Column-Free Purification Methods for Recombinant Proteins Using Self-Cleaving Aggregating Tags
Abstract
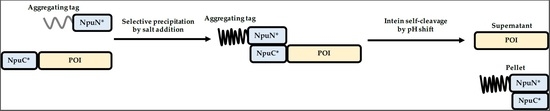
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Plasmid Construction
2.3. Protein Expression
2.4. Lysis and Recovery
2.5. ELP-Mediated Protein Purification
2.6. BRT17-Mediated Protein Purification
2.7. Intein Cleavage Analysis
2.8. Protein Quantification
2.9. Green Fluorescent Protein
2.10. β-Lactamase Activity Assay
2.11. Streptokinase Activity Assay
3. Results
3.1. pH Controllability of Aggragating Tags
3.2. Purification of Recombinant Proteins Using Self-Cleaving Elastin-Like Polypeptide (ELP) Tag
3.3. Purification of Recombinant Proteins Using the Self-Cleaving β-Roll Tag (BRT17)
4. Discussion
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Shukla, A.A.; Wolfe, L.S.; Mostafa, S.S.; Norman, C. Evolving trends in mab production processes. Bioeng. Transl. Med. 2017, 2, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Herzer, S. Downstream processing technologies/capturing and final purification: Opportunities for innovation, change, and improvement. A review of downstream processing developments in protein purification. Adv. Biochem. Eng. Biotechnol. 2017. [Google Scholar] [CrossRef]
- Terpe, K. Overview of tag protein fusions: From molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2003, 60, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Banki, M.R.; Feng, L.A.; Wood, D.W. Simple bioseparations using self-cleaving elastin-like polypeptide tags. Nat. Methods 2005, 2, 659–661. [Google Scholar] [CrossRef] [PubMed]
- Fong, B.A.; Wu, W.Y.; Wood, D.W. Optimization of elp-intein mediated protein purification by salt substitution. Protein Expr. Purif. 2009, 66, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Xing, L.; Zhou, B.; Lin, Z. Active protein aggregates induced by terminally attached self-assembling peptide elk16 in escherichia coli. Microb. Cell Fact. 2011, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Shur, O.; Dooley, K.; Blenner, M.; Baltimore, M.; Banta, S. A designed, phase changing rtx-based peptide for efficient bioseparations. Biotechniques 2013, 54, 197–198. [Google Scholar] [CrossRef] [PubMed]
- Raran-Kurussi, S.; Cherry, S.; Zhang, D.; Waugh, D.S. Removal of affinity tags with tev protease. Methods Mol. Biol. 2017, 1586, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, A.; Dutta, K.; Pascal, S.M. Mbp fusion protein with a viral protease cleavage site: One-step cleavage/purification of insoluble proteins. Biotechniques 2001, 30, 1194–1198. [Google Scholar] [PubMed]
- Block, H.; Maertens, B.; Spriestersbach, A.; Kubicek, J.; Schafer, F. Proteolytic affinity tag cleavage. Methods Enzymol. 2015, 559, 71–97. [Google Scholar] [CrossRef] [PubMed]
- Goh, H.C.; Sobota, R.M.; Ghadessy, F.J.; Nirantar, S. Going native: Complete removal of protein purification affinity tags by simple modification of existing tags and proteases. Protein Expr. Purif. 2017, 129, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Fong, B.A.; Wu, W.Y.; Wood, D.W. The potential role of self-cleaving purification tags in commercial-scale processes. Trends Biotechnol. 2010, 28, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.W.; Wu, W.; Belfort, G.; Derbyshire, V.; Belfort, M. A genetic system yields self-cleaving inteins for bioseparations. Nat. Biotechnol. 1999, 17, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.; Bacolla, A.; Wells, R.D.; Roberts, R.J. Recombinant human DNA (cytosine-5) methyltransferase I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 1999, 274, 33002–33010. [Google Scholar] [CrossRef] [PubMed]
- Babu, K.S.; Antony, A.; Muthukumaran, T.; Meenakshisundaram, S. Construction of intein-mediated hgmcsf expression vector and its purification in pichia pastoris. Protein Expr. Purif. 2008, 57, 201–205. [Google Scholar] [CrossRef]
- Morassutti, C.; De Amicis, F.; Skerlavaj, B.; Zanetti, M.; Marchetti, S. Production of a recombinant antimicrobial peptide in transgenic plants using a modified vma intein expression system. FEBS Lett. 2002, 519, 141–146. [Google Scholar] [CrossRef]
- Jun, Y.; Wickner, W. Assays of vacuole fusion resolve the stages of docking, lipid mixing, and content mixing. Proc. Natl. Acad. Sci. USA 2007, 104, 13010–13015. [Google Scholar] [CrossRef] [PubMed]
- Sands, D.; Whitton, C.M.; Longstaff, C. International collaborative study to establish the 3rd international standard for streptokinase. J. Thromb. Haemost. 2004, 2, 1411–1415. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.W.; Camarero, J.A. Intein applications: From protein purification and labeling to metabolic control methods. J. Biol. Chem. 2014, 289, 14512–14519. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.A.; Taris, J.E.; Shi, C.; Wood, D.W. A convenient split-intein tag method for the purification of tagless target proteins. Curr. Protoc. Protein Sci. 2018, 91. [Google Scholar] [CrossRef]
- Shi, C.; Meng, Q.; Wood, D.W. A dual ELP-tagged split intein system for non-chromatographic recombinant protein purification. Appl. Microbiol. Biotechnol. 2013, 97, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Aranko, A.S.; Wlodawer, A.; Iwai, H. Nature’s recipe for splitting inteins. Protein Eng. Des. Sel. 2014, 27, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Southworth, M.W.; Adam, E.; Panne, D.; Byer, R.; Kautz, R.; Perler, F.B. Control of protein splicing by intein fragment reassembly. EMBO J. 1998, 17, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Otomo, T.; Ito, N.; Kyogoku, Y.; Yamazaki, T. NMR observation of selected segments in a larger protein: Central-segment isotope labeling through intein-mediated ligation. Biochemistry 1999, 38, 16040–16044. [Google Scholar] [CrossRef] [PubMed]
- Zettler, J.; Schutz, V.; Mootz, H.D. The naturally split npu dnae intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 2009, 583, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Ramirez, M.; Chen, Z. Split intein mediated ultra-rapid purification of tagless protein (SIRP). Biotechnol. Bioeng. 2013, 110, 2471–2481. [Google Scholar] [CrossRef] [PubMed]
- Iwai, H.; Zuger, S.; Jin, J.; Tam, P.H. Highly efficient protein trans-splicing by a naturally split dnae intein from nostoc punctiforme. FEBS Lett. 2006, 580, 1853–1858. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Zhou, B.; Gao, X.; Xing, L.; Wang, X.; Lin, Z. A cleavable self-assembling tag strategy for preparing proteins and peptides with an authentic n-terminus. Biotechnol. J. 2017, 12. [Google Scholar] [CrossRef] [PubMed]





| Primer Name | Sequence 5′–3′ |
|---|---|
| BglII-Forward | ATCGAGATCTGACGTCCGATCCC |
| NpuC*-Blac-1 (overlap) | CTCAAAAATGGCTTCATAGCTCATAATATGCACCCAGAAACGCTGGTG |
| NpuC*-Blac-2 (overlap) | CACCAGCGTTTCTGGGTGCATATTATGAGCTATGAAGCCATTTTTGAG |
| Blac-2 (His-Stop-XhoI) | GCGCTCGAGTCAGTGATGATGATGATGATGCCAATGCTTAATCAGTGAGGC |
| NpuC*-sfGFP-1 (overlap) | CAAAAATGGCTTCATAGCTCATAATATGGTGAGCAAGGGCGAGGAGCTG |
| NpuC*-sfGFP-2 (overlap) | CAGCTCCTCGCCCTTGCTCACCATATTATGAGCTATGAAGCCATTTTTG |
| T7 terminator | GCCCCAAGGGGTTATGCTAG |
| NpuC*-MBP-1 (overlap) | CTCAAAAATGGCTTCATAGCTCATAATATGAAAATCGAAGAAGGTAAA |
| NpuC*-MBP-2 (overlap) | TTTACCTTCTTCGATTTTCATATTATGAGCTATGAAGCCATTTTTGAG |
| MBP-2 (His-Stop-XhoI) | GCGCTCGAGTCATTAATGATGATGATGATGATGCGAGCTCGAATTAGTCTGCGC |
| EcoRI-NpuN*-F | GCGGAATTCGGTGACGGTCACGGTGCCTTAAGCTATGAAACGGA |
| XbaI-NpuN*-R | GGCTCTAGATTAATTCGGCAAATTATCAACCCG |
| BRT17-I-F | GACCTAGAGGAGTAATAATAATAACAATAACAACAACCTCGGGATCGAGGGAAGGATTTC |
| BRT17-I-R | GACCTAAAGCTTTTAGTTGTGTACAACAACCCCTTCGGC |
| EcoRI-NpuN* Forward | GAATTCGGTGGAGGCGGGTCTGGTGACGGTCACGGTGCCTTAAG |
| HindIII-His6-NpuN*-R | ATATAAGCTTATGGTGATGGTGATGGTGATTCGGCAAATTATCAACCCGC |
| Aggregating Tag | Product Protein | Yield 1 (μg/mL) | Specific Activity (Unit/mg) | Recovery 2 |
|---|---|---|---|---|
| ELP | sfGFP | 338.5 ± 45.5 | Fluorescent under 485 nm excitation | 28.6 ± 0.1% |
| β-lactamase | 248.1 ± 43.2 | 167.2 ± 13.3 | 79.0 ± 4.4% | |
| Streptokinase | 389.0 ± 65.3 | 22600.5 ± 1851.9 | 30.3 ± 0.5% | |
| MBP | 223.0 ± 62.2 | Binds Maltose resin | NR 3 | |
| BRT17 | sfGFP | 291.5 ± 78.0 | Fluorescent under 485 nm excitation | 49.0 ± 8.9% |
| β-lactamase | 196.5 ± 27.1 | 84.1 ± 10.1 | 85.8 ± 7.8% | |
| Streptokinase | 210.7 ± 40.8 | 5874.7 ± 2234.9 | 11.8 ± 5.1% | |
| MBP | 214.0 ± 44.0 | Binds Maltose resin | NR 3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Y.; Miozzi, J.M.; Stimple, S.D.; Han, T.-C.; Wood, D.W. Column-Free Purification Methods for Recombinant Proteins Using Self-Cleaving Aggregating Tags. Polymers 2018, 10, 468. https://doi.org/10.3390/polym10050468
Fan Y, Miozzi JM, Stimple SD, Han T-C, Wood DW. Column-Free Purification Methods for Recombinant Proteins Using Self-Cleaving Aggregating Tags. Polymers. 2018; 10(5):468. https://doi.org/10.3390/polym10050468
Chicago/Turabian StyleFan, Yamin, Jackelyn M. Miozzi, Samuel D. Stimple, Tzu-Chiang Han, and David W. Wood. 2018. "Column-Free Purification Methods for Recombinant Proteins Using Self-Cleaving Aggregating Tags" Polymers 10, no. 5: 468. https://doi.org/10.3390/polym10050468