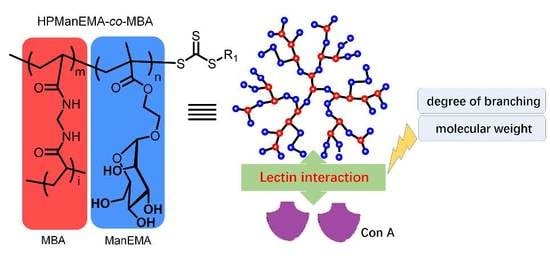
Hyperbranched Glycopolymers of 2-(α-d-Mannopyranose) Ethyl Methacrylate and N,N’-Methylenebisacrylamide: Synthesis, Characterization and Multivalent Recognitions with Concanavalin A
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials
2.2. Measurements
2.3. Synthesis of HPManEMA-co-MBA
3. Results and Discussion
3.1. Reaction Mechanism of HPAcManEMA-co-MBA
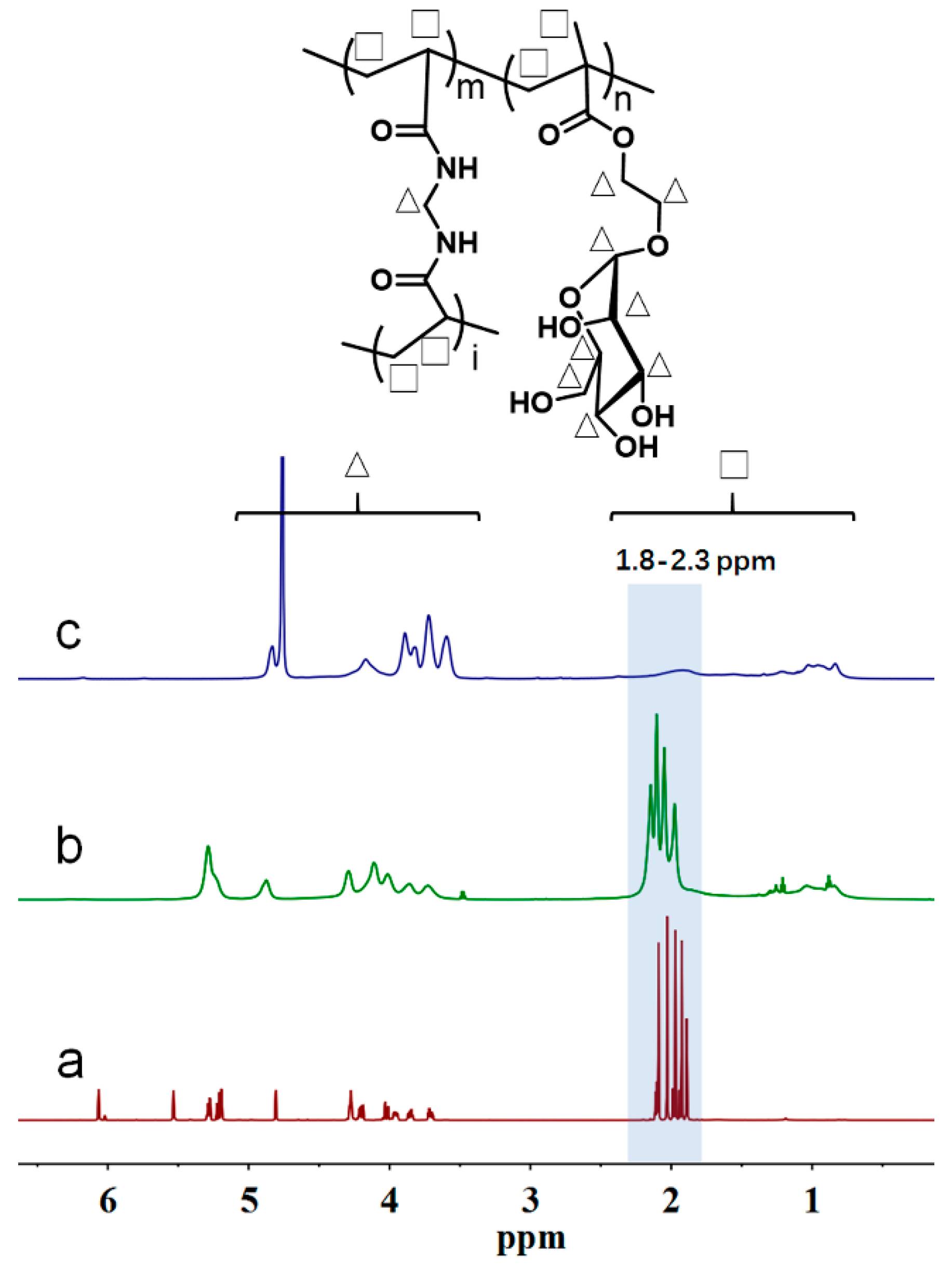
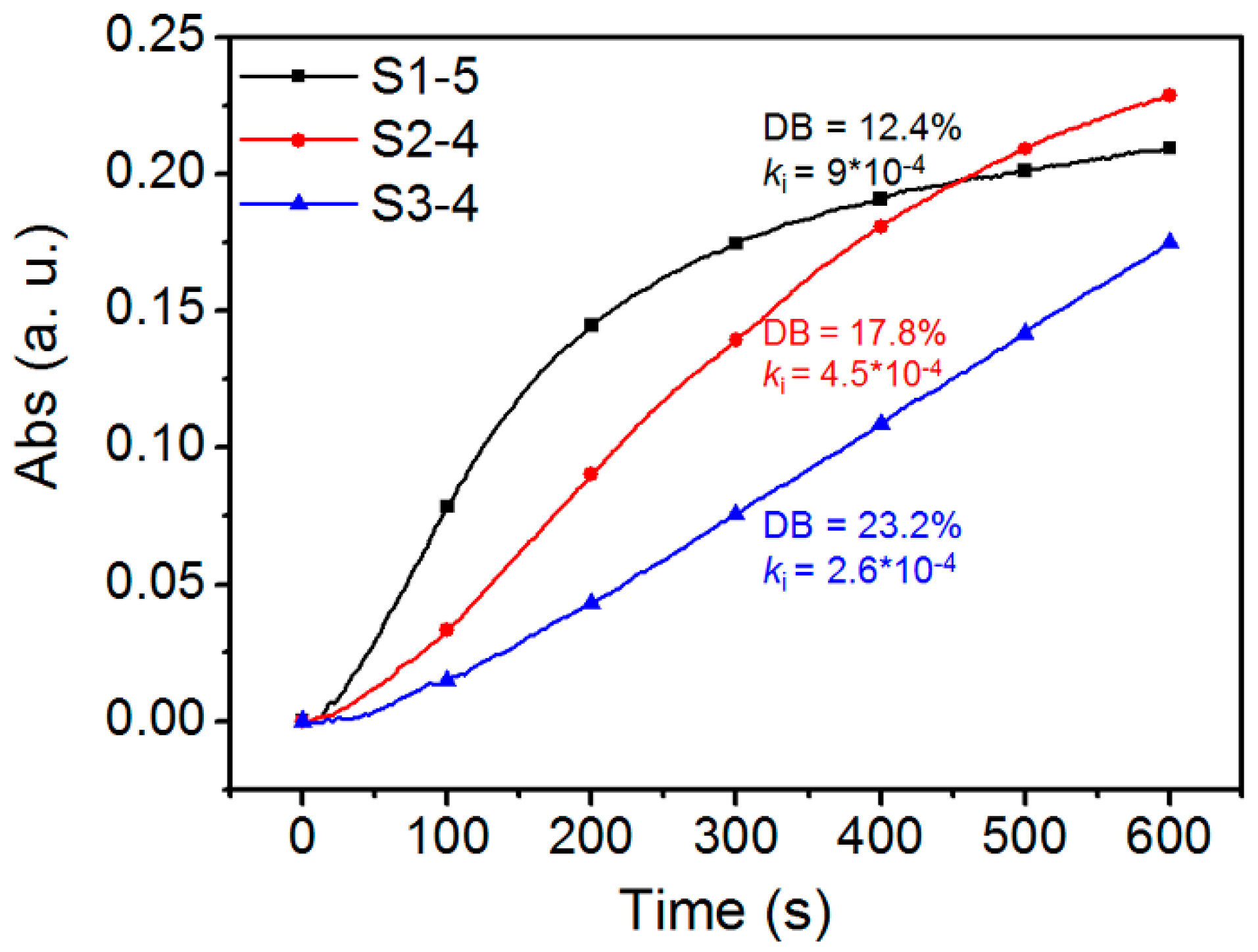
3.2. Determination of DB Value
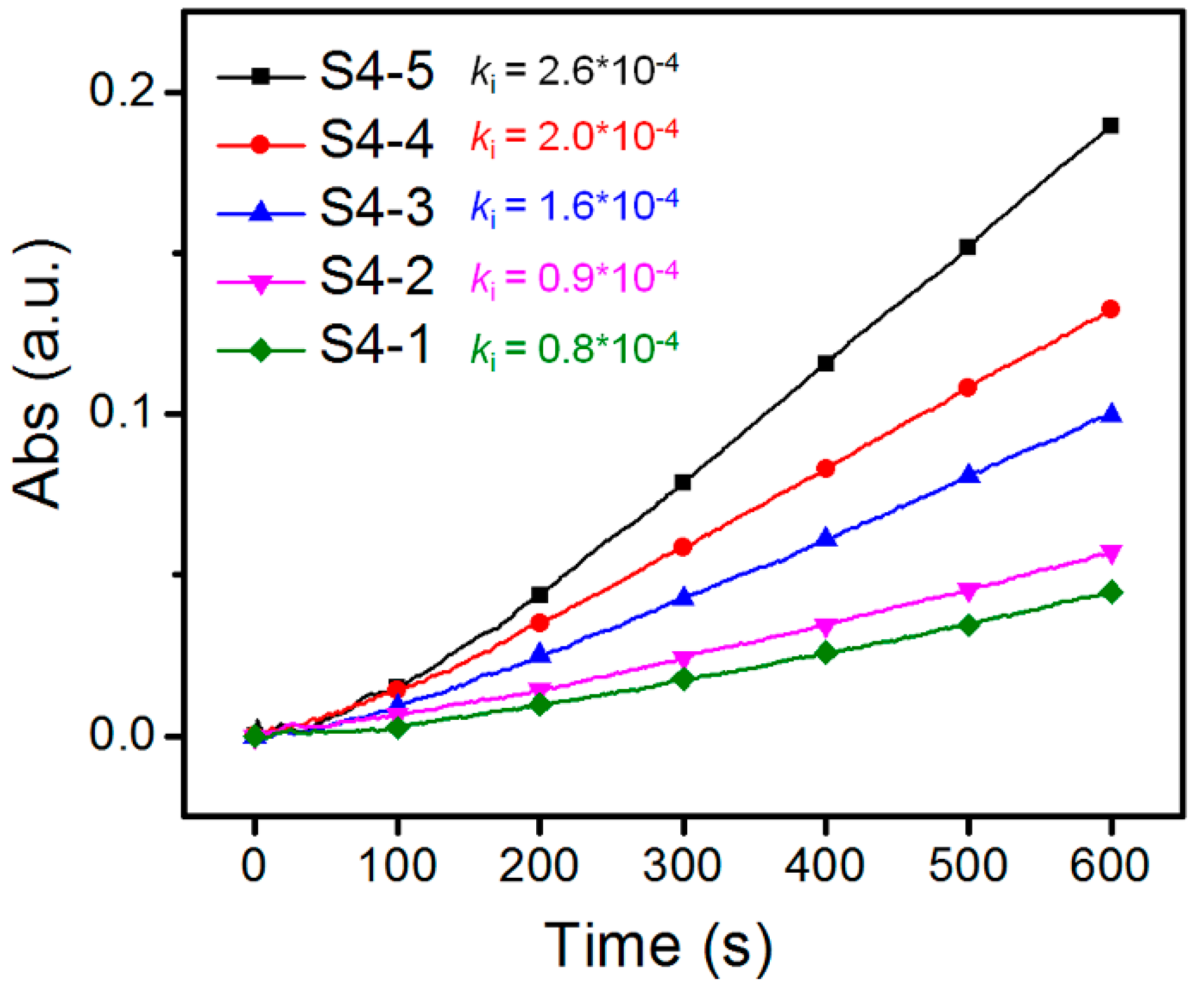
3.3. Effect of DB and Mn on The Binding Behavior between HPManEMA-co-MBA and Con A
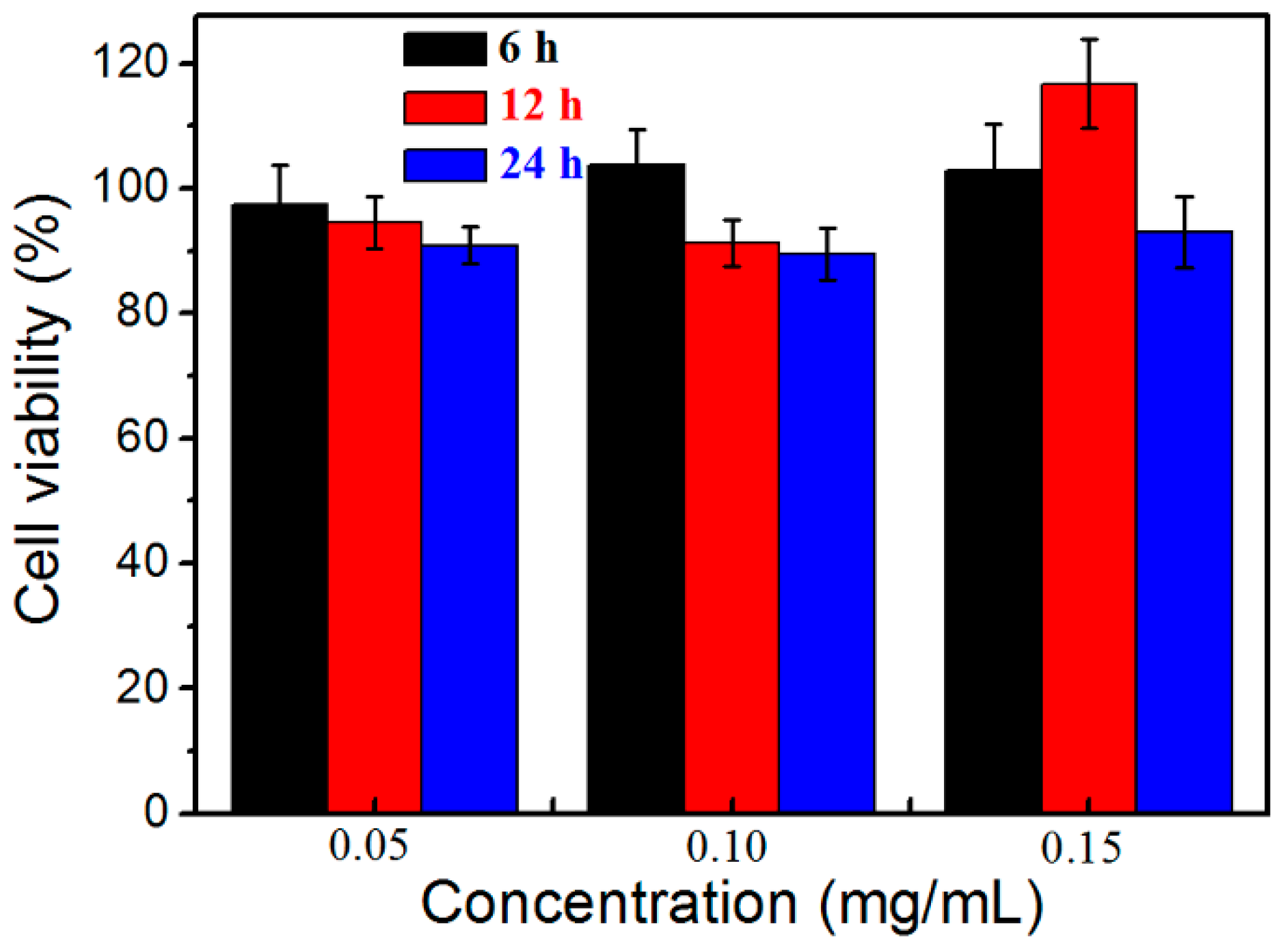
3.4. Cytotoxicity Test of HPManEMA-co-MBA
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cecioni, S.; Imberty, A.; Vidal, S. Glycomimetics versus multivalent glycoconjugates for the design of high affinity lectin ligands. Chem. Rev. 2015, 115, 525–561. [Google Scholar] [CrossRef] [PubMed]
- Moog, K.E.; Barz, M.; Bartneck, M.; Beceren-Braun, F.; Mohr, N.; Wu, Z.; Braun, L.; Dernedde, J.; Liehn, E.A.; Tacke, F.; et al. Polymeric selectin ligands mimicking complex carbohydrates: From selectin binders to modifiers of macrophage migration. Angew. Chem. Int. Ed. 2017, 56, 1416–1421. [Google Scholar] [CrossRef] [PubMed]
- Isono, T.; Miyachi, K.; Satoh, Y.; Nakamura, R.; Zhang, Y.; Otsuka, I.; Tajima, K.; Kakuchi, T.; Borsali, R.; Satoh, T. Self-assembly of maltoheptaose-block-polycaprolactone copolymers: Carbohydrate-decorated nanoparticles with tunable morphology and size in aqueous media. Macromolecules 2016, 49, 4178–4194. [Google Scholar] [CrossRef]
- Spain, S.G.; Cameron, N.R. A spoonful of sugar: The application of glycopolymers in therapeutics. Polym. Chem. 2011, 2, 60–68. [Google Scholar] [CrossRef]
- Chen, K.; Bao, M.; Bonilla, A.M.; Zhang, W.; Chen, G. A biomimicking and electrostatic self-assembly strategy for the preparation of glycopolymer decorated photoactive nanoparticles. Polym. Chem. 2016, 7, 2565–2572. [Google Scholar] [CrossRef]
- Lavilla, C.; Yilmaz, G.; Uzunova, V.; Napier, R.M.; Becer, C.R.; Heise, A. Block-sequence-specific glycopolypeptides with selective lectin binding properties. Biomacromolecules 2017, 18, 1928–1936. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Amajjahe, S.; Stenzel, M.H. Synthesis of thiol-linked neoglycopolymers and thermo-responsive glycomicelles as potential drug carrier. Chem. Commun. 2009, 10, 1198–1200. [Google Scholar] [CrossRef] [PubMed]
- Lavilla, C.; Byrne, M.; Heise, A. Block-sequence-specific polypeptides from α-amino acid N-carboxyanhydrides: Synthesis and influence on polypeptide properties. Macromolecules 2016, 49, 2942–2947. [Google Scholar] [CrossRef]
- Salvadó, M.; Reina, J.J.; Rojo, J.; Castillón, S.; Boutureira, O. Topological defects in hyperbranched glycopolymers enhance binding to lectins. Chem.-Eur. J. 2017, 23, 15790–15794. [Google Scholar] [CrossRef] [PubMed]
- Ting, S.R.S.; Chen, G.; Stenzel, M.H. Synthesis of glycopolymers and their multivalent recognitions with lectins. Polym. Chem. 2010, 1, 1392–1412. [Google Scholar] [CrossRef]
- Ruiz, C.; Sánchez-Chaves, M.; Cerrada, M.L.; Fernández-García, M. Glycopolymers resulting from ethylene-vinyl alcohol copolymers: Synthetic approach, characterization, and interactions with lectins. J. Polym. Sci. Part A Polym. Chem. 2008, 46, 7238–7248. [Google Scholar] [CrossRef]
- Chen, Q.; Xu, Y.; Du, Y.; Han, B.H. Triphenylamine-based fluorescent conjugated glycopolymers: Synthesis, characterization and interactions with lectins. Polymer 2009, 50, 2830–2835. [Google Scholar] [CrossRef]
- Mandal, D.K.; Brewer, C.F. Differences in the binding affinities of dimeric concanavalin A (including acetyl and succinyl derivatives) and tetrameric concanavalin a with large oligomannose-type glycopeptides. Biochemistry 1993, 32, 5116–5120. [Google Scholar] [CrossRef] [PubMed]
- Munoz, E.M.; Correa, J.; Fernandez-Megia, E.; Riguera, R. Probing the relevance of lectin clustering for the rellable evaluation of multivalent carbohydrate recognition. J. Am. Chem. Soc. 2009, 131, 17765–17767. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, J.; Gleinich, A.S.; Zhang, Q.; Whitfield, R.; Kempe, K.; Haddleton, D.M.; Davis, T.P.; Perrier, S.; Mitchell, D.A.; Wilson, P. Specific and differential binding of N-acetylgalactosamine glycopolymers to the human macrophage galactose lectin and asialoglycoprotein receptor. Biomacromolecules 2017, 18, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Nagao, M.; Fujiwara, Y.; Matsubara, T.; Hoshino, Y.; Sato, T.; Miura, Y. Design of glycopolymers carrying sialyl oligosaccharides for controlling the interaction with the influenza virus. Biomacromolecules 2017, 18, 4385–4392. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.; Byrne, A.; Gathergood, N.; Keyes, T.E.; Heuts, J.P.A.; Heise, A. Facile synthesis of fluorescent latex nanoparticles with selective binding properties using amphiphilic glycosylated polypeptide surfactants. Macromolecules 2014, 47, 7303–7310. [Google Scholar] [CrossRef]
- Basuki, J.S.; Esser, L.; Duong, H.T.T.; Zhang, Q.; Wilson, P.; Whittaker, M.R.; Haddleton, D.M.; Boyer, C.; Davis, T.P. Magnetic nanoparticles with diblock glycopolymer shells give lectin concentration-dependent MRI signals and selective cell uptake. Chem. Sci. 2014, 5, 715–726. [Google Scholar] [CrossRef]
- Zheng, Q.; Collins, J.; Anastasaki, A.; Wallis, R.; Mitchell, D.; Becer, C.R.; Haddleton, D.M. Sequence-controlled multi-block glycopolymers to inhibit DC-SIGN-gp120 binding. Angew. Chem. Int. Ed. 2013, 52, 4435–4439. [Google Scholar] [CrossRef] [PubMed]
- Becer, C.R.; Gibson, M.I.; Geng, J.; IIyas, R.; Wallis, R.; Mitchell, D.A.; Haddleton, D.M. High-affinity glycopolymer binding to human DC-SIGN and disruption of DC-SIGN interactions with HIV envelope glycoprotein. J. Am. Chem. Soc. 2010, 132, 15130–15132. [Google Scholar] [CrossRef] [PubMed]
- Von der Ehe, C.; Weber, C.; Gottschaldt, M.; Schubert, U.S. Immobilized glycopolymers: Synthesis, methods and applications. Prog. Polym. Sci. 2016, 57, 64–102. [Google Scholar] [CrossRef]
- Miura, Y.; Hoshino, Y.; Seto, H. Glycopolymer nanobiotechnology. Chem. Rev. 2015, 116, 1673–1692. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gao, H. Recent progress on hyperbranched polymers synthesized via radical-based self-condensing vinyl polymerization. Polymers 2017, 9, 188. [Google Scholar] [CrossRef]
- Ganda, S.; Jiang, Y.; Thomas, D.S.; Eliezar, J.; Stenzel, M.H. Biodegradable glycopolymeric micelles obtained by RAFT-controlled radical ring-opening polymerization. Macromolecules 2016, 49, 4136–4146. [Google Scholar] [CrossRef]
- Hoai, N.T.; Sasaki, A.; Sasaki, M.; Kaga, H.; Kakuchi, T.; Satoh, T. Synthesis, characterization, and lectin recognition of hyperbranched polysaccharide obtained from 1,6-anhydro-D-hexofuranose. Biomacromolecules 2011, 12, 1891–1899. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Kasko, A.M. Effect of branching density on avidity of hyperbranched glycomimetics for mannose binding lectin. Biomacromolecules 2013, 14, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Vandewalle, S.; Wallyn, S.; Chattopadhyay, S.; Becer, C.R.; Du Prez, F. Thermoresponsive hyperbranched glycopolymers: Synthesis, characterization and lectin interaction studies. Eur. Polym. J. 2015, 69, 490–498. [Google Scholar] [CrossRef]
- Ahmed, M.; Lai, B.F.; Kizhakkedathu, J.N.; Narain, R. Hyperbranched glycopolymers for blood biocompatibility. Bioconjugate Chem. 2012, 23, 1050–1058. [Google Scholar] [CrossRef] [PubMed]
- Semsarilar, M.; Ladmiral, V.; Perrier, S. Highly branched and hyperbranched glycopolymers via reversible addition-fragmentation chain transfer polymerization and click chemistry. Macromolecules 2010, 43, 1438–1443. [Google Scholar] [CrossRef]
- Cook, A.B.; Barbey, R.; Burns, J.A.; Perrier, S. Hyperbranched polymers with high degrees of branching and low dispersity values: Pushing the limits of thiol-yne chemistry. Macromolecules 2016, 49, 1296–1304. [Google Scholar] [CrossRef]
- Shi, Y.; Graff, R.W.; Cao, X.; Wang, X.; Gao, H. Chain-growth click polymerization of AB2 monomers for the formation of hyperbranched polymers with low polydispersities in a one-pot process. Angew. Chem. Int. Ed. 2015, 54, 7631–7635. [Google Scholar] [CrossRef] [PubMed]
- Graff, R.W.; Wang, X.; Gao, H. Exploring self-condensing vinyl polymerization of inimers in microemulsion to regulate the structures of hyperbranched polymers. Macromolecules 2015, 48, 2118–2126. [Google Scholar] [CrossRef]
- Wu, W.; Li, Z. Further improvement of the macroscopic NLO coefficient and optical transparency of hyperbranched polymers by enhancing the degree of branching. Polym. Chem. 2014, 5, 5100–5108. [Google Scholar] [CrossRef]
- Pranantyo, D.; Xu, L.Q.; Hou, Z.; Kang, E.T.; Chan-Park, M.B. Increasing bacterial affinity and cytocompatibility with four-arm star glycopolymers and antimicrobial α-polylysine. Polym. Chem. 2017, 8, 3364–3373. [Google Scholar] [CrossRef]
- Ghadban, A.; Reynaud, E.; Rinaudo, M.; Albertin, L. RAFT copolymerization of alginate-derived macromonomers-synthesis of a well-defined poly(HEMAm)-graft-(1→4)-α-l-guluronan copolymer capable of ionotropic gelation. Polym. Chem. 2013, 4, 4578–4583. [Google Scholar] [CrossRef]
- Pröhl, M.; Seupel, S.; Sungur, P.; Höppener, S.; Gottschaldt, M.; Brendel, J.C.; Schubert, U.S. The influence of the grafting density of glycopolymers on the lectin binding affinity of block copolymer micelles. Polymer 2017, 133, 205–212. [Google Scholar] [CrossRef]
- Zhou, S.; Zhang, D.; Bai, L.; Zhao, J.; Wu, Y.; Zhao, H.; Ba, X. The synthesis of backbone thermo and pH responsive hyperbranched poly (bis (N,N-propyl acryl amide))s by RAFT. Polymers 2016, 8, 135. [Google Scholar] [CrossRef]
- Voit, B.I.; Lederer, A. Hyperbranched and highly branched polymer architectures-synthetic strategies and major characterization aspects. Chem. Rev. 2009, 109, 5924–5973. [Google Scholar] [CrossRef] [PubMed]
- Obata, M.; Shimizu, M.; Ohta, T.; Matsushige, A.; Iwai, K.; Hirohara, S.; Tanihara, M. Synthesis, characterization and cellular internalization of poly(2-hydroxyethyl methacrylate) bearing α-d-mannopyranose. Polym. Chem. 2011, 2, 651–658. [Google Scholar] [CrossRef]
- Munoz, E.M.; Correa, J.; Riguera, R.; Fernandez-Megia, E. Real-time evaluation of binding mechanisms in multivalent interactions: A surface plasmon resonance kinetic approach. J. Am. Chem. Soc. 2013, 135, 5966–5969. [Google Scholar] [CrossRef] [PubMed]
- Cairo, C.W.; Gestwicki, J.E.; Kanai, M.; Kiessling, L.L. Control of multivalent interactions by binding epitope density. J. Am. Chem. Soc. 2002, 124, 1615–1619. [Google Scholar] [CrossRef] [PubMed]

| Sample | [AcManEMA]:[MBA] | Yield% | Mn × 104 | Mw × 104 | α |
|---|---|---|---|---|---|
| S1 | 100:3 | 47.8 | 8.8 | 9.8 | 0.70 |
| S2 | 100:5 | 48.2 | 8.5 | 9.6 | 0.63 |
| S3 | 100:10 | 48.3 | 10.2 | 11.5 | 0.55 |
| S4 | 100:20 | 58.6 | 16.2 | 20.8 | 0.40 |
| Elemental | C/% | N/% | H/% | R(ManEMA:MBA) | DB% |
|---|---|---|---|---|---|
| S1 | 44.45 | 0.55 | 6.67 | 15.13 | 12.4 |
| S2 | 44.47 | 0.80 | 6.85 | 10.23 | 17.8 |
| S3 | 44.29 | 1.06 | 6.81 | 7.62 | 23.2 |
| S4 | 43.22 | 1.37 | 5.22 | 5.55 | 30.6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Wang, B.; Zhang, Y.; Zheng, Y.; Wen, X.; Bai, L.; Wu, Y. Hyperbranched Glycopolymers of 2-(α-d-Mannopyranose) Ethyl Methacrylate and N,N’-Methylenebisacrylamide: Synthesis, Characterization and Multivalent Recognitions with Concanavalin A. Polymers 2018, 10, 171. https://doi.org/10.3390/polym10020171
Zhang Y, Wang B, Zhang Y, Zheng Y, Wen X, Bai L, Wu Y. Hyperbranched Glycopolymers of 2-(α-d-Mannopyranose) Ethyl Methacrylate and N,N’-Methylenebisacrylamide: Synthesis, Characterization and Multivalent Recognitions with Concanavalin A. Polymers. 2018; 10(2):171. https://doi.org/10.3390/polym10020171
Chicago/Turabian StyleZhang, Yuangong, Bo Wang, Ye Zhang, Ying Zheng, Xin Wen, Libin Bai, and Yonggang Wu. 2018. "Hyperbranched Glycopolymers of 2-(α-d-Mannopyranose) Ethyl Methacrylate and N,N’-Methylenebisacrylamide: Synthesis, Characterization and Multivalent Recognitions with Concanavalin A" Polymers 10, no. 2: 171. https://doi.org/10.3390/polym10020171