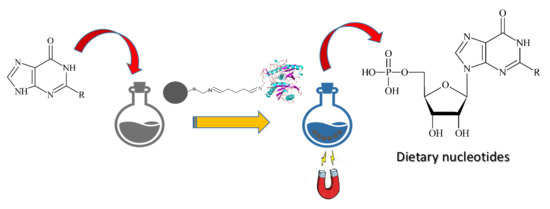
One-Pot, One-Step Production of Dietary Nucleotides by Magnetic Biocatalysts
Abstract
:1. Introduction
2. Results and Discussion
2.1. Covalent Immobilization of TtHGXPRT
2.2. Biochemical Characterization of MTtHGXPRT Derivatives
2.3. Thermal Stability of MTtHGXPRT Derivatives
2.4. Recycling of MTtHGXPRT Derivatives
2.5. Effect of Molar Ratio
2.6. Enzymatic Production of IMP and GMP
3. Materials and Methods
3.1. Chemicals
3.2. Production TtHGXPRT
3.3. Enzyme Immobilization
3.4. Enzyme Activity Assay for Immobilized TtHGXPRT
3.5. Biochemical Characterization of Immobilized Biocatalysts
3.6. Thermal Stability and Reusability of MTtHGXPRT
3.7. Enzymatic Production of Dietary Nucleotides
3.8. Analytical Methods
3.9. Molecular Docking and Surface Analysis of TtHGXPRT
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Behrens, M.; Meyerhof, W.; Hellfritsch, C.; Hofmann, T. Sweet and umami taste: Natural products, their chemosensory targets, and beyond. Angew. Chem. Int. Ed. 2011, 50, 2220–2242. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, J.S.; Gibson, R.A.; Roberton, D.; Makrides, M. Effect of dietary nucleotide supplementation on growth and immune function in term infants: A randomized controlled trial. Eur. J. Clin. Nutr. 2006, 60, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Lapponi, M.J.; Rivero, C.W.; Zinni, M.A.; Britos, C.N.; Trelles, J.A. New developments in nucleoside analogues biosynthesis: A review. J. Mol. Catal. B Enzym. 2016, 133, 218–233. [Google Scholar] [CrossRef]
- Mikhailopulo, I.A. Biotechnology of nucleic acid constituents—State of the art and perspectives. Curr. Org. Chem. 2007, 11, 317–333. [Google Scholar] [CrossRef]
- Fresco-Taboada, A.; de la Mata, I.; Arroyo, M.; Fernández-Lucas, J. New insights on nucleoside 2’-deoxyribosyltransferases: A versatile biocatalyst for one-pot one-step synthesis of nucleoside analogs. Appl. Microbiol. Biotechnol. 2013, 97, 3773–3785. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Kato, T.; Takenishi, T. A novel method for phosphorylation of nucleosides to 5′-nucleotides. Tetrahedron Lett. 1967, 8, 5065–5068. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Kato, T.; Takenishi, T. Studies of phosphorylation. III. Selective phosphorylation of unprotected nucleosides. Bull. Chem. Soc. Jpn. 1969, 42, 3505–3508. [Google Scholar] [CrossRef]
- Del Arco, J.; Fernández-Lucas, J. Purine and Pyrimidine Phosphoribosytransferases: A versatile tool for enzymatic synthesis of nucleoside-5’-monophosphates. Curr. Pharm. Des. 2015, 23, 6898–6912. [Google Scholar] [CrossRef] [PubMed]
- Del Arco, J.; Acosta, J.; Pereira, H.M.; Perona, A.; Lokanath, N.K.; Kunishima, N.; Fernández-Lucas, J. Enzymatic production of non-natural nucleoside-5’-monophosphates by a novel thermostable uracil phosphoribosyltransferase. ChemCatChem 2018, 10, 439–448. [Google Scholar] [CrossRef]
- Del Arco, J.; Martinez, M.; Donday, M.; Clemente-Suarez, V.J.; Fernández-Lucas, J. Cloning, expression and biochemical characterization of xanthine and adenine phosphoribosyltransferases from Thermus thermophilus HB8. Biocatal. Biotransform. 2017, 1–8. [Google Scholar] [CrossRef]
- Del Arco, J.; Cejudo-Sanches, J.; Esteban, I.; Clemente-Suarez, V.J.; Hormigo-Cisneros, D.; Perona, A.; Fernández-Lucas, J. Enzymatic production of dietary nucleotides from low-soluble purine bases by an efficient, thermostable and alkali-tolerant biocatalyst. Food Chem. 2017, 237, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Serra, I.; Conti, S.; Piškur, J.; Clausen, A.R.; Munch-Petersen, B.; Terreni, M.; Ubiali, D. Immobilized Drosophila melanogaster deoxyribonucleoside kinase (DmdNK) as a high performing biocatalyst for the synthesis of purine arabinonucleotides. Adv. Synth. Catal. 2014, 356, 563–570. [Google Scholar] [CrossRef]
- Fernández-Lucas, J. Multienzymatic synthesis of nucleic acid derivatives: A general perspective. Appl. Microbiol. Biotechnol. 2015, 99, 4615–4627. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Ding, Q.; Ou, L.; Yan, B. Efficient production of deoxynucleoside-5′-monophosphates using deoxynucleoside kinase coupled with a GTP-regeneration system. Appl. Microbiol. Biotechnol. 2013, 97, 9389–9395. [Google Scholar] [CrossRef] [PubMed]
- Mori, H.; Iida, A.; Fujio, T.; Teshiba, S. A novel process of inosine 5′-monophosphate production using overexpressed guanosine/inosine kinase. Appl. Microbiol. Biotechnol. 1997, 48, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Gudiño, E.D.; Santillán, J.Y.; Iglesias, L.E.; Iribarren, A.M. An enzymatic alternative for the synthesis of nucleoside 5′-monophosphates. Enzyme Microb. Technol. 2018, 111, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Sheldon, R.A.; van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef] [PubMed]
- Secundo, F. Conformational changes of enzymes upon immobilization. Chem. Soc. Rev. 2013, 42, 6250–6261. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Strategies for the one-step immobilization-purification of enzymes as industrial biocatalysts. Biotechnol. Adv. 2015, 33, 435–456. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in bio-catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Zucca, P.; Sanjust, E. Inorganic materials as supports for covalent enzyme immobilization: Methods and mechanisms. Molecules 2014, 19, 14139–14194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Lucas, J.; Harris, R.; Mata-Casar, I.; Heras, A.; de la Mata, I.; Arroyo, M. Magnetic chitosan beads for covalent immobilization of nucleoside 2′-deoxyribosyltransferase: Application in nucleoside analogues synthesis. J. Ind. Microbiol. Biotechnol. 2013, 40, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Bayramoglu, G.; Yilmaz, M.; Arica, M.Y. Preparation and characterization of epoxy-functionalized magnetic chitosan beads: Laccase immobilized for degradation of reactive dyes. Bioprocess Biosyst. Eng. 2010, 33, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Sheng, W.; Xi, Y.; Zhang, L.; Ye, T.; Zhao, X. Enhanced activity and stability of papain by covalent immobilization on porous magnetic nanoparticles. Int. J. Biol. Macromol. 2018, 114, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Acosta, J.; Arco, J.D.; Martinez-Pascual, S.; Clemente-Suárez, V.J.; Fernández-Lucas, J. One-Pot Multi-Enzymatic Production of Purine Derivatives with Application in Pharmaceutical and Food Industry. Catalysts 2018, 8, 9. [Google Scholar] [CrossRef]
- Scism, R.A.; Bachmann, B.O. Five-component cascade synthesis of nucleotide analogues in an engineered self-immobilized enzyme aggregate. ChemBioChem 2010, 11, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, O.; Torres, R.; Ortiz, C.; Fernandez-Lafuente, R. Modulation of the properties of immobilized CALB by chemical modification with 2, 3, 4-trinitrobenzenesulfonate or ethylendiamine. Advantages of using adsorbed lipases on hydrophobic supports. Process. Biochem. 2012, 47, 1220–1227. [Google Scholar] [CrossRef]
- Hernandez, K.; Fernandez-Lafuente, R. Control of protein immobilization: coupling immobilization and site-directed mutagenesis to improve biocatalyst or biosensor performance. Enzyme Microb. Technol. 2011, 48, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Betancor, L.; López-Gallego, F.; Hidalgo, A.; Alonso-Morales, N.; Dellamora-Ortiz, G.; Mateo, C.; Fernández-Lafuente, R.; Guisán, J.M. Different mechanisms of protein immobilization on glutaraldehyde activated supports: Effect of support activation and immobilization conditions. Enzyme Microb. Technol. 2006, 39, 877–882. [Google Scholar] [CrossRef]
- Adriano, W.S.; Filho, E.H.C.; Silva, J.A.; Giordano, R.L.C.; Gonçalves, L.R.B. Stabilization of penicillin G acylase by immobilization on glutaraldehyde-activated chitosan. Braz. J. Chem. Eng. 2005, 22, 529–538. [Google Scholar] [CrossRef]
- Adriano, W.S.; Filho, E.H.C.; Silva, J.A.; Gonçalves, L.R.B. Optimization of penicillin G acylase multipoint immobilization on to glutaraldehyde–chitosan beads. Biotechnol. Appl. Biochem. 2005, 41, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Adriano, W.S.; Mendonça, D.B.; Rodrigues, D.S.; Mammarella, E.J.; Giordano, R.L.C. Improving the properties of chitosan as support for the covalent multipoint immobilization of chymotrypsin. Biomacromolecules 2008, 9, 2170–2179. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, O.; Torres, R.; Ortiz, C.; Berenguer-Murcia, Á.; Rodrigues, R.C.; Fernandez-Lafuente, R. Heterofunctional supports in enzyme immobilization: From traditional immobilization protocols to opportunities in tuning enzyme properties. Biomacromolecules 2013, 14, 2433–2462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Lafuente, R. Stabilization of multimeric enzymes: Strategies to prevent subunit dissociation. Enzyme Microb. Technol. 2009, 45, 405–418. [Google Scholar] [CrossRef]
- Jordaan, J.; Simpson, C.; Brady, D.; Gardiner, N.S.; Gerber, I.B. Emulsion-derived particles. U.S. Patent No. 9,574,054, 2017. [Google Scholar]
- Kanagawa, M.; Baba, S.; Ebihara, A.; Shinkai, A.; Hirotsu, K.; Mega, R.; Kim, K.; Kuramitsu, S.; Sampei, G.; Kawai, G. Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 893–898. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL molecular graphics system. 2002. Available online: http://www.pymol.org (accessed on 20 April 2018).
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]




| Derivative | Biocatalyst Loading (Mgenz/gsupport) | Immobilization Yield (%) | Activity (IU/gsupport) | Recovery(%) |
|---|---|---|---|---|
| MTtHGXPRT pH 8.5 | ||||
| MTtHGXPRT1 | 102 | 84 ± 1 | 800 ± 33 | 25 ± 2 |
| MTtHGXPRT2 | 226 | 88 ± 4 | 802 ± 29 | 21 ± 1 |
| MTtHGXPRT3 | 322 | 85 ± 3 | 1581 ± 27 | 29 ± 2 |
| MTtHGXPRT pH 10.0 | ||||
| MTtHGXPRT4 | 226 | 67 ± 2 | 783 ± 26 | 21 ± 4 |
| MTtHGXPRT5 | 322 | 71 ± 3 | 1108 ± 21 | 23 ± 1 |
| PRPP (mM) | Hypoxanthine (mM) | MgCl2 (mM) | IMP (mM) | Activity (IU/gsupport) |
|---|---|---|---|---|
| 10 | 10 | 12 | 2.1 ± 0.1 | 1830 ± 24 |
| 10 | 24 | 1.1 ± 0.2 | 962 ± 32 | |
| 20 | 12 | 3.0 ± 0.1 | 2400 ± 57 | |
| 20 | 24 | 1.4 ± 0.1 | 1060 ± 35 |
| PRPP (mM) | Base (mM) | MgCl2 (mM) | Derivative (µg) | IMP (mM) | Activity (IU/gsupport) | |
|---|---|---|---|---|---|---|
| IMP synthesis | ||||||
| 10 | 20 | 12 | 12 | 2.9 ± 0.1 | 2200 ± 24 | |
| 20 | 40 | 24 | 12 | 2.4 ± 0.2 | 1654 ± 29 | |
| 40 | 80 | 48 | 12 | 4.8 ± 0.2 | 3365 ± 45 | |
| 10 | 20 | 12 | 30 | 3.8 ± 0.1 | 2800 ± 69 | |
| 20 | 40 | 24 | 30 | 5.6 ± 0.1 | 4400 ± 100 | |
| 40 | 80 | 48 | 30 | 7.5 ± 0.1 | 5600 ± 49 | |
| GMP synthesis | ||||||
| 10 | 20 | 12 | 12 | 1.6 ± 0.1 | 1149 ± 120 | |
| 20 | 40 | 24 | 12 | 3.7 ± 0.1 | 2722 ± 80 | |
| 40 | 80 | 48 | 12 | 1.4 ± 0.2 | 1000 ± 70 | |
| 10 | 20 | 12 | 30 | 3.8 ± 0.1 | 2835 ± 87 | |
| 20 | 40 | 24 | 30 | 3.2 ± 0.1 | 2335 ± 90 | |
| 40 | 80 | 48 | 30 | 2.4 ± 0.2 | 1790 ± 56 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Del Arco, J.; Martínez-Pascual, S.; Clemente-Suárez, V.J.; Corral, O.J.; Jordaan, J.; Hormigo, D.; Perona, A.; Fernández-Lucas, J. One-Pot, One-Step Production of Dietary Nucleotides by Magnetic Biocatalysts. Catalysts 2018, 8, 184. https://doi.org/10.3390/catal8050184
Del Arco J, Martínez-Pascual S, Clemente-Suárez VJ, Corral OJ, Jordaan J, Hormigo D, Perona A, Fernández-Lucas J. One-Pot, One-Step Production of Dietary Nucleotides by Magnetic Biocatalysts. Catalysts. 2018; 8(5):184. https://doi.org/10.3390/catal8050184
Chicago/Turabian StyleDel Arco, Jon, Sara Martínez-Pascual, Vicente Javier Clemente-Suárez, Octavio Jorge Corral, Justin Jordaan, Daniel Hormigo, Almudena Perona, and Jesús Fernández-Lucas. 2018. "One-Pot, One-Step Production of Dietary Nucleotides by Magnetic Biocatalysts" Catalysts 8, no. 5: 184. https://doi.org/10.3390/catal8050184
APA StyleDel Arco, J., Martínez-Pascual, S., Clemente-Suárez, V. J., Corral, O. J., Jordaan, J., Hormigo, D., Perona, A., & Fernández-Lucas, J. (2018). One-Pot, One-Step Production of Dietary Nucleotides by Magnetic Biocatalysts. Catalysts, 8(5), 184. https://doi.org/10.3390/catal8050184