Dinuclear Molybdenum(VI) Complexes Based on Flexible Succinyl and Adipoyl Dihydrazones
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis
2.1.1. Synthesis of [Mo2O4(MeOH)2(L1)]∙2MeOH
2.1.2. Synthesis of [Mo2O4(MeOH)2(L2)]
2.1.3. Synthesis of [Mo2O4(MeOH)2(L3)]∙2MeOH
2.1.4. Synthesis of [Mo2O4(MeOH)2(L4)]
2.1.5. Synthesis of [Mo2O4(MeOH)2(L5)]
2.1.6. Synthesis of [Mo2O4(MeOH)2(L6)]
2.1.7. Synthesis of [Mo2O4(MeOH)2(L7)]
2.1.8. Synthesis of [Mo2O4(MeOH)2(L8)]∙2MeOH
2.2. Methods
3. Results and Discussion
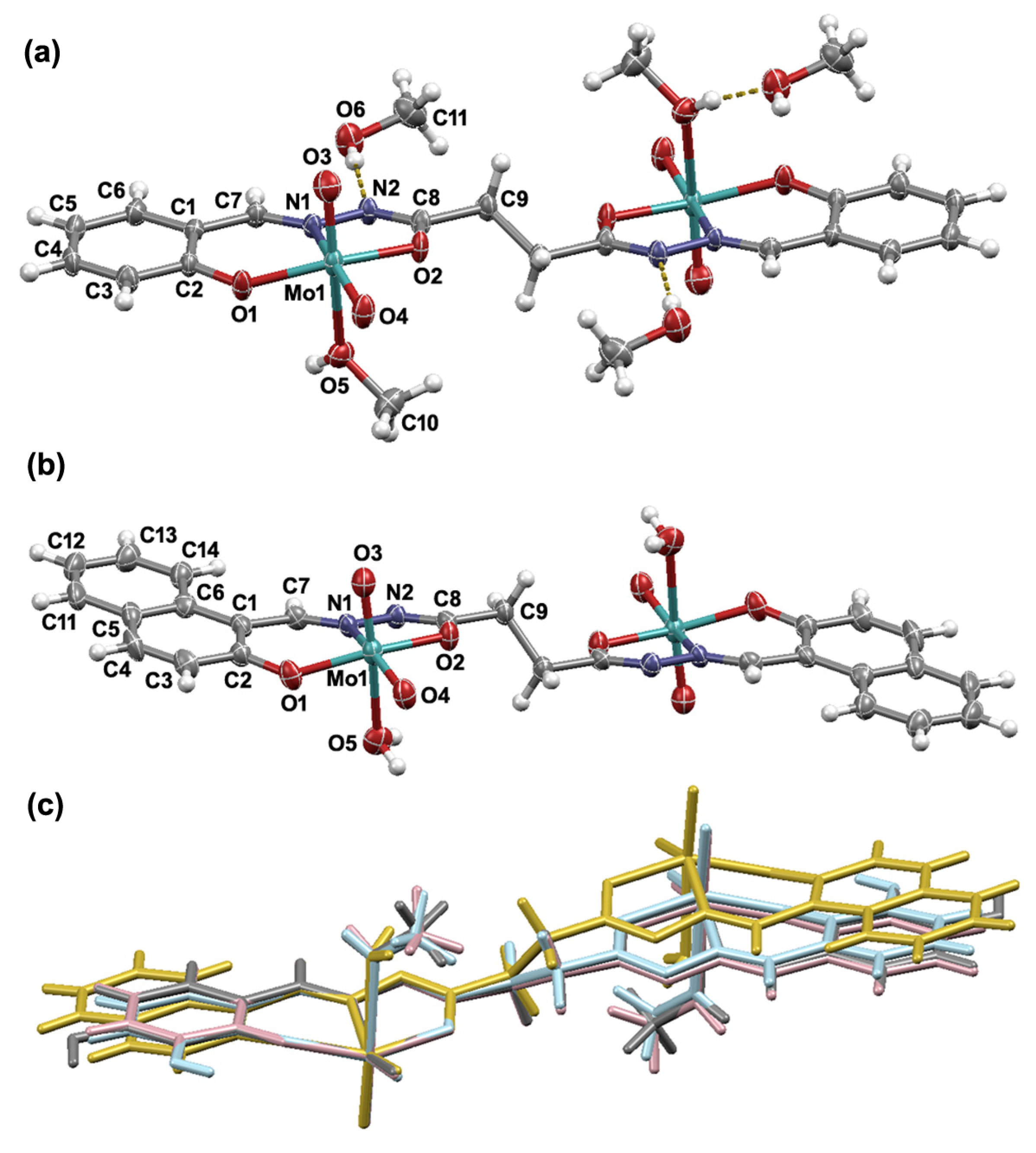
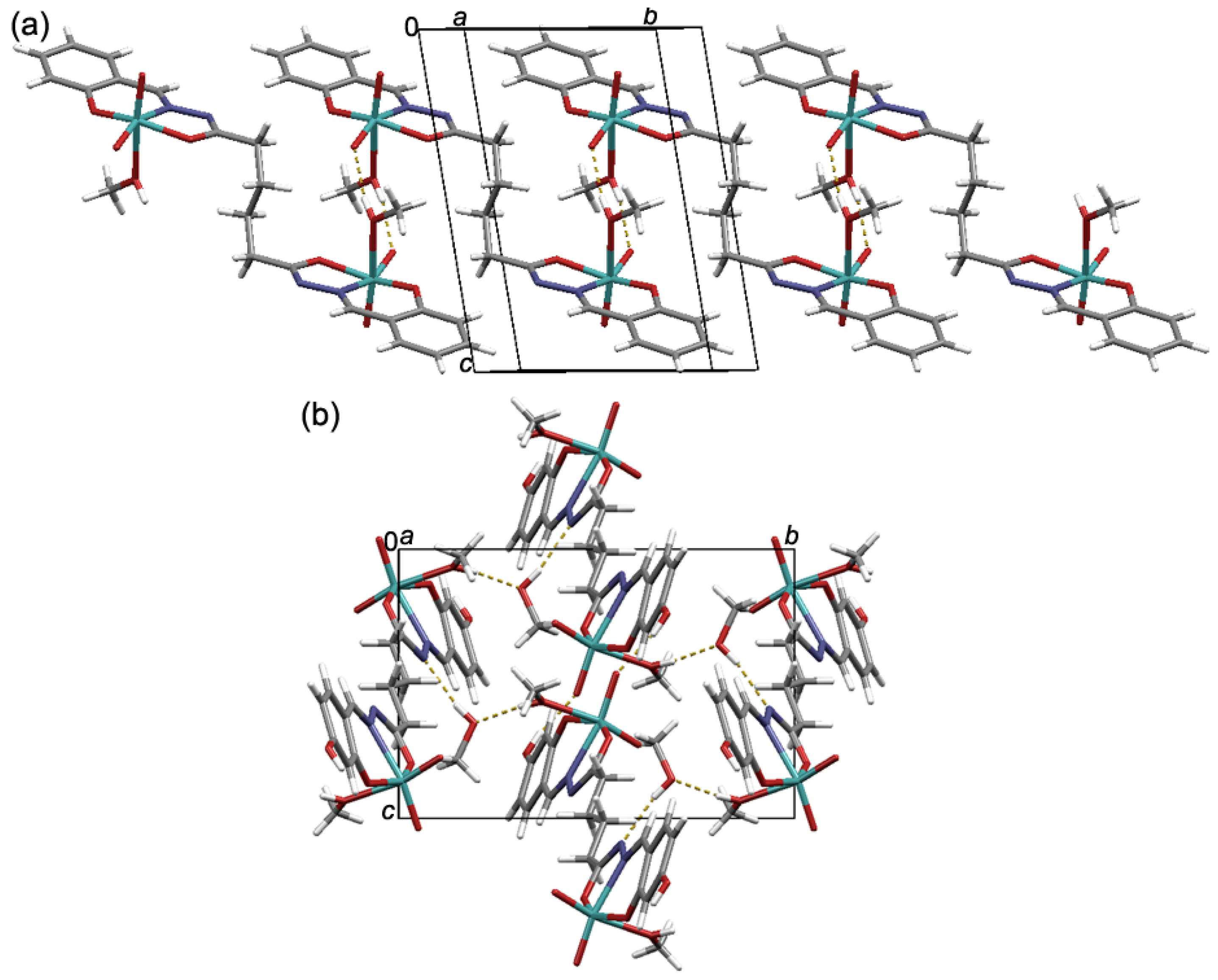
3.1. Synthesis and Solid-State Characterization
3.2. NMR and FT-IR Spectroscopy
3.3. In Vitro Cytotoxic and Antibacterial Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mali, S.N.; Thorat, B.R.; Gupta, D.R.; Pandey, A. Mini-Review of the Importance of Hydrazides and Their Derivatives—Synthesis and Biological Activity. Eng. Proc. 2021, 11, 21. [Google Scholar]
- Tatum, L.A.; Su, X.; Aprahamian, I. Simple Hydrazone Building Blocks for Complicated Functional Materials. Acc. Chem. Res. 2014, 47, 2141–2149. [Google Scholar] [CrossRef] [PubMed]
- Uribe-Romo, F.J.; Doonan, C.J.; Furukawa, H.; Oisaki, K.; Yaghi, O.M. Crystalline covalent organic frameworks with hydrazone linkages. J. Am. Chem. Soc. 2011, 133, 11478–11481. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.K.; Plutenko, M.O.; Schachner, J.A.; Haukka, M.; Mösch-Zanetti, N.C.; Fritsky, I.O. Dioxomolybdenum(VI) complexes of hydrazone phenolate ligands—Syntheses and activities in catalytic oxidation reactions. J. Indian Chem. Soc. 2021, 98, 100006. [Google Scholar] [CrossRef]
- Guskos, N.; Likodimos, V.; Glenis, S.; Typek, J.; Wabia, M.; Paschalidis, D.G.; Tossidis, I.; Lin, C.L. Magnetic properties of rare-earth hydrazone compounds. J. Magn. Magn. Mater. 2004, 272, 1067–1069. [Google Scholar] [CrossRef]
- Liu, R.; Cui, J.; Ding, T.; Liu, Y.; Liang, H. Research Progress on the Biological Activities of Metal Complexes Bearing Polycyclic Aromatic Hydrazones. Molecules 2022, 27, 8393. [Google Scholar] [CrossRef] [PubMed]
- Tupolova, Y.P.; Popov, L.D.; Vlasenko, V.G.; Gishko, K.B.; Kapustina, A.A.; Berejnaya, A.G.; Golubeva, Y.A.; Klyushova, L.S.; Lider, E.V.; Lazarenko, V.A.; et al. Crystal structure and cytotoxic activity of Cu(ii) complexes with bis-benzoxazolylhydrazone of 2,6-diacetylpyridine. New J. Chem. 2023, 47, 14972–14985. [Google Scholar] [CrossRef]
- Tupolova, Y.P.; Shcherbakov, I.N.; Popov, L.D.; Vlasenko, V.G.; Gishko, K.B.; Kapustina, A.A.; Berezhnaya, A.G.; Golubeva, Y.A.; Klyushova, L.S.; Lider, E.V.; et al. Copper coordination compounds based on bis-quinolylhydrazone of 2,6-diacetylpyridine: Synthesis, structure and cytotoxic activity. Polyhedron 2023, 233, 116292. [Google Scholar] [CrossRef]
- Su, X.; Aprahamian, I. Hydrazone-based switches, metallo-assemblies and sensors. Chem. Soc. Rev. 2014, 43, 1963–1981. [Google Scholar] [CrossRef]
- Zavalishin, M.N.; Gamov, G.A.; Pimenov, O.A.; Pogonin, A.E.; Aleksandriiskii, V.V.; Usoltsev, S.D.; Marfin, Y.S. Pyridoxal 5′-phosphate 2-methyl-3-furoylhydrazone as a selective sensor for Zn2+ ions in water and drug samples. J. Photochem. Photobiol. A 2022, 432, 114112. [Google Scholar] [CrossRef]
- Bagherian, N.; Karimi, A.R.; Amini, A. Chemically stable porous crystalline macromolecule hydrazone-linked covalent organic framework for CO2 capture. Colloids Surf. A 2021, 613, 126078. [Google Scholar] [CrossRef]
- Golla, U.; Adhikary, A.; Mondal, A.K.; Tomar, R.S.; Konar, S. Synthesis, structure, magnetic and biological activity studies of bis-hydrazone derived Cu(ii) and Co(ii) coordination compounds. Dalton Trans. 2016, 45, 11849–11863. [Google Scholar] [CrossRef]
- Wang, M.; Cheng, C.; Chunbo, L.; Wu, D.; Song, J.; Wang, J.; Zhou, X.; Xiang, H.; Liu, J. Smart, chiral, and non-conjugated cyclohexane-based bissalicylaldehyde hydrazides: Multi-stimuli-responsive, turn-on, ratiometric, and thermochromic fluorescence, single crystal structures, and DFT calculations. J. Mater. Chem. C 2019, 7, 6767–6778. [Google Scholar] [CrossRef]
- Chen, Z.; Zhou, S.; Shen, Y.; Zou, H.; Liu, D.; Liang, F. Copper(II) Clusters of Two Pairs of 2,3-Dihydroxybutanedioyl Dihydrazones: Synthesis, Structure, and Magnetic Properties. Eur. J. Inorg. Chem. 2014, 2014, 5783–5792. [Google Scholar] [CrossRef]
- Chen, Z.; Shen, Y.; Li, L.; Zou, H.; Fu, X.; Liu, Z.; Wang, K.; Liang, F. High-nuclearity heterometallic clusters with both an anion and a cation sandwiched by planar cluster units: Synthesis, structure and properties. Dalton Trans. 2017, 46, 15032–15039. [Google Scholar] [CrossRef] [PubMed]
- Mondal, A.K.; Jena, H.S.; Malviya, A.; Konar, S. Lanthanide-Directed Fabrication of Four Tetranuclear Quadruple Stranded Helicates Showing Magnetic Refrigeration and Slow Magnetic Relaxation. Inorg. Chem. 2016, 55, 5237–5244. [Google Scholar] [CrossRef] [PubMed]
- Schattschneider, C.; Doniz Kettenmann, S.; Hinojosa, S.; Heinrich, J.; Kulak, N. Biological activity of amphiphilic metal complexes. Coord. Chem. Rev. 2019, 385, 191–207. [Google Scholar] [CrossRef]
- Liang, J.; Sun, D.; Yang, Y.; Li, M.; Li, H.; Chen, L. Discovery of metal-based complexes as promising antimicrobial agents. Eur. J. Med. Chem. 2021, 224, 113696. [Google Scholar] [CrossRef]
- Verma, G.; Marella, A.; Shaquiquzzaman, M.; Akhtar, M.; Ali, M.R.; Alam, M.M. A review exploring biological activities of hydrazones. J. Pharm. Bioallied Sci. 2014, 6, 69–80. [Google Scholar]
- Le Goff, G.; Ouazzani, J. Natural hydrazine-containing compounds: Biosynthesis, isolation, biological activities and synthesis. Bioorg. Med. Chem. 2014, 22, 6529–6544. [Google Scholar] [CrossRef]
- Kumar, P.; Narasimhan, B. Hydrazides/hydrazones as antimicrobial and anticancer agents in the new millennium. Mini Rev. Med. Chem. 2013, 13, 971–987. [Google Scholar] [CrossRef] [PubMed]
- Popiołek, Ł. Hydrazide-hydrazones as potential antimicrobial agents: Overview of the literature since 2010. Med. Chem. Res. 2017, 26, 287–301. [Google Scholar] [CrossRef]
- Arora, T.; Devi, J.; Boora, A.; Taxak, B.; Rani, S. Synthesis and characterization of hydrazones and their transition metal complexes: Antimicrobial, antituberculosis and antioxidant activity. Res. Chem. Intermed. 2023, 49, 4819–4843. [Google Scholar] [CrossRef]
- Ullah, H.; Previtali, V.; Mihigo, H.B.; Twamley, B.; Rauf, M.K.; Javed, F.; Waseem, A.; Baker, R.J.; Rozas, I. Structure-activity relationships of new Organotin(IV) anticancer agents and their cytotoxicity profile on HL-60, MCF-7 and HeLa human cancer cell lines. Eur. J. Med. Chem. 2019, 181, 111544. [Google Scholar] [CrossRef] [PubMed]
- Topić, E.; Damjanović, V.; Pičuljan, K.; Vrdoljak, V.; Rubčić, M. Succinyl and Adipoyl Dihydrazones: A Solid-State, Solution and Antibacterial Study. Crystals 2022, 12, 1175. [Google Scholar] [CrossRef]
- Gherke, H., Jr.; Veal, J. Acetylacetonate complexes of molybdenum (V) and molybdenum (VI). I. Inorg. Chim. Acta 1969, 3, 623–627. [Google Scholar] [CrossRef]
- CrysAlisPro Software System, version 1.171.41.92a; Rigaku Oxford Diffraction: Oxford, UK, 2020.
- Sheldrick, G.M. SHELXT-Integrated space-group and crystal-structure determination. Acta Crystallograph. A 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallograph. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallograph. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Crystallograph. 2020, 53, 226–235. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Rubčić, M.; Pisk, J.; Pičuljan, K.; Damjanović, V.; Lovrić, J.; Vrdoljak, V. Symmetrical disubstituted carbohydrazides: From solid-state structures to cytotoxic and antibacterial activity. J. Mol. Struct. 2019, 1178, 222–228. [Google Scholar] [CrossRef]
- CLSI Document M07-A8; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2009.
- Ngan, N.K.; Lo, K.M.; Wong, C.S.R. Dinuclear and polynuclear dioxomolybdenum(VI) Schiff base complexes: Synthesis, structural elucidation, spectroscopic characterization, electrochemistry and catalytic property. Polyhedron 2012, 33, 235. [Google Scholar] [CrossRef]
- Kurbah, S.D.; Kumar, A.; Shangpung, S.; Syiemlieh, I.; Khongjoh, I.; Lal, R.A. Synthesis, Characterization, and Fluorescence Chemosensor Properties of a cis-Dioxomolybdenum(VI) Complex Containing Multidentate Hydrazone Ligands. Z. Anorg. Allg. Chem. 2017, 643, 794–801. [Google Scholar] [CrossRef]
- Kurbah, S.D.; Kumar, A.; Syiemlieh, I.; Asthana, M.; Lal, R.A. Bimetallic cis-dioxomolybdenum(VI) complex containing hydrazone ligand: Syntheses, crystal structure and catalytic studies. Inorg. Chem. Commun. 2017, 86, 39–43. [Google Scholar] [CrossRef]
- Vrdoljak, V.; Hrenar, T.; Rubčić, M.; Pavlović, G.; Friganović, T.; Cindrić, M. Ligand-Modulated Nuclearity and Geometry in Nickel(II) Hydrazone Complexes: From Mononuclear Complexes to Acetato- and/or Phenoxido-Bridged Clusters. Int. J. Mol. Sci. 2023, 24, 1909. [Google Scholar] [CrossRef]
- Maurya, M.R.; Saini, N.; Avecilla, F. Effect of N-based additive on the optimization of liquid phase oxidation of bicyclic, cyclic and aromatic alcohols catalyzed by dioxidomolybdenum(vi) and oxidoperoxidomolybdenum(vi) complexes. RSC Adv. 2015, 5, 101076. [Google Scholar] [CrossRef]
- Pisk, J.; Rubčić, M.; Kuzman, D.; Cindrić, M.; Agustin, D.; Vrdoljak, V. Molybdenum(vi) complexes of hemilabile aroylhydrazone ligands as efficient catalysts for greener cyclooctene epoxidation: An experimental and theoretical approach. New J. Chem. 2019, 43, 5531–5542. [Google Scholar] [CrossRef]
- Berg, J.M.; Holm, R.H. Structure proofs of ligated and polymeric dioxomolybdenum(VI)-tridentate complexes: MoO2(C5H3N-2,6-(CH2S)2)(C4H8SO) and [MoO2(C5H3N-2,6-(CH2O)2)]n. Inorg. Chem. 1983, 22, 1768–1771. [Google Scholar] [CrossRef]


| Compound | IC50 (μmol L−1) | MIC (μg mL−1) | ||||
|---|---|---|---|---|---|---|
| THP-1 | HepG2 | S. aureus | E. faecalis | E. coli | M. catarrhalis | |
| 1-Mo | 19.58 | >100 | >256 | 64 | >256 | 32 |
| 2-Mo | 7.04 | >100 | 32 | 32 | 32 | 4 |
| 3-Mo | >100 | >100 | 256 | 256 | 128 | 128 |
| 4-Mo | >100 | >100 | >256 | 128 | >256 | >256 |
| 5-Mo | >100 | >100 | >256 | >256 | >256 | >256 |
| 6-Mo | 9.78 | 57.98 | >256 | 128 | 128 | 2 |
| 7-Mo | >100 | >100 | >256 | 32 | 64 | 64 |
| 8-Mo | >100 | >100 | >256 | >256 | >256 | >256 |
| staurosporine | 0.10 | 7.98 | – | – | – | – |
| azithromycin | – | – | 1 | 8 | 0.50 | 0.125 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Topić, E.; Damjanović, V.; Pičuljan, K.; Rubčić, M. Dinuclear Molybdenum(VI) Complexes Based on Flexible Succinyl and Adipoyl Dihydrazones. Crystals 2024, 14, 135. https://doi.org/10.3390/cryst14020135
Topić E, Damjanović V, Pičuljan K, Rubčić M. Dinuclear Molybdenum(VI) Complexes Based on Flexible Succinyl and Adipoyl Dihydrazones. Crystals. 2024; 14(2):135. https://doi.org/10.3390/cryst14020135
Chicago/Turabian StyleTopić, Edi, Vladimir Damjanović, Katarina Pičuljan, and Mirta Rubčić. 2024. "Dinuclear Molybdenum(VI) Complexes Based on Flexible Succinyl and Adipoyl Dihydrazones" Crystals 14, no. 2: 135. https://doi.org/10.3390/cryst14020135






