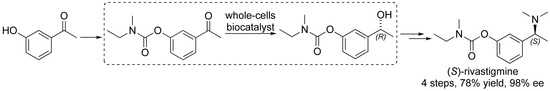
Whole-Cell Biocatalyst for Chemoenzymatic Total Synthesis of Rivastigmine
Abstract
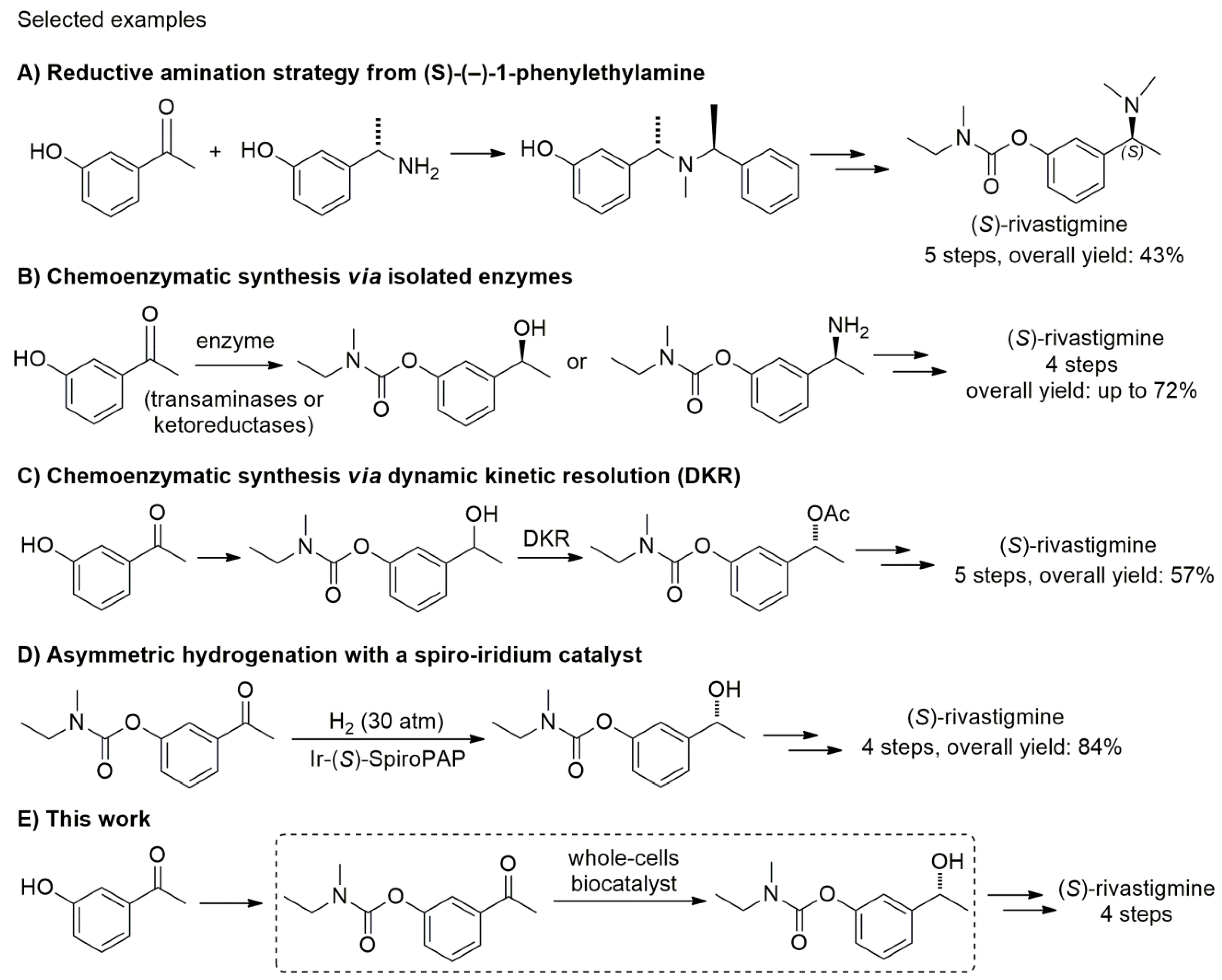
:1. Introduction
2. Results and Discussion
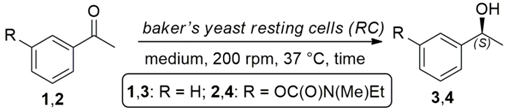
2.1. Bioreduction of Prochiral Ketones 1 and 2 with Baker’s Yeast RC
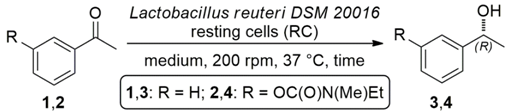
2.2. Bioreduction of Ketones 1 and 2 with Lactobacillus reuteri DSM 20016 Whole Cells
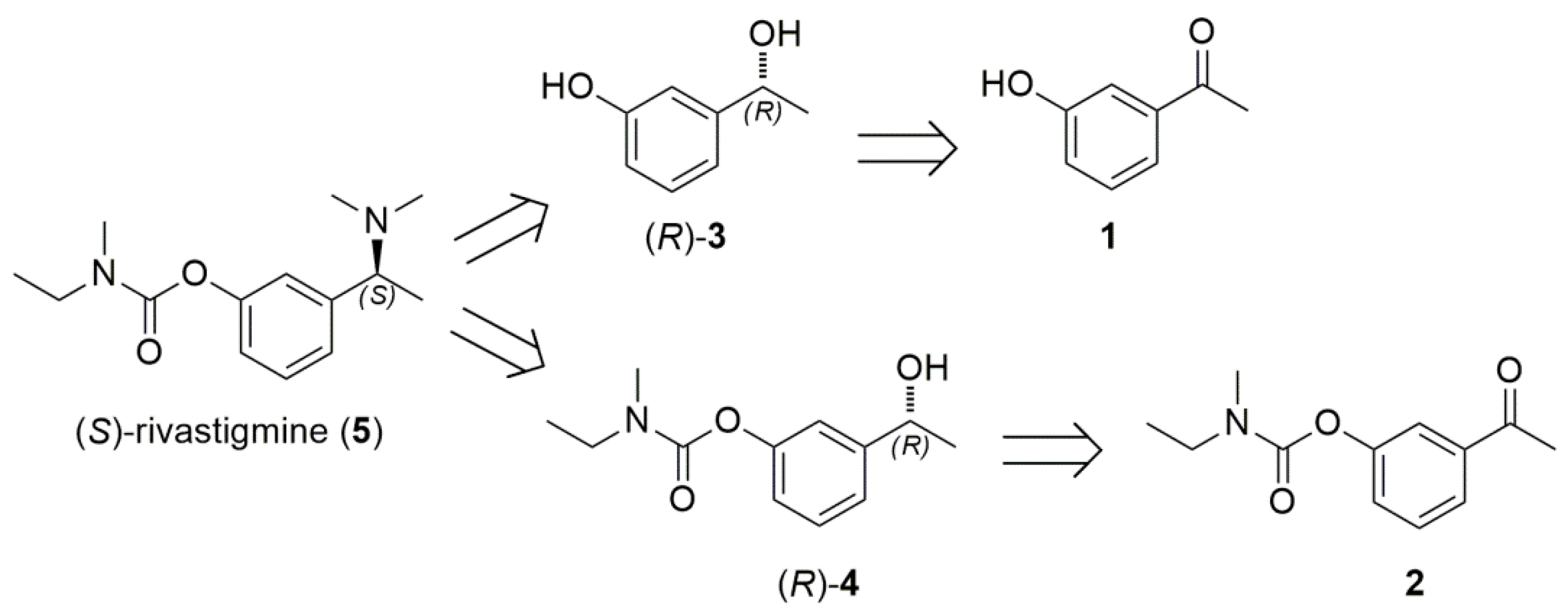
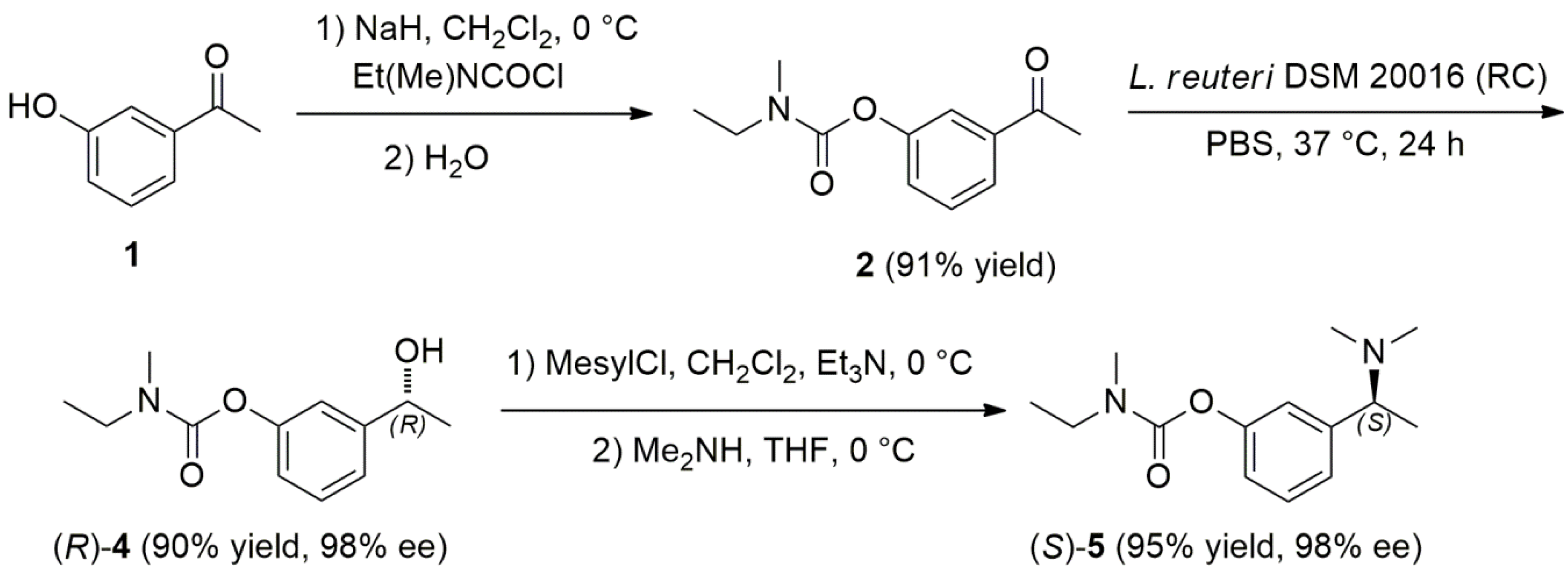
2.3. Chemoenzymatic Total Synthesis of (S)-Rivastigmine
3. Materials and Methods
3.1. General Methods
3.2. Microorganisms and Media
3.3. Blank Experiments
3.4. Bioreduction of Acetophenones 1,2 by Using Baker’s Yeat Cells in Acqueous DESs Mixtures (the General Procedure Described Refers to a ChCl/Gly (1:2 mol/mol) + 10 w% Water Mixture)
3.5. Bioreduction of Ketone 2 by Using Lactobacillus reuteri Growing Cells (Table 2, Entry 2)
3.6. Bioreduction of Acetophenones 1,2 by Using Lactobacillus reuteri Resting Cells: General Procedure (Table 2)
3.7. Bioreduction of Acetophenones 1,2 by Using Lactobacillus reuteri Resting Cells in Bioreactor: General Batch Procedure (Table 2, Entries 7,8)
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Faber, K. Biotransformations. In Organic Chemistry: A Textbook, 6th ed.; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Simon, R.C.; Mutti, F.G.; Kroutil, W. Biocatalytic synthesis of enantiopure building blocks for pharmaceuticals. Drug Discov. Today Technol. 2013, 10, e37–e44. [Google Scholar] [CrossRef] [PubMed]
- Rozzell, D. Tabular Survey of Available Enzymes. In Enzyme Catalysis in Organic Synthesis, 3rd ed.; Drauz, K., Gröger, H., May, O., Eds.; Wiley-VCH Verlag GmbH & Co., KGaA: Weinheim, Germany, 2012; pp. 1847–1938. [Google Scholar]
- Pscheidt, B.; Glieder, A. Yeast cell factories for fine chemical and API production. Microb. Cell Fact. 2008, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.A.R.; Moran, P.J.S.; Conceiçao, G.J.A.; Fardelone, L.C. Recent Advances in the Biocatalytic Asymmetric Reduction of Acetophenones and α,β-Unsaturated Carbonyl Compounds. Food Technol. Biotechnol. 2004, 42, 295–303. [Google Scholar]
- Csuk, R.; Glaenzer, B.I. Baker’s yeast mediated transformations in organic chemistry. Chem. Rev. 1991, 91, 49–97. [Google Scholar] [CrossRef]
- Lamed, R.J.; Keinan, E.; Zeikus, J.G. Potential applications of an alcohol-aldehyde/ketone oxidoreductase from thermophilic bacteria. Enzym. Microb. Technol. 1981, 3, 144–148. [Google Scholar] [CrossRef]
- Pàmies, O.; Bäckvall, J.-E. Combination of Enzymes and Metal Catalysts. A Powerful Approach in Asymmetric Catalysis. Chem. Rev. 2003, 103, 3247–3262. [Google Scholar] [CrossRef] [PubMed]
- García-Urdiales, E.; Rebolledo, F.; Gotor, V. Study of the enantioselectivity of the CAL-B-catalysed transesterification of α-substituted α-propylmethanols and α-substituted benzyl alcohols. Tetrahedron Asymmetry 2001, 12, 3047–3052. [Google Scholar] [CrossRef]
- Francisco, M.; van den Bruinhorst, A.; Kroon, M.C. Low-transition-temperature mixtures (LTTMs): A new generation of designer solvents. Angew. Chem. Int. Ed. 2013, 52, 3074–3085. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.L.; Abbott, A.P.; Ryder, K.S. Deep Eutectic Solvents (DESs) and Their Applications. Chem. Rev. 2014, 114, 11060–11082. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Hao, J.-W.; Mo, L.-P.; Zhang, Z.-H. Recent advances in the application of deep eutectic solvents as sustainable media as well as catalysts in organic reactions. RSC Adv. 2015, 5, 48675–48704. [Google Scholar] [CrossRef]
- Massolo, E.; Palmieri, S.; Benaglia, M.; Capriati, V.; Perna, F.M. Stereoselective organocatalysed reactions in deep eutectic solvents: Highly tunable and biorenewable reaction media for sustainable organic synthesis. Green Chem. 2016, 18, 792–797. [Google Scholar] [CrossRef]
- García-Álvarez, J.; Hevia, E.; Capriati, V. Reactivity of Polar Organometallic Compounds in Unconventional Reaction Media: Challenges and Opportunities. Eur. J. Org. Chem. 2015, 6779–6799. [Google Scholar] [CrossRef] [Green Version]
- Vidal, C.; García-Álvarez, J.; Hernán-Gómez, A.; Kennedy, A.R.; Hevia, E. Exploiting Deep Eutectic Solvents and Organolithium Reagent Partnerships: Chemoselective Ultrafast Addition to Imines and Quinolines Under Aerobic Ambient Temperature Conditions. Angew. Chem. Int. Ed. 2016, 55, 16145–16148. [Google Scholar] [CrossRef] [PubMed]
- Cicco, L.; Sblendorio, S.; Mansueto, R.; Perna, F.M.; Salomone, A.; Florio, S.; Capriati, V. Water opens the door to organolithiums and Grignard reagents: Exploring and comparing the reactivity of highly polar organometallic compounds in unconventional reaction media towards the synthesis of tetrahydrofurans. Chem. Sci. 2016, 7, 1192–1199. [Google Scholar] [CrossRef]
- Alonso, D.A.; Baeza, A.; Chinchilla, R.; Guillena, G.; Pastor, I.M.; Ramón, D.J. Deep Eutectic Solvents: The Organic Reaction Medium of the Century. Eur. J. Org. Chem. 2016, 612–632. [Google Scholar] [CrossRef]
- Xu, P.; Zheng, G.W.; Zong, M.H.; Li, N.; Lou, W.Y. Recent progress on deep eutectic solvents in biocatalysis. Bioresour. Bioprocess. 2017, 4. [Google Scholar] [CrossRef]
- Lan, D.; Wang, X.; Zhou, P.; Hollmann, F.; Wang, Y. Deep eutectic solvents as performance additives in biphasic reactions. RSC Adv. 2017, 7, 40367–40370. [Google Scholar] [CrossRef]
- Sheldon, R.A. Biocatalysis and Biomass Conversion in Alternative Reaction Media. Chem. Eur. J. 2016, 22, 12984–12999. [Google Scholar] [CrossRef] [PubMed]
- Durand, E.; Lecomte, J.; Villeneuve, P. Deep eutectic solvents: Synthesis, application, and focus on lipase-catalyzed reactions. Eur. J. Lipid Technol. 2013, 115, 379–385. [Google Scholar] [CrossRef]
- Mbous, Y.P.; Hayyan, M.; Wong, W.F.; Looi, C.Y.; Hashim, M.A. Unraveling the cytotoxicity and metabolic pathways of binary natural deep eutectic solvent systems. Sci. Rep. 2017, 7, 41257. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, Z.; Domínguez de María, P. Whole-Cell Biocatalysis in Deep-Eutectic-Solvents/Aqueous Mixtures. ChemCatChem 2014, 6, 1535–1537. [Google Scholar] [CrossRef]
- Bubalo, M.C.; Mazur, M.; Radosevic, K.; Redovnikovic, I.R. Baker’s yeast-mediated asymmetric reduction of ethyl 3-oxobutanoate in deep eutectic solvents. Process Biochem. 2015, 50, 1788–1792. [Google Scholar] [CrossRef]
- Vitale, P.; Abbinante, V.M.; Perna, F.M.; Salomone, A.; Cardellicchio, C.; Capriati, V. Unveiling the Hidden Performance of Whole Cells in the Asymmetric Bioreduction of Aryl-containing Ketones in Aqueous Deep Eutectic Solvents. Adv. Synth. Catal. 2017, 359, 1049–1057. [Google Scholar] [CrossRef]
- Vitale, P.; Perna, F.M.; Perrone, M.G.; Scilimati, A. Screening on the use of Kluyveromyces marxianus CBS 6556 growing cells as enantioselective biocatalysts for ketone reductions. Tetrahedron: Asymmetry 2011, 22, 1985–1993. [Google Scholar] [CrossRef]
- Vitale, P.; D’Introno, C.; Perna, F.M.; Perrone, M.G.; Scilimati, A. Kluyveromyces marxianus CBS 6556 growing cells as a new biocatalyst in the asymmetric reduction of substituted acetophenones. Tetrahedron Asymmetry 2013, 24, 389–394. [Google Scholar] [CrossRef]
- Vitale, P.; Perna, F.M.; Agrimi, G.; Scilimati, A.; Salomone, A.; Cardellicchio, C.; Capriati, V. Asymmetric chemoenzymatic synthesis of 1,3-diols and 2,4-disubstituted aryloxetanes by using whole cell biocatalysts. Org. Biomol. Chem. 2016, 14, 11438–11445. [Google Scholar] [CrossRef] [PubMed]
- Vitale, P.; Digeo, A.; Perna, F.M.; Agrimi, G.; Salomone, A.; Scilimati, A.; Cardellicchio, C.; Capriati, V. Stereoselective Chemoenzymatic Synthesis of Optically Active Aryl-Substituted Oxygen-Containing Heterocycles. Catalysts 2017, 7, 37. [Google Scholar] [CrossRef]
- Perna, F.M.; Ricci, M.A.; Scilimati, A.; Mena, M.C.; Pisano, I.; Palmieri, L.; Agrimi, G.; Vitale, P. Cheap and environmentally sustainable stereoselective arylketones reduction by Lactobacillus reuteri whole cells. J. Mol. Catal. B Enzym. 2016, 124, 29–37. [Google Scholar] [CrossRef]
- Kandiah, N.; Pai, M.-C.; Senanarong, V.; Looi, I.; Ampil, E.; Park, K.W.; Karanam, A.K.; Christopher, S. Rivastigmine: The advantages of dual inhibition of acetylcholinesterase and butyrylcholinesterase and its role in subcortical vascular dementia and Parkinson’s disease dementia. Clin. Interv. Aging 2017, 12, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Bhanja, C.; Jena, S. Synthesis Design of ‘Rivastigmine’—A Potent Therapeutic Agent for Alzheimer’s disease using Retrosynthetic Analysis. J. Chem. Pharm. Res. 2012, 4, 3171–3183. [Google Scholar]
- Hu, M.; Zhang, F.-L.; Xie, M.-H. Novel Convenient Synthesis of Rivastigmine. Synth. Commun. 2009, 39, 1527–1533. [Google Scholar] [CrossRef]
- Hana, S.; Josef, H.; Stanislav, S. A Method of Production of (–)-(S)-3-[1-(Dimethylamino)ethyl]phenyl-N-ethyl-N-methylcarbamate. Patent Application No. WO2004037771 A1, 6 May 2004. [Google Scholar]
- Rao, R.; Shewalkar, M.P.; Nandipati, R.; Yadav, J.S.; Khagga, M.; Shinde, D.B. General Strategy for Large-Scale Synthesis of (+)-Rivastigmine and (+)-NPS R-568. Synth. Commun. 2012, 42, 589–598. [Google Scholar] [CrossRef]
- Reddy, V.V.; Rao, M.V.N.B.; Ganesh, V.; Kumar, A.V.; Praveen, C.; Mukkanti, K.; Reddy, G.M.; Reddy, G.M. An Improved Process for the Production of Rivastigmine Tartrate, a Cholinesterase Inhibitor. Lett. Org. Chem. 2010, 7, 149–154. [Google Scholar] [CrossRef]
- Fuchs, M.; Koszelewski, D.; Tauber, K.; Kroutil, W.; Faber, K. Chemoenzymatic asymmetric total synthesis of (S)-Rivastigmine using ϖ-transaminases. Chem. Commun. 2010, 46, 5500–5502. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, M.; Koszelewski, D.; Tauber, K.; Sattler, J.; Banko, W.; Holzer, A.K.; Pickl, M.; Kroutil, W.; Faber, K. Improved chemoenzymatic asymmetric synthesis of (S)-Rivastigmine. Tetrahedron 2012, 68, 7691–7694. [Google Scholar] [CrossRef]
- Mangas-Sánchez, J.; Rodríguez-Mata, M.; Busto, E.; Gotor-Fernández, V.; Gotor, V. Chemoenzymatic Synthesis of Rivastigmine Based on Lipase-Catalyzed Processes. J. Org. Chem. 2009, 74, 5304–5310. [Google Scholar] [CrossRef] [PubMed]
- Sethi, M.K.; Bhandya, S.R.; Maddur, N.; Shukla, R.; Kumar, A.; Mittapalli, V.S.N.J. Asymmetric synthesis of an enantiomerically pure rivastigmine. Tetrahedron Asymmetry 2013, 24, 374–379. [Google Scholar] [CrossRef]
- Sethi, M.K.; Bhandya, S.R.; Kumar, A.; Maddur, N.; Shukla, R.; Mittapalli, V.S.N.J. Chemo-enzymatic synthesis of optically pure rivastigmine intermediate using alcohol dehydrogenase from baker’s yeast. J. Mol. Catal. B Enzym. 2013, 91, 87–92. [Google Scholar] [CrossRef]
- Nagai, T.; Sakurai, S.; Natori, N.; Hataoka, M.; Kinoshita, T.; Inoue, H.; Hanaya, K.; Shoji, M.; Sugai, T. Synthesis of enantiomerically enriched drug precursors and an insect pheromone via reduction of ketones using commercially available carbonyl reductase screening kit “Chiralscreen® OH”. Bioorg. Med. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Park, J.; Kim, M.J. Chemoenzymatic Synthesis of Rivastigmine via Dynamic Kynetic Resolution as a Key Step. J. Org. Chem. 2010, 75, 3105–3108. [Google Scholar] [CrossRef] [PubMed]
- Yan, P.-C.; Zhu, G.-L.; Xie, J.-H.; Zhang, X.-D.; Zhou, Q.-L.; Li, Y.-Q.; Shen, W.-H.; Che, D.-Q. Industrial Scal-Up of Enantioselective Hydrogenation for the Asymmetric Synthesis of Rivastigmine. Org. Proc. Dev. 2013, 17, 307–312. [Google Scholar] [CrossRef]
- De Man, J.C.; Rogosa, M.; Sharpe, M.E. A Medium for the Cultivation of Lactobacilli. J. Appl. Bacteriol. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Ricci, M.A.; Russo, A.; Pisano, I.; Palmieri, L.; de Angelis, M.; Agrimi, G. Improved 1,3-propanediol synthesis from glycerol by the robust Lactobacillus reuteri strain DSM 20016. J. Microbiol. Biotechnol. 2015, 6, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Kišic, A.; Stephan, M.; Mohar, B. ansa-Ruthenium(II) Complexes of R2NSO2DPEN-(CH2)n(η6-Aryl) Conjugate Ligands for Asymmetric Transfer Hydrogenation of Aryl Ketones. Adv. Synth. Catal. 2015, 357, 2540–2546. [Google Scholar] [CrossRef]
- Nasário, F.D.; Cazetta, T.; Moran, P.J.S.; Rodrigues, J.A.R. Deracemization of 1-phenylethanol via tandem biocatalytic oxidation and reduction. Tetrahedron Asymmetry 2016, 27, 404–409. [Google Scholar] [CrossRef]
- Baker’s Yeast LIEVITAL (25gx2—Cod. 100001181). Available online: https://lesaffre.ovh/ (accessed on 16 January 2018).
| Entry | Ketone | t (h) | Reaction Medium | Product (Yield %) 2 | ee (%) 3 | Absolute Configuration 4 |
|---|---|---|---|---|---|---|
| 1 | 1 | 15 | H2O | 3 (5) 5 | N.D. | N.D. |
| 2 | 1 | 24 | H2O | 3 (8) 5 | N.D. | N.D. |
| 3 | 1 | 40 | H2O | 3 (25) | >98 | S |
| 4 | 1 | 120 | H2O | 3 (27) | 98 | S |
| 5 | 1 | 120 | ChCl/Gly 2:1 | N.R. | N.D. | N.D. |
| 6 | 1 | 120 | L-(+)-lactic acid/ChCl 2:1 | N.R. | N.D. | N.D. |
| 7 | 1 | 120 | ChCl/D-fructose 2:1 | N.R. | N.D. | N.D. |
| 8 | 1 | 120 | ChCl/Gly 2:1 + 10 w% H2O | N.R. | N.D. | N.D. |
| 9 | 1 | 120 | L-(+)-lactic acid/ChCl 2:1 + 10 w% H2O | N.R. | N.D. | N.D. |
| 10 | 1 | 120 | ChCl/D-fructose 2:1 + 10 w% H2O | N.R. | N.D. | N.D. |
| 11 | 1 | 120 | ChCl/Gly 2:1 + 20 w% H2O | 3 (8) | >98 | S |
| 12 | 1 | 120 | ChCl/Gly 2:1 + 40 w% H2O | 3 (32) | >98 | S |
| 13 | 2 | 24 | H2O | 4 (12) | >98 | S |
| 14 | 2 | 120 | ChCl/Gly 2:1 + 20 w% H2O | 4 (5) | 95 | S |
| Entry | Ketone | t (h) | Reaction Medium | Cells Concentration(g CDW/L) | Product (Yield %) 2 | ee (%) 3 | Absolute Configuration 4 |
|---|---|---|---|---|---|---|---|
| 1 | 2 | 24 | PBS | 1.1 (RC) | 4 (60) | 98 | R |
| 2 | 2 | 24 | MRS | 1.1 (GC) 1 | 4 (47) | 98 | R |
| 3 | 2 | 48 | PBS | 1.1 (RC) | 4 (64) | 98 | R |
| 4 | 2 | 72 | PBS | 1.1 (RC) | 4 (74) | 98 | R |
| 5 | 1 | 72 | PBS | 1.1 (RC) | 3 (16) | >98 | R |
| 6 | 2 | 72 | PBS | 1.1 (RC) 5 | 4 (68) | 98 | R |
| 7 | 2 | 72 | PBS | 1.1 (RC) 6 | 4 (71) | 98 | R |
| 8 | 2 | 72 | PBS | 1.1 (RC) 7 | 4 (63) | 98 | R |
| 9 | 2 | 24 | PBS | 4.4 (RC) | 4 (90) | 98 | R |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitale, P.; Perna, F.M.; Agrimi, G.; Pisano, I.; Mirizzi, F.; Capobianco, R.V.; Capriati, V. Whole-Cell Biocatalyst for Chemoenzymatic Total Synthesis of Rivastigmine. Catalysts 2018, 8, 55. https://doi.org/10.3390/catal8020055
Vitale P, Perna FM, Agrimi G, Pisano I, Mirizzi F, Capobianco RV, Capriati V. Whole-Cell Biocatalyst for Chemoenzymatic Total Synthesis of Rivastigmine. Catalysts. 2018; 8(2):55. https://doi.org/10.3390/catal8020055
Chicago/Turabian StyleVitale, Paola, Filippo Maria Perna, Gennaro Agrimi, Isabella Pisano, Francesco Mirizzi, Roberto Vito Capobianco, and Vito Capriati. 2018. "Whole-Cell Biocatalyst for Chemoenzymatic Total Synthesis of Rivastigmine" Catalysts 8, no. 2: 55. https://doi.org/10.3390/catal8020055