Immobilization/Stabilization of Ficin Extract on Glutaraldehyde-Activated Agarose Beads. Variables That Control the Final Stability and Activity in Protein Hydrolyses
Abstract
:1. Introduction
2. Results and Discussion
2.1. Immobilization of Ficin Extract in MANAE-Agarose
2.2. Modification of the Ionically Exchanged Enzyme with Glutaraldehyde
2.3. Immobilization of the Enzyme in Glutaraldehyde Pre-Activated MANAE Agarose Beads
2.4. Stability of the Enzyme Immobilized at pH 7 on Glutaraldehyde Pre-Activated MANAE Agarose Beads
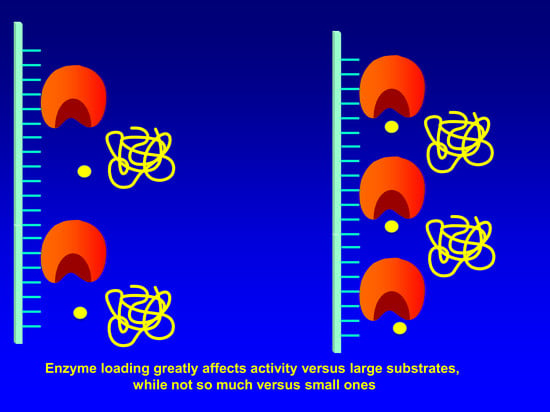
2.5. Determination of Loading Capacity
3. Materials and Methods
3.1. Materials
3.2. Preparation of Glutaraldehyde Agarose Beads
3.3. Preparation of Ficin Extract
3.4. Enzymatic Assays
3.5. Immobilization of Ficin Extract
3.6. Enzyme Inactivation Studies
3.7. Reuse of the Immobilized Ficin in the Hydrolysis of Casein
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tavano, O.L.; Berenguer-Murcia, A.; Secundo, F.; Fernandez-Lafuente, R. Biotechnological Applications of Proteases in Food Technology. Compr. Rev. Food Sci. Food Saf. 2018, 17, 412–436. [Google Scholar] [CrossRef]
- Tavano, O.L. Protein hydrolysis using proteases: An important tool for food biotechnology. J. Mol. Catal. B Enzym. 2013, 90, 1–11. [Google Scholar] [CrossRef]
- Clemente, A. Enzymatic protein hydrolysates in human nutrition. Trends Food Sci. Technol. 2000, 11, 254–262. [Google Scholar] [CrossRef]
- Maurer, K.H. Detergent proteases. Curr. Opin. Biotechnol. 2004, 15, 330–334. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, U.C.; Sani, R.K.; Azmi, W.; Soni, R. Thermostable alkaline protease from Bacillus brevis and its characterization as a laundry detergent additive. Process Biochem. 1999, 35, 213–219. [Google Scholar] [CrossRef]
- Phadatare, S.U.; Deshpande, V.V.; Srinivasan, M.C. High activity alkaline protease from Conidiobolus coronatus (NCL 86.8.20): Enzyme production and compatibility with commercial detergents. Enzyme Microb. Technol. 1993, 15, 72–76. [Google Scholar] [CrossRef]
- David, A.; Singh, C.P.; Kumar, A.; Angural, S.; Kumar, D.; Puri, N.; Gupta, N. Coproduction of protease and mannanase from Bacillus nealsonii PN-11 in solid state fermentation and their combined application as detergent additives. Int. J. Biol. Macromol. 2018, 108, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Cuq, J.C.; Vi, M.; Cheftel, J.C. Tryptophan degradation during heat treatments: Part 2—Degradation of protein-bound tryptophan. Food Chem. 1983, 12, 73–88. [Google Scholar] [CrossRef]
- Masuda, A.; Dohmae, N. Automated protein hydrolysis delivering sample to a solid acid catalyst for amino acid analysis. Anal. Chem. 2010, 82, 8939–8945. [Google Scholar] [CrossRef] [PubMed]
- Feijoo-Siota, L.; Villa, T.G. Native and Biotechnologically Engineered Plant Proteases with Industrial Applications. Food Bioprocess Technol. 2011, 4, 1066–1088. [Google Scholar] [CrossRef]
- Shah, M.A.; Mir, S.A.; Paray, M.A. Plant proteases as milk-clotting enzymes in cheesemaking: A review. Dairy Sci. Technol. 2014, 94, 5–16. [Google Scholar] [CrossRef]
- Faccia, M.; Picariello, G.; Trani, A.; Loizzo, P.; Gambacorta, G.; Lamacchia, C.; Di Luccia, A. Proteolysis of Cacioricotta cheese made from goat milk coagulated with caprifig (Ficus carica sylvestris) or calf rennet. Eur. Food Res. Technol. 2012, 234, 527–533. [Google Scholar] [CrossRef]
- Esmaeilpour, M.; Ehsani, M.R.; Aminlari, M.; Shekarforoush, S.; Hoseini, E. Antimicrobial activity of peptides derived from enzymatic hydrolysis of goat milk caseins. Comp. Clin. Pathol. 2016, 25, 599–605. [Google Scholar] [CrossRef]
- Di Pierro, G.; O’Keeffe, M.B.; Poyarkov, A.; Lomolino, G.; Fitzgerald, R.J. Antioxidant activity of bovine casein hydrolysates produced by Ficus carica L.-derived proteinase. Food Chem. 2014, 156, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Bekhit, A.A.; Hopkins, D.L.; Geesink, G.; Bekhit, A.A.; Franks, P. Exogenous Proteases for Meat Tenderization. Crit. Rev. Food Sci. Nutr. 2014, 54, 1012–1031. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, G.A.; Calkins, C.R. Application of exogenous enzymes to beef muscle of high and low-connective tissue. Meat Sci. 2010, 85, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Mariant, M.; Camagna, M.; Tarditi, L.; Seccamani, E. A new enzymatic method to obtain high-yield F(ab)2 suitable for clinical use from mouse IgGl. Mol. Immunol. 1991, 28, 69–77. [Google Scholar] [CrossRef]
- Sham, J.G.; Kievit, F.M.; Grierson, J.R.; Chiarelli, P.A.; Miyaoka, R.S.; Zhang, M.; Yeung, R.S.; Minoshima, S.; Park, J.O. Glypican-3-targeting F(ab′)2 for 89Zr PET of hepatocellular carcinoma. J. Nucl. Med. 2014, 55, 2032–2037. [Google Scholar] [CrossRef] [PubMed]
- Saczyńska, V.; Bierczyńska-Krzysik, A.; Cecuda-Adamczewska, V.; Baran, P.; Porȩbska, A.; Florys, K.; Zieliński, M.; Płucienniczak, G. Production of highly and broad-range specific monoclonal antibodies against hemagglutinin of H5-subtype avian influenza viruses and their differentiation by mass spectrometry. Virol. J. 2018, 15, 13. [Google Scholar] [CrossRef] [PubMed]
- Haesaerts, S.; Rodriguez Buitrago, J.A.; Loris, R.; Baeyens-Volant, D.; Azarkan, M. Crystallization and preliminary X-ray analysis of four cysteine proteases from Ficus carica latex. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2015, 71, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Zare, H.; Moosavi-Movahedi, A.A.; Salami, M.; Mirzaei, M.; Saboury, A.A.; Sheibani, N. Purification and autolysis of the ficin isoforms from fig (Ficus carica cv. Sabz) latex. Phytochemistry 2013, 87, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, K.B.; Kumar, P.R.; Prakash, V. Purification, characterization, and solvent-induced thermal stabilization of ficin from Ficus carica. J. Agric. Food Chem. 2008, 56, 11417–11423. [Google Scholar] [CrossRef] [PubMed]
- Di Cosimo, R.; McAuliffe, J.; Poulose, A.J.; Bohlmann, G. Industrial use of immobilized enzymes. Chem. Soc. Rev. 2013, 42, 6437–6474. [Google Scholar] [CrossRef]
- Cantone, S.; Ferrario, V.; Corici, L.; Ebert, C.; Fattor, D.; Spizzo, P.; Gardossi, L. Efficient immobilisation of industrial biocatalysts: Criteria and constraints for the selection of organic polymeric carriers and immobilisation methods. Chem. Soc. Rev. 2013, 42, 6262–6276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheldon, R.A.; van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed]
- Liese, A.; Hilterhaus, L. Evaluation of immobilized enzymes for industrial applications. Chem. Soc. Rev. 2013, 42, 6236–6249. [Google Scholar] [CrossRef] [PubMed]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Strategies for the one-step immobilization-purification of enzymes as industrial biocatalysts. Biotechnol. Adv. 2015, 33, 435–456. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.C.S.D.; Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Importance of the Support Properties for Immobilization or Purification of Enzymes. ChemCatChem 2015, 7, 2413–2432. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, A.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 35, 2885–2904. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef] [PubMed]
- Cipolatti, E.P.; Valério, A.; Henriques, R.O.; Moritz, D.E.; Ninow, J.L.; Freire, D.M.G.; Manoel, E.A.; Fernandez-Lafuente, R.; De Oliveira, D. Nanomaterials for biocatalyst immobilization-state of the art and future trends. RSC Adv. 2016, 6, 104675–104692. [Google Scholar] [CrossRef]
- Wang, M.; Qi, W.; Yu, Q.; Su, R.; He, Z. Kinetically controlled enzymatic synthesis of dipeptide precursor of l-alanyl-l-glutamine. Biotechnol. Appl. Biochem. 2011, 58, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Bhalla, T.C. Microbial proteases in peptide synthesis: Approaches and applications. Appl. Microbiol. Biotechnol. 2015, 68, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Lombard, C.; Saulnier, J.; Wallach, J.M. Recent trends in protease-catalyzed peptide synthesis. Protein Pept. Lett. 2005, 12, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Ikada, Y. Protease immobilization onto porous chitosan beads. J. Appl. Polym. Sci. 1991, 42, 85–92. [Google Scholar] [CrossRef]
- Hayashi, T.; Hyon, S.-H.; Cha, W.; Ikada, Y. Immobilization of thiol proteases onto porous poly(vinyl alcohol) beads. Polym. J. 1993, 25, 489–497. [Google Scholar] [CrossRef]
- Tai, D.F.; Huang, H.Y.; Huang, C.C. Immobilized ficin catalyzed synthesis of peptides in organic solvent. Bioorg. Med. Chem. Lett. 1995, 5, 1475–1478. [Google Scholar] [CrossRef]
- Jasim, M.A.; Hall, G.M.; Mann, J.; Taylor, K.D.A. A comparison of immobilised protease activities. J. Chem. Technol. Biotechnol. 1987, 40, 251–258. [Google Scholar] [CrossRef]
- Siar, E.-H.; Zaak, H.; Kornecki, J.F.; Zidoune, M.N.; Barbosa, O.; Fernandez-Lafuente, R. Stabilization of ficin extract by immobilization on glyoxyl agarose. Preliminary characterization of the biocatalyst performance in hydrolysis of proteins. Process Biochem. 2017, 58, 98–104. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in bio-catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Betancor, L.; López-Gallego, F.; Hidalgo, A.; Alonso-Morales, N.; Dellamora-Ortiz, G.; Mateo, C.; Fernández-Lafuente, R.; Guisán, J.M. Different mechanisms of protein immobilization on glutaraldehyde activated supports: Effect of support activation and immobilization conditions. Enzyme Microb. Technol. 2006, 39, 877–882. [Google Scholar] [CrossRef]
- López-Gallego, F.; Betancor, L.; Mateo, C.; Hidalgo, A.; Alonso-Morales, N.; Dellamora-Ortiz, G.; Guisán, J.M.; Fernández-Lafuente, R. Enzyme stabilization by glutaraldehyde crosslinking of adsorbed proteins on aminated supports. J. Biotechnol. 2005, 119, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, O.; Torres, R.; Ortiz, C.; Fernandez-Lafuente, R. Versatility of glutaraldehyde to immobilize lipases: Effect of the immobilization protocol on the properties of lipase B from Candida antárctica. Process Biochem. 2012, 47, 1220–1227. [Google Scholar] [CrossRef]
- Zaak, H.; Peirce, S.; de Albuquerque, T.L.; Sassi, M.; Fernandez-Lafuente, R. Exploiting the versatility of aminated supports activated with glutaraldehyde to immobilize β-galactosidase from Aspergillus oryzae. Catalysts 2017, 7, 250. [Google Scholar] [CrossRef]
- Vazquez-Ortega, P.G.; Alcaraz-Fructuoso, M.T.; Rojas-Contreras, J.A.; López-Miranda, J.; Fernandez-Lafuente, R. Stabilization of dimeric β-glucosidase from Aspergillus niger via glutaraldehyde immobilization under different conditions. Enzyme Microb. Technol. 2018, 110, 38–45. [Google Scholar] [CrossRef] [PubMed]
- De Albuquerque, T.L.; Peirce, S.; Rueda, N.; Marzocchella, A.; Gonçalves, L.R.B.; Rocha, M.V.P.; Fernandez-Lafuente, R. Ion exchange of β-galactosidase: The effect of the immobilization pH on enzyme stability. Process Biochem. 2016, 51, 875–880. [Google Scholar] [CrossRef]
- Fernandez-Lopez, L.; Pedrero, S.G.; Lopez-Carrobles, N.; Gorines, B.C.; Virgen-Ortíz, J.J.; Fernandez-Lafuente, R. Effect of protein load on stability of immobilized enzymes. Enzyme Microb. Technol. 2017, 98, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Zaak, H.; Siar, E.H.; Kornecki, J.F.; Fernandez-Lopez, L.; Pedrero, S.G.; Virgen-Ortíz, J.J.; Fernandez-Lafuente, R. Effect of immobilization rate and enzyme crowding on enzyme stability under different conditions. The case of lipase from Thermomyces lanuginosus immobilized on octyl agarose beads. Process Biochem. 2017, 56, 117–123. [Google Scholar] [CrossRef]
- Virgen-Ortíz, J.J.; Pedrero, S.G.; Fernandez-Lopez, L.; Lopez-Carrobles, N.; Gorines, B.C.; Otero, C.; Fernandez-Lafuente, R. Desorption of lipases immobilized on octyl-agarose beads and coated with ionic polymers after thermal inactivation. Stronger adsorption of polymers/unfolded protein composites. Molecules 2017, 22, 91. [Google Scholar] [CrossRef]
- Virgen-Ortíz, J.J.; Peirce, S.; Tacias-Pascacio, V.G.; Cortes-Corberan, V.; Marzocchella, A.; Russo, M.E.; Fernandez-Lafuente, R. Reuse of anion exchangers as supports for enzyme immobilization: Reinforcement of the enzyme-support multiinteraction after enzyme inactivation. Process Biochem. 2016, 51, 1391–1396. [Google Scholar] [CrossRef]
- Fernandez-Lafuente, R.; Rosell, C.M.; Rodriguez, V.; Guisan, J.M. Strategies for enzyme stabilization by intramolecular crosslinking with bifunctional reagents. Enzyme Microb. Technol. 1995, 17, 517–523. [Google Scholar] [CrossRef]
- Sanchez, A.; Cruz, J.; Rueda, N.; Dos Santos, J.C.S.; Torres, R.; Ortiz, C.; Villalonga, R.; Fernandez-Lafuente, R. Inactivation of immobilized trypsin under dissimilar conditions produces trypsin molecules with different structures. RSC Adv. 2016, 6, 27329–27334. [Google Scholar] [CrossRef]
- Mateo, C.; Abian, O.; Bernedo, M.; Cuenca, E.; Fuentes, M.; Fernandez-Lorente, G.; Palomo, J.M.; Grazu, V.; Pessela, B.C.C.; Giacomini, C.; et al. Some special features of glyoxyl supports to immobilize proteins. Enzyme Microb. Technol. 2005, 37, 456–462. [Google Scholar] [CrossRef]
- Grazu, V.; Betancor, L.; Montes, T.; Lopez-Gallego, F.; Guisan, J.M.; Fernandez-Lafuente, R. Glyoxyl agarose as a new chromatographic matrix. Enzyme Microb. Technol. 2006, 38, 960–966. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fuentes, M.; Betancor, L.; Grazú, V.; Lopez-Gallego, F.; Pessela, B.C.C.; Hidalgo, A.; Fernández-Lorente, G.; Fernández-Lafuente, R. Glyoxyl agarose: A fully inert and hydrophilic support for immobilization and high stabilization of proteins. Enzyme Microb. Technol. 2006, 39, 274–280. [Google Scholar] [CrossRef]
- Fernández-Lafuente, R.; Rosell, C.M.; Rodriguez, V.; Santana, C.; Soler, G.; Bastida, A.; Guisán, J.M. Preparation of activated supports containing low pK amino groups. A new tool for protein immobilization via the carboxyl coupling method. Enzyme Microb. Technol. 1993, 15, 546–550. [Google Scholar] [CrossRef]
- Corma, A.; Fornés, V.; Jordá, J.L.; Rey, F.; Fernandez-Lafuente, R.; Guisan, J.M.; Mateo, C. Electrostatic and covalent immobilisation of enzymes on ITQ-6 delaminated zeolitic materials. Chem. Commun. 2001, 5, 419–420. [Google Scholar] [CrossRef]
- Betancor, L.; López-Gallego, F.; Alonso-Morales, N.; Dellamora, G.; Mateo, C.; Fernandez-Lafuente, R.; Guisan, J.M. Glutaraldehyde in Protein Immobilization. A Versatile Reagent. In Immobilization of Enzymes and Cells, 2nd ed.; Guisan, J.M., Ed.; Humana Press Inc.: New York, NY, USA, 2006; pp. 57–64. ISBN 1-58829-290-8. [Google Scholar]
- Nouani, A.; Dako, E.; Morsli, A.; Belhamiche, N.; Belbraouet, S.; Bellal, M.M.; Dadie, A. Characterization of the purified coagulant extracts derived from artichoke flowers (Cynaras scolymus) and from the fig tree latex (Ficus carica) in light of their use in the manufacture of traditional cheeses in Algeria. J. Food Technol. 2009, 7, 20–29. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Kunitz, M.J. Crystalline soybean trypsin inhibitor: II. General properties. J. Gen. Physiol. 1947, 30, 291–310. [Google Scholar] [CrossRef] [PubMed]











© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siar, E.-H.; Arana-Peña, S.; Barbosa, O.; Zidoune, M.N.; Fernandez-Lafuente, R. Immobilization/Stabilization of Ficin Extract on Glutaraldehyde-Activated Agarose Beads. Variables That Control the Final Stability and Activity in Protein Hydrolyses. Catalysts 2018, 8, 149. https://doi.org/10.3390/catal8040149
Siar E-H, Arana-Peña S, Barbosa O, Zidoune MN, Fernandez-Lafuente R. Immobilization/Stabilization of Ficin Extract on Glutaraldehyde-Activated Agarose Beads. Variables That Control the Final Stability and Activity in Protein Hydrolyses. Catalysts. 2018; 8(4):149. https://doi.org/10.3390/catal8040149
Chicago/Turabian StyleSiar, El-Hocine, Sara Arana-Peña, Oveimar Barbosa, Mohammed Nasreddine Zidoune, and Roberto Fernandez-Lafuente. 2018. "Immobilization/Stabilization of Ficin Extract on Glutaraldehyde-Activated Agarose Beads. Variables That Control the Final Stability and Activity in Protein Hydrolyses" Catalysts 8, no. 4: 149. https://doi.org/10.3390/catal8040149