Biocatalytic Oxidations of Substrates through Soluble Methane Monooxygenase from Methylosinus sporium 5
Abstract
:1. Introduction
2. Results and Discussion
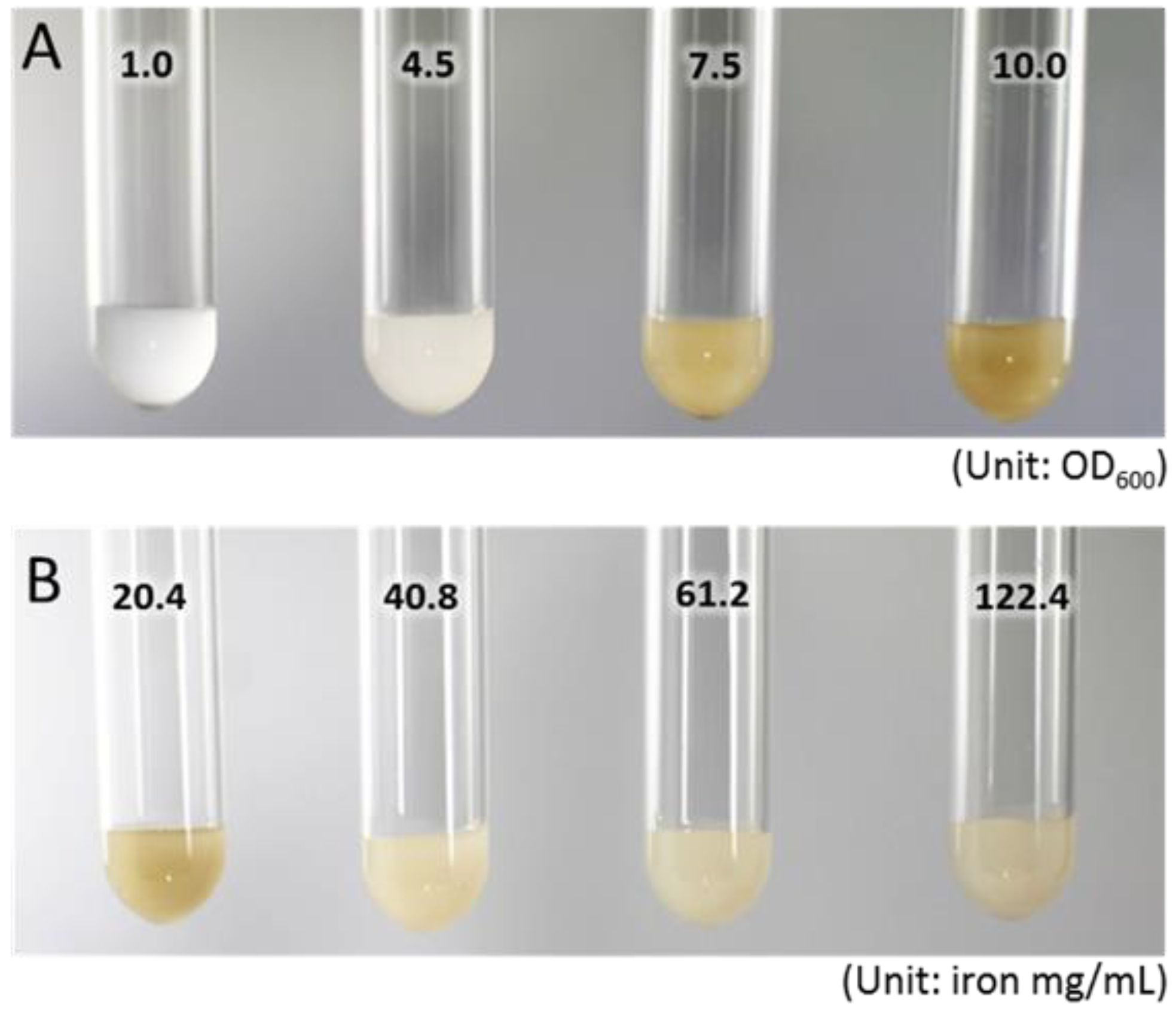
2.1. Cultures of M. sporium 5 and Iron Concentration in NMS Medium
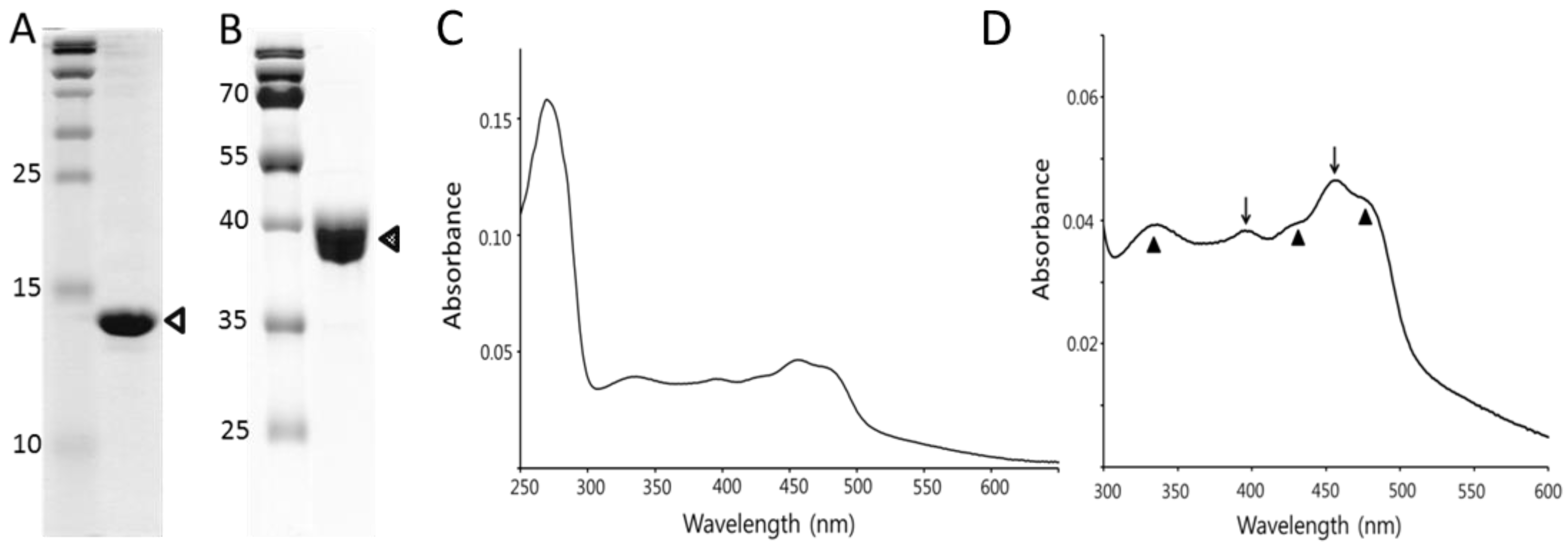
2.2. Expression and Purification of Hydroxylase from M. sporium 5
2.3. Expression and Purification of MMOB and MMOR from E. coli
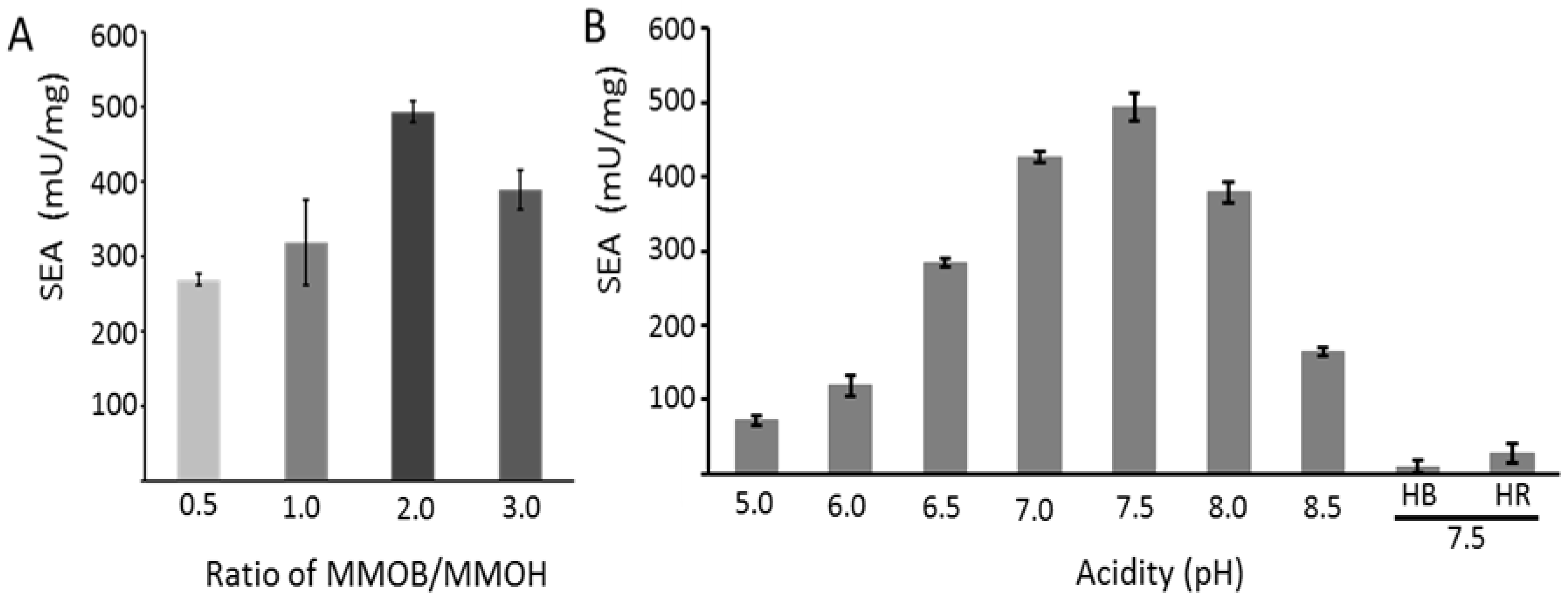
2.4. Activities of Essential Enzymes from M. sporium 5
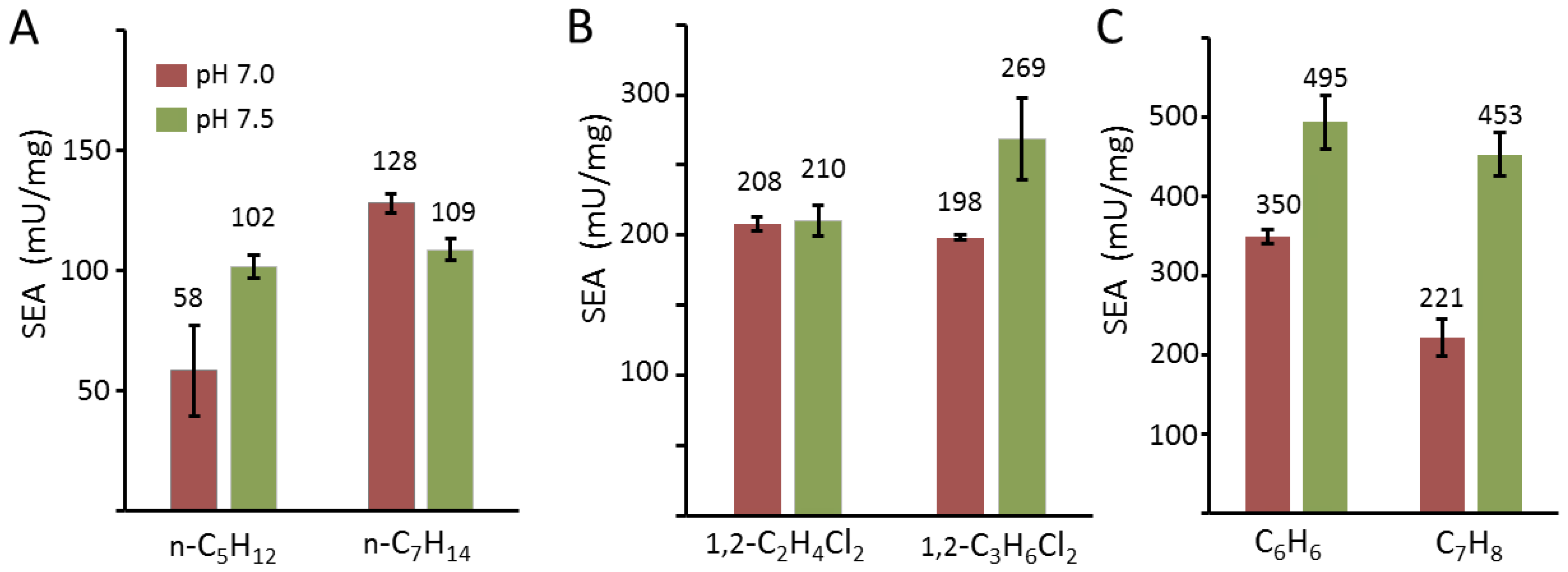
2.5. Oxidation of Alkane, Halogen, and Aromatic Compounds
3. Material and Methods
3.1. General Materials and Chemicals
3.2. Culturing M. sporium 5 and MMOH Purification
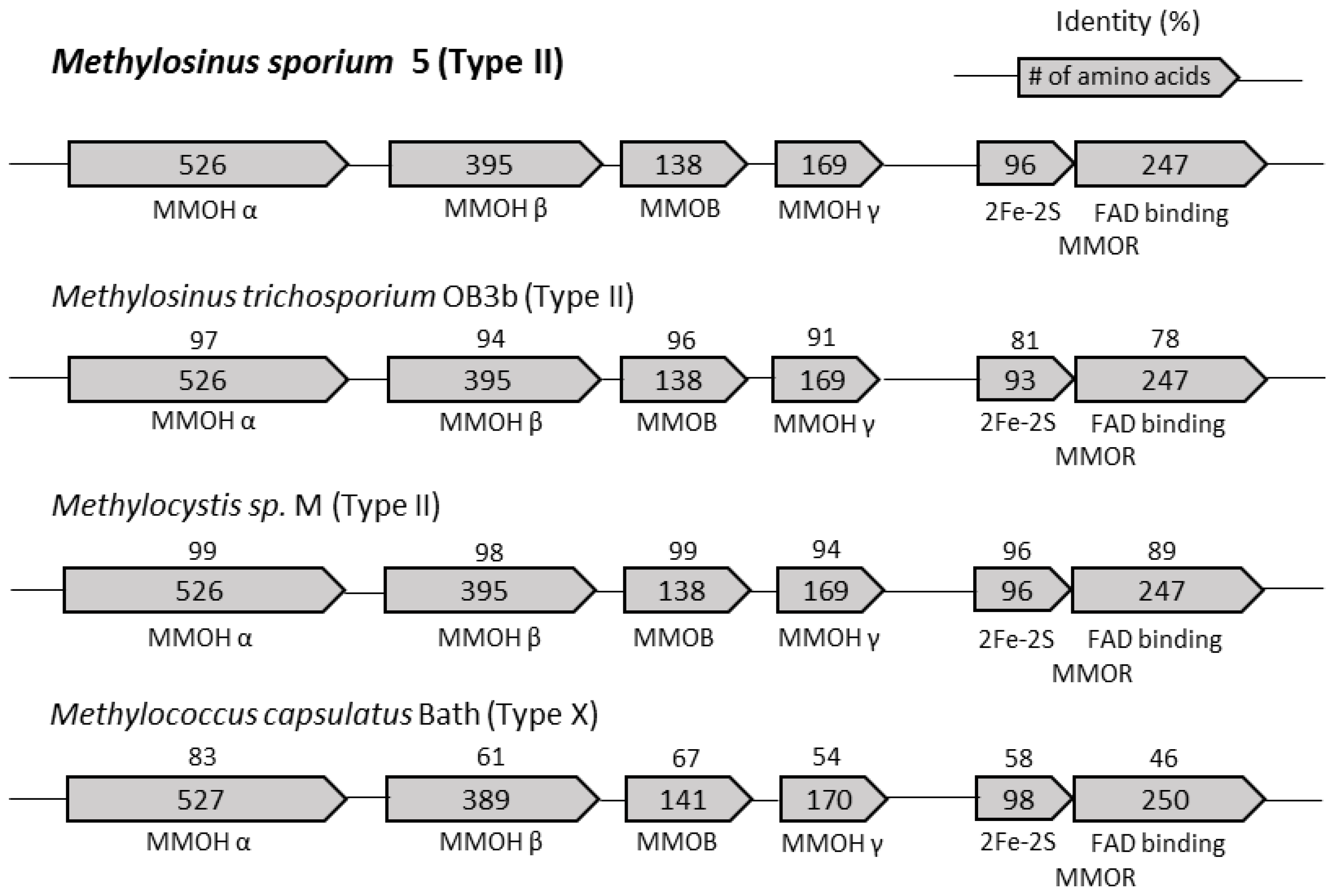
3.3. Construction of mmoB and mmoC from M. sporium 5 and M. species M
3.4. Expression and Purification of MMOB
3.5. Expression and Purification of MMOR and MMOR-FAD
3.6. Measurement of SEA with Various Substrates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [PubMed]
- Lawton, T.J.; Rosenzweig, A.C. Methane-oxidizing enzymes: An upstream problem in biological gas-to-liquids conversion. J. Am. Chem. Soc. 2016, 138, 9327–9340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Semrau, J.D.; DiSpirito, A.A.; Yoon, S. Methanotrophs and copper. FEMS. Microbiol. Rev. 2010, 34, 496–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieberman, R.L.; Rosenzweig, A.C. Biological methane oxidation: Regulation, biochemistry, and active site structure of particulate methane monooxygenase. Crit. Rev. Biochem. Mol. 2004, 39, 147–164. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.P.; Dickinson, D.; Chase, H.A. Methanotrophs, Methylosinus trichosporium OB3b, sMMO, and their application to bioremediation. Crit. Rev. Microbiol. 1998, 24, 335–373. [Google Scholar] [CrossRef] [PubMed]
- Leahy, J.G.; Batchelor, P.J.; Morcomb, S.M. Evolution of the soluble diiron monooxygenases. FEMS Microbiol. Rev. 2003, 27, 449–479. [Google Scholar] [CrossRef] [Green Version]
- Warmuzinski, K. Harnessing methane emissions from coal mining. Process Saf. Environ. 2008, 86, 315–320. [Google Scholar] [CrossRef]
- Haynes, C.A.; Gonzalez, R. Rethinking biological activation of methane and conversion to liquid fuels. Nat. Chem. Biol. 2014, 10, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Sirajuddin, S.; Barupala, D.; Helling, S.; Marcus, K.; Stemmler, T.L.; Rosenzweig, A.C. Effects of zinc on particulate methane monooxygenase activity and structure. J. Biol. Chem. 2014, 289, 21782–21794. [Google Scholar] [CrossRef] [PubMed]
- Sirajuddin, S.; Rosenzweig, A.C. Enzymatic oxidation of methane. Biochemistry 2015, 54, 2283–2294. [Google Scholar] [CrossRef] [PubMed]
- Wang, V.C.C.; Maji, S.; Chen, P.R.Y.; Lee, H.K.; Yu, S.S.F.; Chan, S.I. Alkane oxidation: Methane monooxygenases, related enzymes, and their biomimetics. Chem. Rev. 2017, 117, 8574–8621. [Google Scholar] [CrossRef] [PubMed]
- Hakemian, A.S.; Rosenzweig, A.C. The biochemistry of methane oxidation. Annu. Rev. Biochem. 2007, 76, 223–241. [Google Scholar] [CrossRef] [PubMed]
- Gassner, G.T.; Lippard, S.J. Component interactions in the soluble methane monooxygenase system from Methylococcus capsulatus (Bath). Biochemistry 1999, 38, 12768–12785. [Google Scholar] [CrossRef] [PubMed]
- Merkx, M.; Kopp, D.A.; Sazinsky, M.H.; Blazyk, J.L.; Muller, J.; Lippard, S.J. Dioxygen activation and methane hydroxylation by soluble methane monooxygenase: A tale of two irons and three proteins. Angew. Chem. Int. Ed. 2001, 40, 2782–2807. [Google Scholar] [CrossRef]
- Rosenzweig, A.C.; Nordlund, P.; Takahara, P.M.; Frederick, C.A.; Lippard, S.J. Geometry of the soluble methane monooxygenase catalytic diiron center in two oxidation states. Chem. Biol. 1995, 2, 409–418. [Google Scholar] [CrossRef]
- Sazinsky, M.H.; Lippard, S.J. Correlating structure with function in bacterial multicomponent Monooxygenases and related diiron proteins. Acct. Chem. Res. 2006, 39, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Fox, B.G.; Froland, W.A.; Dege, J.E.; Lipscomb, J.D. Methane monooxygenase from Methylosinus trichosporium OB3b. J. Biol. Chem. 1989, 264, 10023–10033. [Google Scholar] [PubMed]
- Hanson, R.S.; Wattenberg, E.V. Ecology of methylotrophic bacteria. Biotechnology 1991, 18, 325–348. [Google Scholar] [PubMed]
- Banerjee, R.; Proshlyakov, Y.; Lipscomb, J.D.; Proshlyakov, D.A. Structure of the key species in the enzymatic oxidation of methane to methanol. Nature 2015, 518, 431–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elango, N.; Radhakrishnan, R.; Froland, W.A.; Wallar, B.J.; Earhart, C.A.; Lipscomb, J.D.; Ohlendorf, D.H. Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b. Protein Sci. 1997, 6, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, A.C.; Frederick, C.A.; Lippard, S.J.; Nordlund, P. Crystal structure of a bacterial nonheme iron hydroxylase that catalyzes the biological oxidation of methane. Nature 1993, 366, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Froland, W.A.; Andersson, K.K.; Lee, S.K.; Liu, Y.; Lipscomb, J.D. Methane monooxygenase component B and reductase alter the regioselectivity of the hydroxylase component-catalyzed reactions. A novel role for protein-protein interactions in an oxygenase mechanism. J. Biol. Chem. 1992, 267, 17588–17597. [Google Scholar] [PubMed]
- Lee, S.J.; McCormick, M.S.; Lippard, S.J.; Cho, U.S. Control of substrate access to the active site in methane monooxygenase. Nature 2013, 494, 380–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, H.; Scanlan, J.; Dumont, M.G.; Murrell, J.C. Duplication of the mmoX gene in Methylosinus sporium: Cloning, sequencing and mutational analysis. Microbiology 2006, 152, 2931–2942. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Uchiyama, H.; Yagi, O.; Nakahara, T. Purification and properties of a aoluble methane monooxygenase from Methylocystis sp. M. Biosci. Biotechnol. Biochem. 1992, 56, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Pilkington, S.J.; Dalton, H. Purification and characterization of the soluble methane monooxygenase from Methylosinus sporium 5 demonstrates the highly conserved nature of this enzyme in methanotrophs. FEMS Microbiol. Lett. 1991, 78, 103–108. [Google Scholar] [CrossRef]
- Whittenbury, R.; Phillips, K.C.; Wilkinson, J.F. Enrichment, isolation and some properties of methane utilizing bacteria. J. Gen. Microbiol. 1970, 61, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Blazyk, J.L.; Gassner, G.T.; Lippard, S.J. Intermolecular electron transfer reactions in soluble methane monooxygenase: A role for hysteresis in protein function. J. Am. Chem. Soc. 2005, 127, 17364–17376. [Google Scholar] [CrossRef] [PubMed]
- Blazyk, J.L.; Lippard, S.J. Expression and characterization of ferredoxin and flavin adenine dinucleotide binding domains of the reductase component of soluble methane monooxygenase from Methylococcus capsulatus (Bath). Biochemistry 2002, 41, 15780–15794. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Nesheim, J.C.; Paulsen, K.E.; Stankovich, M.T.; Lipscomb, J.D. Roles of the methane monooxygenase reductase component in the regulation of catalysis. Biochemistry 1997, 36, 5223–5233. [Google Scholar] [CrossRef] [PubMed]
- Tinberg, C.E.; Lippard, S.J. Revisiting the mechanism of dioxygen activation in soluble methane monooxygenase from M. capsulatus (Bath): Evidence for a multi-step, proton-dependent reaction pathway. Biochemistry 2009, 48, 12145–12158. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Iacob, R.E.; Luoh, R.P.; Engen, J.R.; Lippard, S.J. Electron transfer control in soluble methane monooxygenase. J. Am. Chem. Soc. 2014, 136, 9754–9762. [Google Scholar] [CrossRef] [PubMed]
- Matsen, J.B.; Yang, S.; Stein, L.Y.; Beck, D.; Kalyuzhnaya, M.G. Global molecular analyses of methane metabolism in methanotrophic alphaproteobacterium, Methylosinus trichosporium OB3b. Part I: Transcriptomic study. Front. Microbiol. 2013, 4, 40. [Google Scholar] [CrossRef] [PubMed]
- Semrau, J.D.; Jagadevan, S.; DiSpirito, A.A.; Khalifa, A.; Scanlan, J.; Bergman, B.H.; Freemeier, B.C.; Baral, B.S.; Bandow, N.L.; Vorobev, A.; et al. Methanobactin and MmoD work in concert to act as the ‘copper-switch’ in methanotrophs. Environ. Microbiol. 2013, 15, 3077–3086. [Google Scholar] [CrossRef] [PubMed]
- Grosse, S.; Laramee, L.; Wendlandt, K.D.; McDonald, I.R.; Miguez, C.B.; Kleber, H.P. Purification and characterization of the soluble methane monooxygenase of the type II methanotrophic bacterium Methylocystis sp strain WI 14. Appl. Environ. Microbiol. 1999, 65, 3929–3935. [Google Scholar] [PubMed]
- Merkx, M.; Lippard, S.J. Why OrfY? Characterization of MMOD, a long overlooked component of the soluble methane monooxygenase from Methylococcus capsulatus (Bath). J. Biol. Chem. 2002, 277, 5858–5865. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Liang, A.D.; Lippard, S.J. Coupling oxygen consumption with hydrocarbon oxidation in bacterial multicomponent monooxygenases. Acct. Chem. Res. 2015, 48, 2632–2639. [Google Scholar] [CrossRef] [PubMed]
- Shaofeng, H.; Shuben, L.; Jiayin, X.; Jianzhong, N.; Chungu, X.; Haidong, T.; Wei, T. Purification and biochemical characterization of soluble methane monooxygenase hydroxylase from Methylosinus trichosporium IMV 3011. Biosci. Biotechnol. Biochem. 2007, 71, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, A.C.; Brandstetter, H.; Whittington, D.A.; Nordlund, P.; Lippard, S.J.; Frederick, C.A. Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): Implications for substrate gating and component interactions. Proteins Struct. Funct. Genet. 1997, 29, 141–152. [Google Scholar] [CrossRef]
- Atkin, C.L.; Thelande, L.; Reichard, P.; Lang, G. Iron and free-radical in ribonucleotide reductase—Exchange of iron and Mössbauer spectroscopy of protein B2 subunit of Escherichia coli Enzyme. J. Biol. Chem. 1973, 248, 7464–7472. [Google Scholar] [PubMed]
- Brandstetter, H.; Whittington, D.A.; Lippard, S.J.; Frederick, C.A. Mutational and structural analyses of the regulatory protein B of soluble methane monooxygenase from Methylococcus capsulatus (Bath). Chem. Biol. 1999, 6, 441–449. [Google Scholar] [CrossRef]
- Colby, J.; Stirling, D.I.; Dalton, H. The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath): Its ability to oxygenate n-alkanes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem. J. 1977, 165, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Walters, K.J.; Gassner, G.T.; Lippard, S.J.; Wagner, G. Structure of the soluble methane monooxygenase regulatory protein B. Proc. Natl. Acad. Sci. USA 1999, 96, 7877–7882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lund, J.; Dalton, H. Further Characterization of the FAD and Fe2S2 redox centers of component C, the NADH—Acceptor reductase of the soluble methane monooxygenase of Methylococcus capsulatus (Bath). Eur. J. Biochem. 1985, 147, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Lund, J.; Woodland, M.P.; Dalton, H. Electron transfer reactions in the soluble methane monooxygenase of Methylococcus capsulatus (Bath). Eur. J. Biochem. 1985, 147, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Kopp, D.A.; Gassner, G.T.; Blazyk, J.L.; Lippard, S.J. Electron-transfer reactions of the reductase component of soluble methane monooxygenase from Methylococcus capsulatus (Bath). Biochemistry 2001, 40, 14932–14941. [Google Scholar] [CrossRef] [PubMed]
- Fox, B.G.; Liu, Y.; Dege, J.E.; Lipscomb, J.D. Complex formation between the protein components of methane monooxygenase from Methylosinus trichosporium Ob3b. Identification of sites of component interaction. J. Biol. Chem. 1991, 266, 540–550. [Google Scholar] [PubMed]
- Liu, K.E.; Lippard, S.J. Redox properties of the hydroxylase component of methane monooxygenase from Methylococcus capsulatus (Bath). Effects of protein B, reductase, and substrate. J. Biol. Chem. 1991, 266, 12836–12839. [Google Scholar] [PubMed]
- Green, J.; Prior, S.D.; Dalton, H. Copper ions as inhibitors of protein C of soluble methane monooxygenase of Methylococcus capsulatus (Bath). Eur. J. Biochem. 1985, 153, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Lipscomb, J.D. Biochemistry of the soluble methane monooxygenase. Annu. Rev. Microbiol. 1994, 48, 371–399. [Google Scholar] [CrossRef] [PubMed]
- Murrell, J.C.; Gilbert, B.; McDonald, I.R. Molecular biology and regulation of methane monooxygenase. Arch. Microbiol. 2000, 173, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Pulver, S.; Froland, W.A.; Fox, B.G.; Lipscomb, J.D.; Solomon, E.I. Spectroscopic studies of the coupled binuclear non-heme iron active site in the fully reduced hydroxylase component of methane monooxygenase: Comparison to deoxy and deoxy-azide hemerythrin. J. Am. Chem. Soc. 1994, 116, 4529–4529. [Google Scholar] [CrossRef]
- McCormick, M.S.; Lippard, S.J. Analysis of substrate access to active sites in bacterial multicomponent monooxygenase hydroxylases: X-ray crystal structure of xenon-pressurized phenol hydroxylase from Pseudomonas sp. OX1. Biochemistry 2011, 50, 11058–11069. [Google Scholar] [CrossRef] [PubMed]
- Stirling, D.I.; Dalton, H. Properties of the methane monooxygenase from extracts of Methylosinus trichosporium OB3b and evidence for its similarity to the enzyme from Methylococcus capsulatus (Bath). Eur. J. Biochem. 1979, 96, 205–212. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, Y.R.; Yoo, H.S.; Song, M.Y.; Lee, D.-H.; Lee, S.J. Biocatalytic Oxidations of Substrates through Soluble Methane Monooxygenase from Methylosinus sporium 5. Catalysts 2018, 8, 582. https://doi.org/10.3390/catal8120582
Park YR, Yoo HS, Song MY, Lee D-H, Lee SJ. Biocatalytic Oxidations of Substrates through Soluble Methane Monooxygenase from Methylosinus sporium 5. Catalysts. 2018; 8(12):582. https://doi.org/10.3390/catal8120582
Chicago/Turabian StylePark, Yeo Reum, Hee Seon Yoo, Min Young Song, Dong-Heon Lee, and Seung Jae Lee. 2018. "Biocatalytic Oxidations of Substrates through Soluble Methane Monooxygenase from Methylosinus sporium 5" Catalysts 8, no. 12: 582. https://doi.org/10.3390/catal8120582



