Cloning, Expression, and Characterization of a Novel Thermostable and Alkaline-stable Esterase from Stenotrophomonas maltophilia OUC_Est10 Catalytically Active in Organic Solvents
Abstract
:1. Introduction
2. Results and Discussion
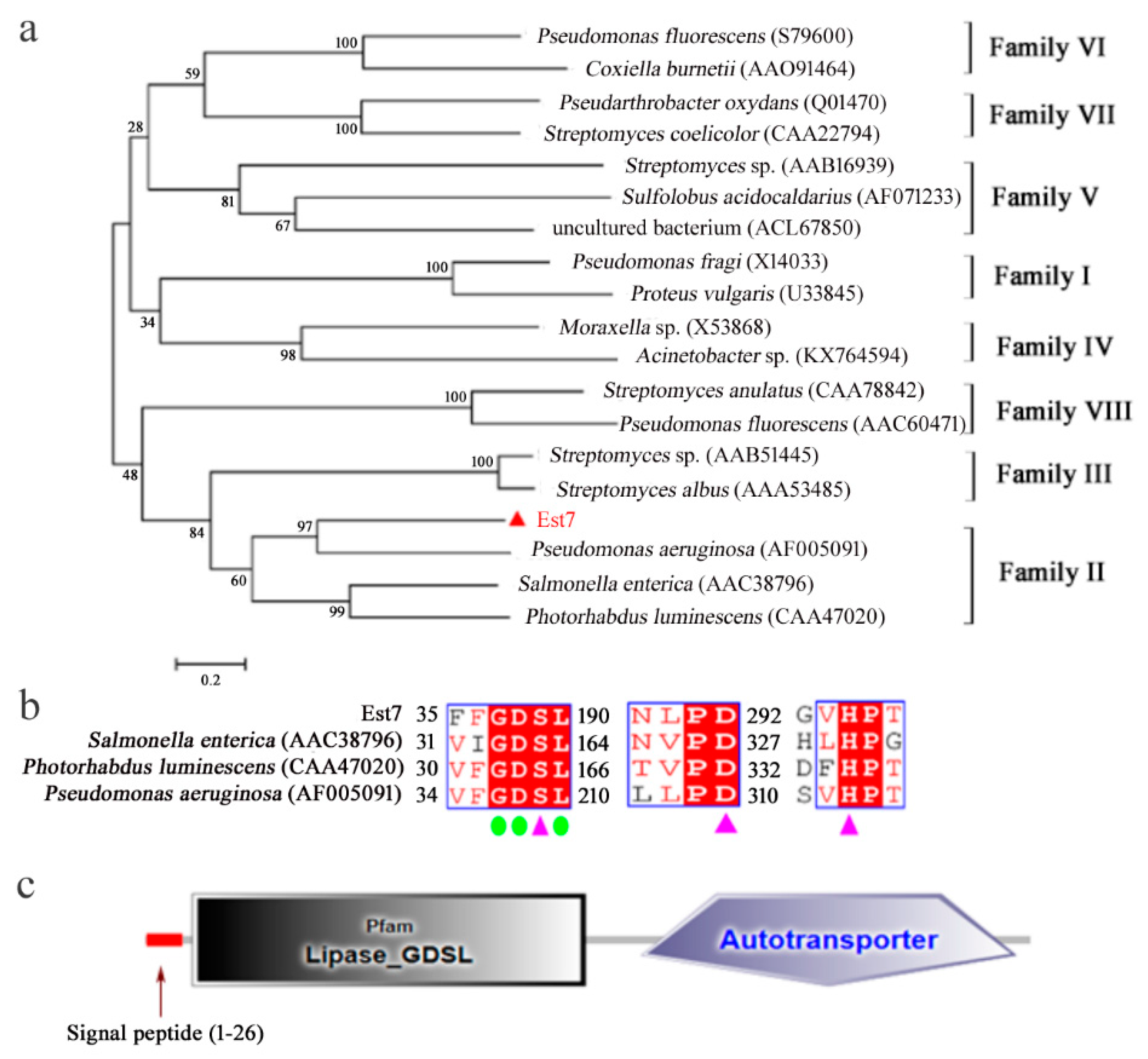
2.1. Esterase Gene Est7 Sequence and Protein Structure Analysis
2.2. Heterologous Expression and Purification of Est7
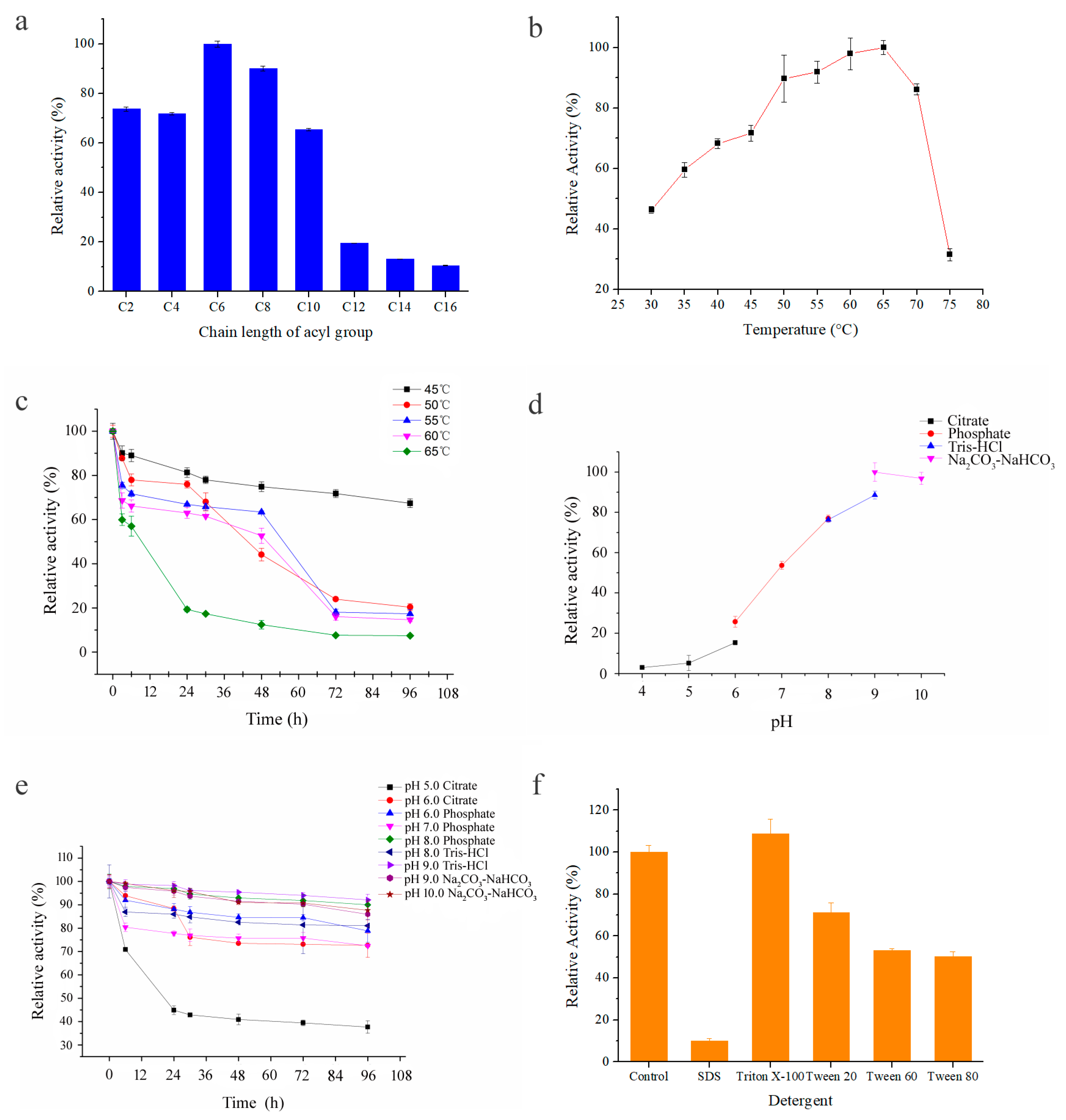
2.3. Enzyme Characterization of Purified Esterase Est7
2.4. Catalytic Activity of Esterase Est7 in Organic Solvents
3. Materials and Methods
3.1. Materials
3.2. Bacterial Strains, Plasmids, and Sequence Analysis of Lipolytic Genes
3.3. Heterologous Expression and Purification of Esterase Est7
3.4. Enzyme Characterization of Purified Esterase Est7
3.5. Esterase Est7 Catalyzed Transesterification
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ramnath, L.; Sithole, B.; Govinden, R. Identification of lipolytic enzymes isolated from bacteria indigenous to Eucalyptus wood species for application in the pulping industry. Biotechnol. Rep. 2017, 15, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Fojan, P.; Jonson, P.H.; Petersen, M.T.N.; Petersen, S.B. What distinguishes an esterase from a lipase: A novel structural approach. Biochimie 2000, 82, 1033–1041. [Google Scholar] [CrossRef]
- Khudary, R.; Venkatachalam, R.; Katzer, M.; Elleuche, S.; Antranikian, G. A cold-adapted esterase of a novel marine isolate, Pseudoalteromonas arctica: Gene cloning, enzyme purification and characterization. Extremophiles 2010, 14, 273–285. [Google Scholar] [CrossRef]
- Jaeger, K.E.; Ransac, S.; Dijkstra, B.W.; Colson, C.; Van, H.M.; Misset, O. Bacterial lipases. FEMS Microbiol. Rev. 1994, 15, 29–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrer, M.; Bargiela, R.; Martínez-Martínez, M.; Mir, J.; Koch, R.; Golyshina, O.V.; Golyshin, P.N. Biodiversity for biocatalysis: A review of the α/β-hydrolase fold superfamily of esterases-lipases discovered in metagenomes. Biocatal. Biotransform. 2016, 33, 1–15. [Google Scholar] [CrossRef]
- Salihu, A.; Alam, M.Z. Solvent tolerant lipases: A review. Process Biochem. 2015, 50, 86–96. [Google Scholar] [CrossRef]
- Persson, M.; Bornscheuer, U.T. Increased stability of an esterase from Bacillus stearothermophilus in ionic liquids as compared to organic solvents. J. Mol. Catal. B Enzym. 2003, 22, 21–27. [Google Scholar] [CrossRef]
- Gao, W.; Wu, K.; Chen, L.; Fan, H.; Zhao, Z.; Gao, B.; Wang, H.; Wei, D. A novel esterase from a marine mud metagenomic library for biocatalytic synthesis of short-chain flavor esters. Microb. Cell Fact. 2016, 15, 41. [Google Scholar] [CrossRef] [PubMed]
- Alfonso, I.; Gotor, V. Biocatalytic and biomimetic aminolysis reactions: Useful tools for selective transformations on polyfunctional substrates. Chem. Soc. Rev. 2004, 33, 201–209. [Google Scholar] [CrossRef]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzyme Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. Microbial carboxyl esterases: Classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 2002, 26, 73–81. [Google Scholar] [CrossRef]
- Shafqet, M.A.; Brown, T.V.; Sharma, R. Normal lipase drug-induced pancreatitis: A novel finding. Am. J. Emerg. Med. 2015, 33, 476.e5–476.e6. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, X.; Zhang, L.; Li, J.; Yang, L.; Gao, P.; Zhou, Z. Lipase-catalyzed synthesis of biodiesel in a hydroxyl-functionalized ionic liquid. Chem. Eng. Res. Des. 2018, 132, 199–207. [Google Scholar] [CrossRef]
- Gupta, R.; Gupta, N.; Rathi, P. Bacterial lipases: An overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 2004, 64, 763–781. [Google Scholar] [CrossRef]
- Enzymatic Reactions in Organic Media; Koskinen, A.M.P.; Klibanov, A.M. (Eds.) Blackie Academic and Professional: London, UK, 1996. [Google Scholar] [CrossRef]
- Mannino, S.; Cosio, M.S.; Wang, J. Determination of peroxide in vegetable oils by an organic-phase enzyme electrode. Anal. Lett. 1994, 27, 299–308. [Google Scholar] [CrossRef]
- Secundo, F.; Spadaro, S.; Carrea, G.; Overbeeke, P.L. Optimization of Pseudomonas cepacia lipase preparations for catalysis in organic solvent. Biotechnol. Bioeng. 1999, 62, 554–561. [Google Scholar] [CrossRef]
- Secundo, F.; Carrea, G.; Soregaroli, C.; Varinelli, D.; Morrone, R. Activity of different Candida antarctica lipase B formulations in organic solvents. Biotechnol. Bioeng. 2001, 73, 157–163. [Google Scholar] [CrossRef]
- Carrea, G.; Ottolina, G.; Riva, S. Role of solvents in the control of enzyme selectivity in organic media. Trends Biotechnol. 1995, 13, 63–70. [Google Scholar] [CrossRef]
- Dong, H.; Secundo, F.; Xue, C.; Mao, X. Whole-cell biocatalytic synthesis of cinnamyl acetate with a novel esterase from the DNA library of Acinetobacter haemolyticus. J. Agric. Food Chem. 2017, 65, 2120–2128. [Google Scholar] [CrossRef]
- Belafriekh, A.; Secundo, F.; Serra, S.; Djeghaba, Z. Enantioselective enzymatic resolution of racemic alcohols by lipases in green organic solvents. Tetrahedron: Asymmetry 2017, 28, 473–478. [Google Scholar] [CrossRef]
- Dong, H.; Li, X.; Xue, C.; Mao, X. Astaxanthin preparation by fermentation of esters from Haematococcus pluvialis algal extracts with Stenotrophomonas species. Biotechnol. Prog. 2016, 32, 649–656. [Google Scholar] [CrossRef]
- Dong, H.; Rui, J.; Sun, J.; Li, X.; Mao, X. Complete genome sequencing and diversity analysis of lipolytic enzymes in Stenotrophomonas maltophilia OUC_Est10. Acta Microbiol. Sin. 2017, 57, 1716–1721. [Google Scholar] [CrossRef]
- Fu, C.; Hu, Y.; Xie, F.; Guo, H.; Ashforth, E.J.; Polyak, S.W.; Zhu, B.; Zhang, L. Molecular cloning and characterization of a new cold-active esterase from a deep-sea metagenomic library. Appl. Microbiol. Biotechnol. 2011, 90, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Gao, X.; Dong, H.; Liu, Z.; Secundo, F.; Xue, C.; Mao, X. Identification of a novel esterase from marine environmental genomic DNA libraries and its application in production of free all-trans-astaxanthin. J. Agric. Food Chem. 2018, 66, 2812–2821. [Google Scholar] [CrossRef]
- Laane, C.; Boeren, S.; Vos, K.; Veeger, C. Rules for optimization of biocatalysis in organic solvents. Biotechnol. Bioeng. 2009, 102, 2–8. [Google Scholar] [CrossRef]
- Öztürk, D.C.; Kazan, D.; Denizci, A.A.; Grimoldi, D.; Secundo, F.; Erarslan, A. Water miscible mono alcohols effect on the structural conformation of Bacillus clausii, GMBAE 42 serine alkaline protease. J. Mol. Catal. B Enzym. 2010, 64, 184–188. [Google Scholar] [CrossRef]
- Secundo, F.; Fialà, S.; Fraaije, M.W.; De Gonzalo, G.; Meli, M.; Zambianchi, F.; Ottolina, G. Effects of water miscible organic solvents on the activity and conformation of the baeyer-villiger monooxygenases from Thermobifida fusca and Acinetobacter calcoaceticus: A comparative study. Biotechnol. Bioeng. 2011, 108, 491–499. [Google Scholar] [CrossRef]
- Zaks, A.; Klibanov, A.M. Enzymatic catalysis in nonaqueous solvents. J. Biol. Chem. 1988, 263, 3194–3201. [Google Scholar] [PubMed]
- Wang, G.; Wang, Q.; Lin, X.; Ng, T.B.; Yan, R.; Lin, J.; Ye, X. A novel cold-adapted and highly salt-tolerant esterase from Alkalibacterium sp. SL3 from the sediment of a soda lake. Sci. Rep. 2016, 6, 19494. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Ma, J.; Wei, D.Z.; Lin, J.P.; Wei, W. Functional expression of a novel alkaline-adapted lipase of Bacillus amyloliquefaciens from stinky tofu brine and development of immobilized enzyme for biodiesel production. Antonie Van Leeuwenhoek 2014, 106, 1049–1060. [Google Scholar] [CrossRef]
- Arpigny, J.L.; Jaeger, K.E. Bacterial lipolytic enzymes: Classification and properties. Biochem. J. 1999, 343, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]


| Ion | Relative Activity (%) a | |
|---|---|---|
| 1 mM | 10 mM | |
| Control | 100 ± 2.8 | 100 ± 1.2 |
| Na2-EDTA | 96.7 ± 1.5 | 85.3 ± 2.1 |
| Co2+ | 78.1 ± 1.7 | 67.6 ± 2.8 |
| K+ | 150.2 ± 2.1 | 112.2 ± 2.8 |
| Fe3+ | 92.3 ± 3.2 | 55.1 ± 2.8 |
| Ca2+ | 121.3 ± 1.8 | 109.2 ± 1.7 |
| Mg2+ | 98.7 ± 2.3 | 76.5 ± 2.7 |
| Zn2+ | 98.5 ± 2.3 | 87.5 ± 1.2 |
| Ni2+ | 95.7 ± 1.4 | 85.1 ± 0.6 |
| Cu2+ | 93.8 ± 2.4 | 56.6 ± 2.7 |
| Organic Solvents | log P a | Residual Activity (%) b | ||
|---|---|---|---|---|
| 25 c | 50 c | 100 c | ||
| Control | 100 ± 3.4 | 100 ± 2.1 | 100 ± 3.2 | |
| DMSO | −1.3 | 76.6 ± 1.8 | 56.9 ± 2.7 | 32.1 ± 0.9 |
| Methanol | −0.76 | 93.6 ± 4.3 | 70.7 ± 1.2 | 38.8 ± 1.7 |
| Ethanol | −0.24 | 92.38 ± 3.5 | 71.3 ± 0.8 | 55.3 ± 4.1 |
| Acetone | −0.23 | 88.4 ± 1.9 | 66.9 ± 3.2 | 80.5 ± 3.3 |
| Acetonitrile | −0.15 | 90.2 ± 0.3 | 64.7 ± 2.1 | 97.2 ± 0.2 |
| Isopropyl alcohol | 0.1 | 94.5 ± 0.6 | 78.2 ± 1.2 | 41.2 ± 0.4 |
| n-Propyl alcohol | 0.28 | 97.6 ± 0.4 | 86.3 ± 0.7 | 50.8 ± 1.5 |
| Chloroform d | 2.0 | 70.5 ± 1.7 | 60.5 ± 1.7 | 24.3 ± 0.8 |
| Cyclohexane d | 3.2 | 89.2 ± 1.4 | 77.8 ± 1.4 | 87.4 ± 1.6 |
| n-Hexane d | 3.5 | 75.3 ± 2.4 | 66.7 ± 2.1 | 83.8 ± 1.8 |
| Isooctane d | 4.5 | 74.7 ± 2.5 | 62.1 ± 2.3 | 73.4 ± 2.6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Mao, X.; Lu, P.; Secundo, F.; Xue, C.; Sun, J. Cloning, Expression, and Characterization of a Novel Thermostable and Alkaline-stable Esterase from Stenotrophomonas maltophilia OUC_Est10 Catalytically Active in Organic Solvents. Catalysts 2019, 9, 401. https://doi.org/10.3390/catal9050401
Gao X, Mao X, Lu P, Secundo F, Xue C, Sun J. Cloning, Expression, and Characterization of a Novel Thermostable and Alkaline-stable Esterase from Stenotrophomonas maltophilia OUC_Est10 Catalytically Active in Organic Solvents. Catalysts. 2019; 9(5):401. https://doi.org/10.3390/catal9050401
Chicago/Turabian StyleGao, Xinwei, Xiangzhao Mao, Ping Lu, Francesco Secundo, Changhu Xue, and Jianan Sun. 2019. "Cloning, Expression, and Characterization of a Novel Thermostable and Alkaline-stable Esterase from Stenotrophomonas maltophilia OUC_Est10 Catalytically Active in Organic Solvents" Catalysts 9, no. 5: 401. https://doi.org/10.3390/catal9050401






