macromolecules 2012, 45, 7179–7185. [Google Scholar] [CrossRef]
- Wang, X.; McHale, R. Metal-containing polymers: Building blocks for functional (nano)materials. Macromol. Rapid Commun. 2010, 31, 331–350. [Google Scholar] [CrossRef] [PubMed]
- Whittell, G.R.; Hager, M.D.; Schubert, U.S.; Manners, I. Functional soft materials from metallopolymers and metallosupramolecular polymers. Nat. Mater. 2011, 10, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-X.; Liao, P.-Q.; Lin, R.-B.; Wei, Y.-S.; Zeng, M.-H.; Chen, X.-M. Metal cluster-based functional porous coordination polymers. Coord. Chem. Rev. 2015, 293–294, 263–278. [Google Scholar] [CrossRef]
- Tan, H.; Zhang, L.; Ma, C.; Song, Y.; Xu, F.; Chen, S.; Wang, L. Terbium-based coordination polymer nanoparticles for detection of ciprofloxacin in tablets and biological fluids. ACS Appl. Mater. Interfaces 2013, 5, 11791–11796. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.M.; Jin, A.; Lin, W. Surfactant-assisted synthesis of nanoscale gadolinium metal–organic frameworks for potential multimodal imaging. Angew. Chem. Int. Ed. 2008, 47, 7722–7725. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Huxford, R.C.; Lin, W. Phosphorescent nanoscale coordination polymers as contrast agents for optical imaging. Angew. Chem. Int. Ed. 2011, 50, 3696–3700. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.D.; Feldblyum, J.I.; Wong-Foy, A.G.; Matzger, A.J. Filling pore space in a microporous coordination polymer to improve methane storage performance. Langmuir 2015, 31, 2211–2217. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.F.; Zheng, L.L.; Liang, L.J.; Yang, X.X.; Li, Y.F.; Huang, C.Z. A new type of ph-responsive coordination polymer sphere as a vehicle for targeted anticancer drug delivery and sustained release. J. Mater. Chem. B 2013, 1, 3202–3208. [Google Scholar] [CrossRef]
- Liu, Y.; Tang, Z. Nanoscale biocoordination polymers: Novel materials from an old topic. Chemistry 2012, 18, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Zhang, Z.; Yuan, Q.; Liu, J. Self-healing metal-coordinated hydrogels using nucleotide ligands. Chem. Commun. 2015, 51, 15196–15199. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Ge, J.; Liu, W.; Lan, M.; Zhang, H.; Wang, P.; Wang, Y.; Niu, Z. Multi-enzyme co-embedded organic-inorganic hybrid nanoflowers: Synthesis and application as a colorimetric sensor. Nanoscale 2014, 6, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Mantion, A.; Massüger, L.; Rabu, P.; Palivan, C.; McCusker, L.B.; Taubert, A. Metal-peptide frameworks (mpfs): “Bioinspired” metal organic frameworks. J. Am. Chem. Soc. 2008, 130, 2517–2526. [Google Scholar] [CrossRef] [PubMed]
- Imaz, I.; Rubio-Martínez, M.; Saletra, W.J.; Amabilino, D.B.; Maspoch, D. Amino acid based metal-organic nanofibers. J. Am. Chem. Soc. 2009, 131, 18222–18223. [Google Scholar] [CrossRef] [PubMed]
- Moussatova, A.; Vázquez, M.-V.; Martínez, A.; Dolgounitcheva, O.; Zakrzewski, V.G.; Ortiz, J.V.; Pedersen, D.B.; Simard, B. Theoretical study of the structure and bonding of a metal—DNA Base complex: Al-guanine. J. Phys. Chem. A 2003, 107, 9415–9421. [Google Scholar] [CrossRef]
- Wang, F.; Liu, B.; Huang, P.-J.J.; Liu, J. Rationally designed nucleobase and nucleotide coordinated nanoparticles for selective DNA adsorption and detection. Anal. Chem. 2013, 85, 12144–12151. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Mishra, A.K.; Kumar, J. The many facets of adenine: Coordination, crystal patterns, and catalysis. Acc. Chem. Res. 2010, 43, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Bakkali, H.E.; Castiñeiras, A.; Garcíasantos, I.; Gonzálezpérez, J.M.; Niclósgutiérrez, J. Metallo-supramolecular structures by self-assembly through weak interactions in mixed ligand metal complexes of adenine and malonate. Cryst. Growth Des. 2015, 14, 249–260. [Google Scholar] [CrossRef]
- Navarro, J.A.R.; Lippert, B. Molecular architecture with metal ions, nucleobases and other heterocycles. Coord. Chem. Rev. 1999, 185, 653–667. [Google Scholar] [CrossRef]
- Singh, P. The family of n 9 -adenine: New entry for adenine-benzamide conjugates linked via versatile spacers. J. Chem. Sci. 2014, 126, 159–167. [Google Scholar] [CrossRef]
- An, J.; Geib, S.J.; Rosi, N.L. Cation-triggered drug release from a porous zinc-adeninate metal-organic framework. J. Am. Chem. Soc. 2009, 131, 8376–8377. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Li, B.; Du, Y.; Shaojun Dong, A.; Wang, E. Nucleobase-metal hybrid materials: Preparation of submicrometer-scale, spherical colloidal particles of adenine-gold(iii) via a supramolecular hierarchical self-assembly approach. Chem. Mater. 2007, 21, 2987–2993. [Google Scholar] [CrossRef]
- And, C.S.P.; Verma, S. A luminescent silver-adenine metallamacrocyclic quartet. J. Am. Chem. Soc. 2006, 128, 400–401. [Google Scholar]
- Burneo, I.; Stylianou, K.C.; Rodríguez-Hermida, S.; Juanhuix, J.; Fontrodona, X.; Imaz, I.; Maspoch, D. Two new adenine-based co(II) coordination polymers: Synthesis, crystal structure, coordination modes, and reversible hydrochromic behavior. Cryst. Growth Des. 2015, 15, 3182–3189. [Google Scholar] [CrossRef]
- Garcíaterán, J.P.; Castillo, O.; Luque, A.; Garcíacouceiro, U.; Pascual Román, A.; Lezama, L. An unusual 3d coordination polymer based on bridging interactions of the nucleobase adenine. Inorg. Chem. 2004, 43, 4549–4551. [Google Scholar] [CrossRef] [PubMed]
- Ge, J. Protein-inorganic hybrid nanoflowers. Nat. Nanotechnol. 2012, 7, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Lu, D.; Liu, Z.; Liu, Z. Recent advances in nanostructured biocatalysts. Biochem. Eng. J. 2009, 44, 53–59. [Google Scholar] [CrossRef]
- Kim, J.; Grate, J.W.; Wang, P. Nanobiocatalysis and its potential applications. Top. Catal. 2012, 108, 639–646. [Google Scholar] [CrossRef]
- Wu, X.; Ge, J.; Yang, C.; Hou, M.; Liu, Z. Facile synthesis of multiple enzyme-containing metal-organic frameworks in a biomolecule-friendly environment. Chem. Commun. 2015, 51, 13408–13411. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Wang, Y.; Zhao, M.; Zhang, L. Intrinsic peroxidase-like activity and catalase-like activity of CO3O4 nanoparticles. Chem. Commun. 2012, 48, 2540–2542. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, H.; Sun, S.; Zhou, Y.; Liu, Y. In-Situ Self-Assembly of Zinc/Adenine Hybrid Nanomaterials for Enzyme Immobilization. Catalysts 2017, 7, 327. https://doi.org/10.3390/catal7110327
Liang H, Sun S, Zhou Y, Liu Y. In-Situ Self-Assembly of Zinc/Adenine Hybrid Nanomaterials for Enzyme Immobilization. Catalysts. 2017; 7(11):327. https://doi.org/10.3390/catal7110327
Chicago/Turabian StyleLiang, Hao, Shanshan Sun, Yan Zhou, and Yanhui Liu. 2017. "In-Situ Self-Assembly of Zinc/Adenine Hybrid Nanomaterials for Enzyme Immobilization" Catalysts 7, no. 11: 327. https://doi.org/10.3390/catal7110327