Improving the Stability of Cold-Adapted Enzymes by Immobilization
Abstract
:1. Introduction
2. Protein Engineering to Improve the Stability of Cold-Adapted Enzymes
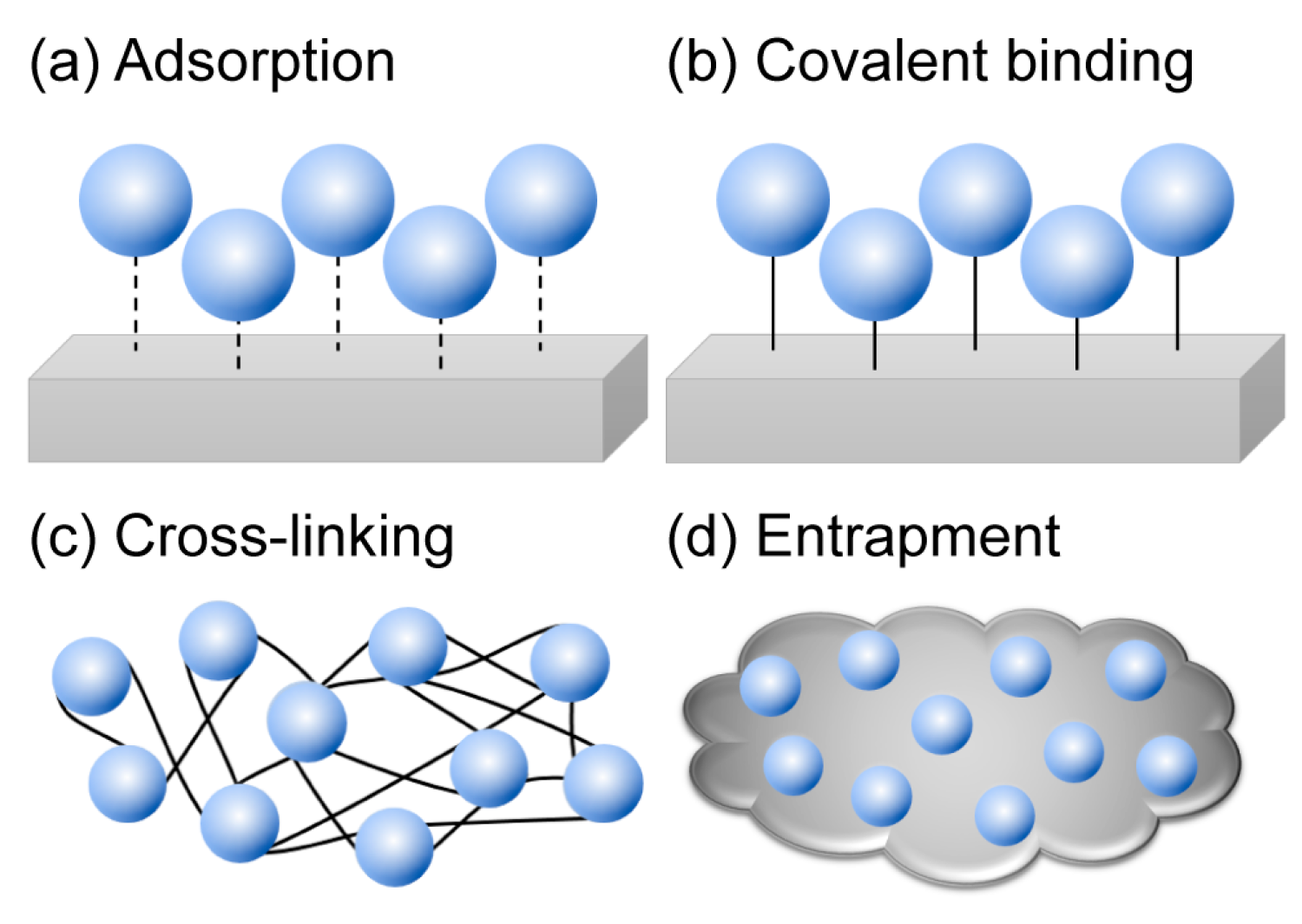
3. Immobilization of Cold-Adapted Enzymes
3.1. Adsorption
3.2. Covalent Binding
3.3. Cross-Linking and Entrapment
4. Stability in Organic Solvent
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Lee, Y.M.; Kim, G.; Jung, Y.J.; Choe, C.D.; Yim, J.H.; Lee, H.K.; Hong, S.G. Polar and Alpine Microbial Collection (PAMC): A culture collection dedicated to polar and alpine microorganisms. Polar Biol. 2012, 35, 1433–1438. [Google Scholar] [CrossRef]
- Nichols, D.; Bowman, J.; Sanderson, K.; Nichols, C.M.; Lewis, T.; McMeekin, T.; Nichols, P.D. Developments with Antarctic microorganisms: Culture collections, bioactivity screening, taxonomy, PUFA production and cold-adapted enzymes. Curr. Opin. Biotechnol. 1999, 10, 240–246. [Google Scholar] [CrossRef]
- Helmke, E.; Weyland, H. Psychrophilic versus psychrotolerant bacteria-occurrence and significance in polar and temperate marine habitats. Cell. Mol. Biol. 2004, 50, 553–561. [Google Scholar] [PubMed]
- Cowan, D.A.; Casanueva, A.; Stafford, W. Ecology and biodiversity of cold-adapted microorganisms. In Physiology and Biochemistry of Extremophiles; American Society of Microbiology: Washington, DC, USA, 2007. [Google Scholar]
- D’Amico, S.; Collins, T.; Marx, J.C.; Feller, G.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef] [PubMed]
- De Maayer, P.; Anderson, D.; Cary, C.; Cowan, D.A. Some like it cold: Understanding the survival strategies of psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Struvay, C.; Feller, G. Optimization to low temperature activity in psychrophilic enzymes. Int. J. Mol. Sci. 2012, 13, 11643–11665. [Google Scholar] [CrossRef] [PubMed]
- Gerday, C. Catalysis and Protein Folding in Psychrophiles. In Cold-Adapted Microorganisms; Yumoto, I., Ed.; Caister Academic Press: Norfolk, UK, 2013; pp. 137–160. [Google Scholar]
- Siddiqui, K.S.; Cavicchioli, R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Fields, P.A.; Somero, G.N. Hot spots in cold adaptation: Localized increases in conformational flexibility in lactate dehydrogenase A4 orthologs of Antarctic notothenioid fishes. Proc. Natl. Acad. Sci. USA 1998, 95, 11476–11481. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.A.; Motoshima, H.; Watanabe, K. Fluorescence studies on the stability, flexibility and substrate-induced conformational changes of acetate kinases from psychrophilic and mesophilic bacteria. Protein J. 2012, 31, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S.; Feller, G.; D’Amico, S.; Gerday, C.; Giaquinto, L.; Cavicchioli, R. The active site is the least stable structure in the unfolding pathway of a multidomain cold-adapted alpha-amylase. J. Bacteriol. 2005, 187, 6197–6205. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, E.; Riccardi, L.; Villa, C.; Fantucci, P.; De Gioia, L. Flexibility and enzymatic cold-adaptation: A comparative molecular dynamics investigation of the elastase family. Biochim. Biophys. Acta 2006, 1764, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Merlino, A.; Russo Krauss, I.; Castellano, I.; De Vendittis, E.; Rossi, B.; Conte, M.; Vergara, A.; Sica, F. Structure and flexibility in cold-adapted iron superoxide dismutases: The case of the enzyme isolated from Pseudoalteromonas haloplanktis. J. Struct. Biol. 2010, 172, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, E.; Pasi, M.; Riccardi, L.; Sambi, I.; Fantucci, P.; De Gioia, L. Protein flexibility in psychrophilic and mesophilic trypsins. Evidence of evolutionary conservation of protein dynamics in trypsin-like serine-proteases. FEBS Lett. 2008, 582, 1008–1018. [Google Scholar] [CrossRef] [PubMed]
- Lonhienne, T.; Gerday, C.; Feller, G. Psychrophilic enzymes: Revisiting the thermodynamic parameters of activation may explain local flexibility. Biochim. Biophys. Acta 2000, 1543, 1–10. [Google Scholar] [CrossRef]
- Casanueva, A.; Tuffin, M.; Cary, C.; Cowan, D.A. Molecular adaptations to psychrophily: The impact of ‘omic’ technologies. Trends Microbiol. 2010, 18, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Ayala-del-Rio, H.L.; Chain, P.S.; Grzymski, J.J.; Ponder, M.A.; Ivanova, N.; Bergholz, P.W.; Di Bartolo, G.; Hauser, L.; Land, M.; Bakermans, C.; et al. The genome sequence of Psychrobacter arcticus 273–274, a psychroactive Siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl. Environ. Microbiol. 2010, 76, 2304–2312. [Google Scholar] [CrossRef] [PubMed]
- Methe, B.A.; Nelson, K.E.; Deming, J.W.; Momen, B.; Melamud, E.; Zhang, X.; Moult, J.; Madupu, R.; Nelson, W.C.; Dodson, R.J.; et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl. Acad. Sci. USA 2005, 102, 10913–10918. [Google Scholar] [CrossRef] [PubMed]
- Mavromatis, K.; Tsigos, I.; Tzanodaskalaki, M.; Kokkinidis, M.; Bouriotis, V. Exploring the role of a glycine cluster in cold adaptation of an alkaline phosphatase. Eur. J. Biochem. 2002, 269, 2330–2335. [Google Scholar] [CrossRef] [PubMed]
- Kulakova, L.; Galkin, A.; Nakayama, T.; Nishino, T.; Esaki, N. Cold-active esterase from Psychrobacter sp. Ant300: Gene cloning, characterization, and the effects of Gly-->Pro substitution near the active site on its catalytic activity and stability. Biochim. Biophys. Acta 2004, 1696, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, M.; Matsuzaki, M.; Niimiya, K.; Seino, J.; Sugahara, Y.; Kawakita, M. Role of proline residues in conferring thermostability on aqualysin I. J. Biochem. 2007, 141, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Vieille, C.; Zeikus, G.J. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 2001, 65, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Zanphorlin, L.M.; de Giuseppe, P.O.; Honorato, R.V.; Tonoli, C.C.; Fattori, J.; Crespim, E.; de Oliveira, P.S.; Ruller, R.; Murakami, M.T. Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7. Sci. Rep. 2016, 6, 23776. [Google Scholar] [CrossRef] [PubMed]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Microbiol. 2003, 1, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Georlette, D.; Damien, B.; Blaise, V.; Depiereux, E.; Uversky, V.N.; Gerday, C.; Feller, G. Structural and functional adaptations to extreme temperatures in psychrophilic, mesophilic, and thermophilic DNA ligases. J. Biol. Chem. 2003, 278, 37015–37023. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, S.; Marx, J.C.; Gerday, C.; Feller, G. Activity-stability relationships in extremophilic enzymes. J. Biol. Chem. 2003, 278, 7891–7896. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S. Defying the activity-stability trade-off in enzymes: Taking advantage of entropy to enhance activity and thermostability. Crit. Rev. Biotechnol. 2017, 37, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Truongvan, N.; Jang, S.H.; Lee, C. Flexibility and stability trade-off in active site of cold-adapted Pseudomonas mandelii esterase EstK. Biochemistry 2016, 55, 3542–3549. [Google Scholar] [CrossRef] [PubMed]
- Santiago, M.; Ramírez-Sarmiento, C.A.; Zamora, R.A.; Parra, L.P. Discovery, molecular mechanisms, and industrial applications of cold-active enzymes. Front. Microbiol. 2016, 7, 1408. [Google Scholar] [CrossRef] [PubMed]
- Kirk, O.; Christensen, M.W. Lipases from Candida antarctica: Unique biocatalysts from a unique origin. Org. Process Res. Dev. 2002, 6, 446–451. [Google Scholar] [CrossRef]
- Uppenberg, J.; Hansen, M.T.; Patkar, S.; Jones, T.A. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure 1994, 2, 293–308. [Google Scholar] [CrossRef]
- Anderson, E.M.; Larsson, K.M.; Kirk, O. One biocatalyst–many applications: The use of Candida antarctica B-lipase in organic yynthesis. Biocatal. Biotransform. 1998, 16, 181–204. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Charlton, T.; Ertan, H.; Mohd Omar, S.; Siddiqui, K.S.; Williams, T.J. Biotechnological uses of enzymes from psychrophiles. Microb. Biotechnol. 2011, 4, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Joseph, B.; Ramteke, P.W.; Thomas, G. Cold active microbial lipases: Some hot issues and recent developments. Biotechnol. Adv. 2008, 26, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Margesin, R.; Feller, G. Biotechnological applications of psychrophiles. Environ. Technol. 2010, 31, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento, F.; Peralta, R.; Blamey, J.M. Cold and hot extremozymes: Industrial relevance and current trends. Front. Bioeng. Biotechnol. 2015, 3, 148. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S. Some like it hot, some like it cold: Temperature dependent biotechnological applications and improvements in extremophilic enzymes. Biotechnol. Adv. 2015, 33, 1912–1922. [Google Scholar] [CrossRef] [PubMed]
- Lane, M.D.; Seelig, B. Advances in the directed evolution of proteins. Curr. Opin. Chem. Biol. 2014, 22, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Porter, J.L.; Rusli, R.A.; Ollis, D.L. Directed evolution of enzymes for industrial biocatalysis. Chembiochem 2016, 17, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Le, Q.A.T.; Kim, Y.H. Development of thermostable lipase B from Candida antarctica (CalB) through in silico design employing B-factor and RosettaDesign. Enzyme Microb. Technol. 2010, 47, 1–5. [Google Scholar] [CrossRef]
- Narasimhan, D.; Nance, M.R.; Gao, D.; Ko, M.C.; Macdonald, J.; Tamburi, P.; Yoon, D.; Landry, D.M.; Woods, J.H.; Zhan, C.G.; et al. Structural analysis of thermostabilizing mutations of cocaine esterase. Protein Eng. Des. Sel. 2010, 23, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Chow, K.M.; Hou, S.; Xue, L.; Chen, X.; Rodgers, D.W.; Zheng, F.; Zhan, C.G. Rational design, preparation, and characterization of a therapeutic enzyme mutant with improved stability and function for cocaine detoxification. ACS Chem. Biol. 2014, 9, 1764–1772. [Google Scholar] [CrossRef] [PubMed]
- Eijsink, V.G.H.; Gåseidnes, S.; Borchert, T.V.; van den Burg, B. Directed evolution of enzyme stability. Biomol. Eng. 2005, 22, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K.; Wintrode, P.L.; Grayling, R.A.; Rubingh, D.N.; Arnold, F.H. Directed evolution study of temperature adaptation in a psychrophilic enzyme. J. Mol. Biol. 2000, 297, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, M.; Wyss, M. Engineering proteins for thermostability: The use of sequence alignments versus rational design and directed evolution. Curr. Opin. Biotechnol. 2001, 12, 371–375. [Google Scholar] [CrossRef]
- Boyineni, J.; Kim, J.; Kang, B.S.; Lee, C.; Jang, S.H. Enhanced catalytic site thermal stability of cold-adapted esterase EstK by a W208Y mutation. Biochim. Biophys. Acta 2014, 1844, 1076–1082. [Google Scholar] [CrossRef] [PubMed]
- Perl, D.; Mueller, U.; Heinemann, U.; Schmid, F.X. Two exposed amino acid residues confer thermostability on a cold shock protein. Nat. Struct. Biol. 2000, 7, 380–383. [Google Scholar] [PubMed]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Homaei, A.A.; Sariri, R.; Vianello, F.; Stevanato, R. Enzyme immobilization: An update. J. Chem. Biol. 2013, 6, 185–205. [Google Scholar] [CrossRef] [PubMed]
- Cao, L. Carrier-bound Immobilized Enzymes: Principles, Applications and Design; Wiley-VCH: Weinheim, Germany, 2005. [Google Scholar]
- Guisan, J.M. Immobilization of Enzymes and Cells, 3rd ed.; Humana Press: New York, NY, USA, 2013. [Google Scholar]
- Hermanson, G.T. Bioconjugate Techniques, 3rd ed.; Elsevier/AP: London, UK, 2013. [Google Scholar]
- Cowan, D.A.; Fernandez-Lafuente, R. Enhancing the functional properties of thermophilic enzymes by chemical modification and immobilization. Enzyme Microb. Technol. 2011, 49, 326–346. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A.; van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Szczesna-Antczak, M.; Kaminska, J.; Florczak, T.; Turkiewicz, M. Cold-active yaeast lipases: Recent issues and future prospects. In Cold-Adapted Yeasts; Buzzini, P., Margesin, R., Eds.; Springer: Heidelberg, Germany, 2014; pp. 353–375. [Google Scholar]
- Brígida, A.I.S.; Amaral, P.F.F.; Coelho, M.A.Z.; Gonçalves, L.R.B. Lipase from Yarrowia lipolytica: Production, characterization and application as an industrial biocatalyst. J. Mol. Catal. B Enzym. 2014, 101, 148–158. [Google Scholar] [CrossRef]
- Idris, A.; Bukhari, A. Immobilized Candida antarctica lipase B: Hydration, stripping off and application in ring opening polyester synthesis. Biotechnol. Adv. 2012, 30, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Hu, J.; Miller, E.M.; Xie, W.; Cai, M.; Gross, R.A. Candida antarctica lipase B chemically immobilized on epoxy-activated micro- and nanobeads: Catalysts for polyester synthesis. Biomacromolecules 2008, 9, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Culsum, U.; Kumar, A.; Gao, H.; Hu, N. Immobilization of a novel cold active esterase onto Fe3O4~cellulose nano-composite enhances catalytic properties. Int. J. Biol. Macromol. 2016, 87, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.; Geueke, B.; Delgado, O.; Coleman, J.; Hatti-Kaul, R. Beta-galactosidase from a cold-adapted bacterium: Purification, characterization and application for lactose hydrolysis. Appl. Microbiol. Biotechnol. 2002, 58, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Fresco-Taboada, A.; Serra, I.; Fernandez-Lucas, J.; Acebal, C.; Arroyo, M.; Terreni, M.; de la Mata, I. Nucleoside 2′-deoxyribosyltransferase from psychrophilic bacterium Bacillus psychrosaccharolyticus-preparation of an immobilized biocatalyst for the enzymatic synthesis of therapeutic nucleosides. Molecules 2014, 19, 11231–11249. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, A.; Bhattacharyya, T.; Dasgupta, A.K.; Chakrabarti, K. Nanotechnology based activation-immobilization of psychrophilic pectate lyase: A novel approach towards enzyme stabilization and enhanced activity. J. Mol. Catal. B Enzym. 2015, 119, 54–63. [Google Scholar] [CrossRef]
- Mukhopadhyay, A.; Dasgupta, A.K.; Chakrabarti, K. Enhanced functionality and stabilization of a cold active laccase using nanotechnology based activation-immobilization. Bioresour. Technol. 2015, 179, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Rajaei, S.; Noghabi, K.A.; Sadeghizadeh, M.; Zahiri, H.S. Characterization of a pH and detergent-tolerant, cold-adapted type I pullulanase from Exiguobacterium sp. SH3. Extremophiles 2015, 19, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
- Vaghari, H.; Jafarizadeh-Malmiri, H.; Mohammadlou, M.; Berenjian, A.; Anarjan, N.; Jafari, N.; Nasiri, S. Application of magnetic nanoparticles in smart enzyme immobilization. Biotechnol. Lett. 2016, 38, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Pavlidis, I.V.; Vorhaben, T.; Tsoufis, T.; Rudolf, P.; Bornscheuer, U.T.; Gournis, D.; Stamatis, H. Development of effective nanobiocatalytic systems through the immobilization of hydrolases on functionalized carbon-based nanomaterials. Bioresour. Technol. 2012, 115, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, F.; Yang, H.; Huang, X.; Liu, H.; Guo, S. Graphene oxide as a matrix for enzyme immobilization. Langmuir 2010, 26, 6083–6085. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, C.; Guo, S.; Zhang, J. Interactions of graphene and graphene oxide with proteins and peptides. Nanotechnol. Rev. 2013, 2, 27–45. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Huang, X.; Wu, H.; Guo, S. Assembly of graphene oxide-enzyme conjugates through hydrophobic interaction. Small 2012, 8, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Jeong, H.K.; Han, J.; Chung, H.S.; Jang, S.H.; Lee, C. Increased thermal stability of cold-adapted esterase at ambient temperatures by immobilization on graphene oxide. Bioresour. Technol. 2013, 148, 620–623. [Google Scholar] [CrossRef] [PubMed]
- Makowski, K.; Bialkowska, A.; Szczesna-Antczak, M.; Kalinowska, H.; Kur, J.; Cieslinski, H.; Turkiewicz, M. Immobilized preparation of cold-adapted and halotolerant Antarctic beta-galactosidase as a highly stable catalyst in lactose hydrolysis. FEMS Microbiol. Ecol. 2007, 59, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.B.; Gao, C.; Zheng, Z.; Liu, F.M.; Zang, J.Y.; Miao, J.L. Immobilization of cold-active cellulase from Antarctic bacterium and its use for kelp cellulose thanol fermentation. BioResources 2015, 10, 1757–1772. [Google Scholar]
- Fernandez-Lafuente, R.; Armisén, P.; Sabuquillo, P.; Fernández-Lorente, G.; Guisán, J.M. Immobilization of lipases by selective adsorption on hydrophobic supports. Chem. Phys. Lipids 1998, 93, 185–197. [Google Scholar] [CrossRef]
- Issa, B.; Obaidat, I.M.; Albiss, B.A.; Haik, Y. Magnetic nanoparticles: Surface effects and properties related to biomedicine applications. Int. J. Mol. Sci. 2013, 14, 21266–21305. [Google Scholar] [CrossRef] [PubMed]
- Fresco-Taboada, A.; Serra, I.; Arroyo, M.; Fernández-Lucas, J.; de la Mata, I.; Terreni, M. Development of an immobilized biocatalyst based on Bacillus psychrosaccharolyticus NDT for the preparative synthesis of trifluridine and decytabine. Catal. Today 2016, 259, 197–204. [Google Scholar] [CrossRef]
- Fernandez-Lafuente, R. Stabilization of multimeric enzymes: Strategies to prevent subunit dissociation. Enzyme Microb. Technol. 2009, 45, 405–418. [Google Scholar] [CrossRef]
- Sheldon, R.A. Cross-linked enzyme aggregates (CLEAs): Stable and recyclable biocatalysts. Biochem. Soc. Trans. 2007, 35, 1583–1587. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A. Cross-linked enzyme aggregates as industrial biocatalysts. Org. Process Res. Dev. 2011, 15, 213–223. [Google Scholar] [CrossRef]
- Stepankova, V.; Bidmanova, S.; Koudelakova, T.; Prokop, Z.; Chaloupkova, R.; Damborsky, J. Strategies for stabilization of enzymes in organic solvents. ACS Catal. 2013, 3, 2823–2836. [Google Scholar] [CrossRef]
- Adlercreutz, P. Immobilisation and application of lipases in organic media. Chem. Soc. Rev. 2013, 42, 6406–6436. [Google Scholar] [CrossRef] [PubMed]
- Gorman, L.A.; Dordick, J.S. Organic solvents strip water off enzymes. Biotechnol. Bioeng. 1992, 39, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Zaks, A.; Klibanov, A.M. Enzyme-catalyzed processes in organic solvents. Proc. Natl. Acad. Sci. USA 1985, 82, 3192–3196. [Google Scholar] [CrossRef] [PubMed]
- Sellek, G.A.; Chaudhuri, J.B. Biocatalysis in organic media using enzymes from extremophiles. Enzyme Microb. Technol. 1999, 25, 471–482. [Google Scholar] [CrossRef]
- Ogino, H.; Miyamoto, K.; Ishikawa, H. Organic-solvent-tolerant bacterium which secretes organic-solvent-stable lipolytic enzyme. Appl. Environ. Microbiol. 1994, 60, 3884–3886. [Google Scholar] [PubMed]
- Gupta, A.; Khare, S.K. Enzymes from solvent-tolerant microbes: Useful biocatalysts for non-aqueous enzymology. Crit. Rev. Biotechnol. 2009, 29, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Doukyu, N.; Ogino, H. Organic solvent-tolerant enzymes. Biochem. Eng. J. 2010, 48, 270–282. [Google Scholar] [CrossRef]
- Song, J.K.; Rhee, J.S. Enhancement of stability and activity of phospholipase A(1) in organic solvents by directed evolution. Biochim. Biophys. Acta 2001, 1547, 370–378. [Google Scholar] [CrossRef]
- Reetz, M.T. Changing the enantioselectivity of enzymes by directed evolution. Methods Enzymol. 2004, 388, 238–256. [Google Scholar] [PubMed]
- Dror, A.; Shemesh, E.; Dayan, N.; Fishman, A. Protein engineering by random mutagenesis and structure-guided consensus of Geobacillus stearothermophilus Lipase T6 for enhanced stability in methanol. Appl. Environ. Microbiol. 2014, 80, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Kawata, T.; Ogino, H. Enhancement of the organic solvent-stability of the LST-03 lipase by directed evolution. Biotechnol. Prog. 2009, 25, 1605–1611. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, Y.; Shao, Y.; Chen, W.; Chen, F.; Li, M. Cloning, expression and characterization of a novel cold-active and organic solvent-tolerant esterase from Monascus ruber M7. Extremophiles 2016, 20, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhang, X.; Wei, L.; Wu, G.; Kumar, A.; Mao, T.; Liu, Z. A cold-adapted, solvent and salt tolerant esterase from marine bacterium Psychrobacter pacificensis. Int. J. Biol. Macromol. 2015, 81, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jang, S.H.; Lee, C. An organic solvent-tolerant alkaline lipase from cold-adapted Pseudomonas mandelii: Cloning, expression, and characterization. Biosci. Biotechnol. Biochem. 2013, 77, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Ganasen, M.; Yaacob, N.; Rahman, R.N.Z.R.A.; Leow, A.T.C.; Basri, M.; Salleh, A.B.; Ali, M.S.M. Cold-adapted organic solvent tolerant alkalophilic family I.3 lipase from an Antarctic Pseudomonas. Int. J. Biol. Macromolec. 2016, 92, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, L.-R.; Xu, G.; Wu, J.-P. Screening, purification and characterization of a novel cold-active and organic solvent-tolerant lipase from Stenotrophomonas maltophilia CGMCC 4254. Bioresour. Technol. 2013, 148, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Dachuri, V.; Boyineni, J.; Choi, S.; Chung, H.-S.; Jang, S.-H.; Lee, C. Organic solvent-tolerant, cold-adapted lipases PML and LipS exhibit increased conformational flexibility in polar organic solvents. J. Mol. Catal. B Enzym. 2016, 131, 73–78. [Google Scholar] [CrossRef]
- Kamal, M.Z.; Yedavalli, P.; Deshmukh, M.V.; Rao, N.M. Lipase in aqueous-polar organic solvents: Activity, structure, and stability. Protein Sci. 2013, 22, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, P.A.; Ringe, D.; Klibanov, A.M. X-ray crystal structure of cross-linked subtilisin Carlsberg in water vs. acetonitrile. Biochem. Biophys. Res. Commun. 1994, 198, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Allen, K.N.; Bellamacina, C.R.; Ding, X.; Jeffery, C.J.; Mattos, C.; Petsko, G.A.; Ringe, D. An experimental approach to mapping the binding surfaces of crystalline proteins. J. Phys. Chem. 1996, 100, 2605–2611. [Google Scholar] [CrossRef]
| Enzyme Name | Species | Support | Chemistry | Comments | Reference | |
|---|---|---|---|---|---|---|
| Adsorption | Esterase (EstH) | Zunongwangia sp. | Fe3O4-cellulose | Hydrogen bonding | 48% activity after 30 min at 50 °C | [61] |
| β-Galactosidase | Pseudoalteromonas sp. | DEAE-Sepharose | Ionic interaction | 87%–89% storage stability after 1 week at 4 °C | [62] | |
| Nucleoside 2′-deoxyribosyltransferase | Bacillus psychrosaccharolyticus | PEI-coated agarose | Ionic interaction | Unstable; lost activity within 2 h | [63] | |
| Covalent binding | Pullulanase | Exiguobacterium sp. | Epoxy-functionalized silica | Epoxyl group | Maintained thermal stability at 50 °C | [66] |
| Esterase (EstK) | Pseudomonas mandelii | Graphene oxide | Sulfo-NHS and EDC | Enhanced thermal stability at 40 °C; catalytic efficiency reduced to 40% of free enzyme | [72] | |
| β-Galactosidase | Pseudoalteromonas sp. | Epoxy-activated Sepharose | Epoxyl group | 87%–89% storage stability after 1 week at 4 °C | [62] | |
| β-Galactosidase | Pseudoalteromonas sp. | PEI-coated Sepharose | Glutaraldehyde | 98% storage stability after 1 week at 4 °C | [62] | |
| β-Galactosidase | Pseudoalteromonas sp. | Glutaraldehyde-treated chitosan | Glutaraldehyde | Enhanced themal stability at 50 °C; longer shelf life over 12 months | [73] | |
| Nucleoside 2′-deoxyribosyltransferase | Bacillus psychrosaccharolyticus | PEI-coated agarose | Aldehyde-dextran | Operational stability at 37 °C with 75% activity after 30 cycles | [63] | |
| Entrapment | Cellulase | Pseudoalteromonas sp. | Sodium alginate beads | Glutaraldehyde cross-linking entrapment | 58% activity after seven cycles | [74] |
| Pectate lyase | Bacillus subtilis | Lipid-functionalized SWCNT | - | Thermal stability at 4–80 °C | [64] | |
| Laccase | Pseudomonas putida | Lipid-functionalized SWCNT | - | Thermal stability at 4–80 °C | [65] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, C.; Jang, S.-H.; Chung, H.-S. Improving the Stability of Cold-Adapted Enzymes by Immobilization. Catalysts 2017, 7, 112. https://doi.org/10.3390/catal7040112
Lee C, Jang S-H, Chung H-S. Improving the Stability of Cold-Adapted Enzymes by Immobilization. Catalysts. 2017; 7(4):112. https://doi.org/10.3390/catal7040112
Chicago/Turabian StyleLee, ChangWoo, Sei-Heon Jang, and Hye-Shin Chung. 2017. "Improving the Stability of Cold-Adapted Enzymes by Immobilization" Catalysts 7, no. 4: 112. https://doi.org/10.3390/catal7040112





