Co-Encapsulation and Co-Delivery of Peptide Drugs via Polymeric Nanoparticles
Abstract
:1. Introduction
2. Materials & Methods
2.1. Materials
2.2. Synthesis of mPEG-b-PCL, Allyl-PEG-b-PCL, TGN-PEG-b-PCL, COG133-PEG-b-PCL, and RXR-PEG-b-PCL
2.3. Preparation of NPs by Double Emulsion
2.4. Preparation of NPs by Nanoprecipitation in the Presence of POPG
2.5. Nanoparticle Characterization
2.6. Drug Encapsulation Efficiency and Release Study
2.7. Cell Uptake Study
2.8. Lactate Dehydrogenase (LDH) Cytotoxicity Assay
3. Results
3.1. Parameters of Double Emulsion (DE) Method Were Optimised to Achieve Suitable Size and Low Polydispersity Index (PDI)
3.2. Functionalisation of Polymers Affects Nanoparticle Size, PDI, Drug Encapsulation Efficiency (E.E.), and Drug Release Profile
3.3. Incorporation of Negatively-Charged POPG Lipid Increased Drug E.E. to Almost 100%
3.4. Functionalised Polymer Increased Cell Penetration of DE and POPG Nanoparticles
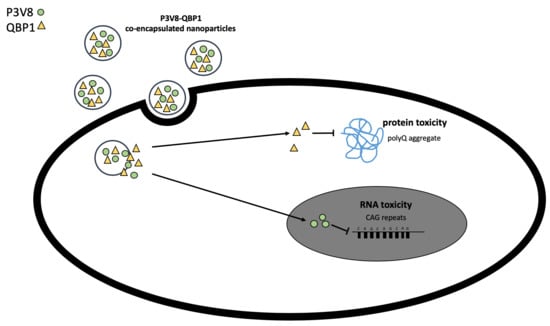
3.5. Encapsulation of Drugs into Either Functionalised DE or POPG NPs Enabled Cell Rescue from CAG RNA Toxicity
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Sun, T.-M.; Du, J.-Z.; Yao, Y.-D.; Mao, C.-Q.; Dou, S.; Huang, S.-Y.; Zhang, P.-Z.; Leong, K.W.; Song, E.-W.; Wang, J. Simultaneous Delivery of siRNA and Paclitaxel via a “Two-in-One” Micelleplex Promotes Synergistic Tumor Suppression. ACS Nano 2011, 5, 1483–1494. [Google Scholar] [CrossRef]
- Kato, T.; Natsume, A.; Toda, H.; Iwamizu, H.; Sugita, T.; Hachisu, R.; Watanabe, R.; Yuki, K.; Motomura, K.; Bankiewicz, K.; et al. Efficient delivery of liposome-mediated MGMT-siRNA reinforces the cytotoxity of temozolomide in GBM-initiating cells. Gene Ther. 2010, 17, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tsoi, H.; Peng, S.; Li, P.P.; Lau, K.-F.; Rudnicki, D.D.; Ngo, J.C.-K.; Chan, H.Y.E. Assessing a peptidylic inhibitor-based therapeutic approach that simultaneously suppresses polyglutamine RNA- and protein-mediated toxicities in patient cells and Drosophila. Dis. Model. Mech. 2016, 9, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Tucker, T.; Ren, H.; Kenan, D.J.; Henderson, B.S.; Keene, J.D.; Strittmatter, W.J.; Burke, J.R. Inhibition of Polyglutamine Protein Aggregation and Cell Death by Novel Peptides Identified by Phage Display Screening. J. Biol. Chem. 2000, 275, 10437–10442. [Google Scholar] [CrossRef]
- Popiel, H.A.; Burke, J.R.; Strittmatter, W.J.; Oishi, S.; Fujii, N.; Takeuchi, T.; Toda, T.; Wada, K.; Nagai, Y. The Aggregation Inhibitor Peptide QBP1 as a Therapeutic Molecule for the Polyglutamine Neurodegenerative Diseases. J. Amino Acids 2011, 2011, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Manning, M.C.; Chou, D.K.; Murphy, B.M.; Payne, R.W.; Katayama, D.S. Stability of Protein Pharmaceuticals: An Update. Pharm. Res. 2010, 27, 544–575. [Google Scholar] [CrossRef] [PubMed]
- Wardwell, P.R.; Bader, R.A. Challenges of Drug Delivery. In Engineering Polymer Systems for Improved Drug Delivery; Bader, R.A., Putnam, D.A., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014; pp. 29–54. ISBN 9781118747896. [Google Scholar]
- Kreuter, J. Nanoparticulate systems for brain delivery of drugs. Adv. Drug Deliv. Rev. 2012, 64, 213–222. [Google Scholar] [CrossRef]
- Tan, M.L.; Choong, P.F.M.; Dass, C.R. Recent developments in liposomes, microparticles and nanoparticles for protein and peptide drug delivery. Peptides 2010, 31, 184–193. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Hu, Y.; Yin, L.; Tang, C.; Yin, C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials 2010, 31, 3657–3666. [Google Scholar] [CrossRef]
- Alexis, F.; Pridgen, E.; Molnar, L.K.; Farokhzad, O.C. Factors Affecting the Clearance and Biodistribution of Polymeric Nanoparticles. Mol. Pharm. 2008, 5, 505–515. [Google Scholar] [CrossRef]
- Marin, E.; Briceño, M.I.; Caballero-George, C. Critical evaluation of biodegradable polymers used in nanodrugs. Int. J. Nanomed. 2013, 8, 3071–3090. [Google Scholar]
- Manavitehrani, I.; Fathi, A.; Badr, H.; Daly, S.; Negahi Shirazi, A.; Dehghani, F. Biomedical Applications of Biodegradable Polyesters. Polymers 2016, 8, 20. [Google Scholar] [CrossRef]
- Zhang, Q.; Yang, M.; Sørensen, K.K.; Madsen, C.S.; Boesen, J.T.; An, Y.; Peng, S.H.; Wei, Y.; Wang, Q.; Jensen, K.J.; et al. A brain-targeting lipidated peptide for neutralizing RNA-mediated toxicity in Polyglutamine Diseases. Sci. Rep. 2017, 7, 12077. [Google Scholar] [CrossRef] [PubMed]
- Ni, R.; Chau, Y. Structural Mimics of Viruses through Peptide/DNA Co-Assembly. J. Am. Chem. Soc. 2014, 136, 17902–17905. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chau, Y. Different oligoarginine modifications alter endocytic pathways and subcellular trafficking of polymeric nanoparticles. Biomater. Sci. 2016, 4, 1462–1472. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Feng, L.; Fan, L.; Zha, Y.; Guo, L.; Zhang, Q.; Chen, J.; Pang, Z.; Wang, Y.; Jiang, X.; et al. Targeting the brain with PEG–PLGA nanoparticles modified with phage-displayed peptides. Biomaterials 2011, 32, 4943–4950. [Google Scholar] [CrossRef] [PubMed]
- Zensi, A.; Begley, D.; Pontikis, C.; Legros, C.; Mihoreanu, L.; Wagner, S.; Büchel, C.; von Briesen, H.; Kreuter, J. Albumin nanoparticles targeted with Apo E enter the CNS by transcytosis and are delivered to neurones. J. Controll. Release 2009, 137, 78–86. [Google Scholar] [CrossRef]
- Du, L.; Kayali, R.; Bertoni, C.; Fike, F.; Hu, H.; Iversen, P.L.; Gatti, R.A. Arginine-rich cell-penetrating peptide dramatically enhances AMO-mediated ATM aberrant splicing correction and enables delivery to brain and cerebellum. Hum. Mol. Genet. 2011, 20, 3151–3160. [Google Scholar] [CrossRef]
- Tsoi, H.; Lau, C.K.; Lau, K.F.; Chan, H.Y.E. Perturbation of U2AF65/NXF1-mediated RNA nuclear export enhances RNA toxicity in polyQ diseases. Hum. Mol. Genet. 2011, 20, 3787–3797. [Google Scholar] [CrossRef]
- Huang, M.-H.; Li, S.; Hutmacher, D.W.; Schantz, J.-T.; Vacanti, C.A.; Braud, C.; Vert, M. Degradation and cell culture studies on block copolymers prepared by ring opening polymerization ofε-caprolactone in the presence of poly(ethylene glycol). J. Biomed. Mater. Res. 2004, 69A, 417–427. [Google Scholar] [CrossRef]
- Huang, M.-H.; Li, S.; Hutmacher, D.W.; Coudane, J.; Vert, M. Degradation characteristics of poly(ε-caprolactone)-based copolymers and blends. J. Appl. Polym. Sci. 2006, 102, 1681–1687. [Google Scholar] [CrossRef]
- Zheng, X.; Zhang, C.; Guo, Q.; Wan, X.; Shao, X.; Liu, Q.; Zhang, Q. Dual-functional nanoparticles for precise drug delivery to Alzheimer’s disease lesions: Targeting mechanisms, pharmacodynamics and safety. Int. J. Pharm. 2017, 525, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Royle, S.J. The cellular functions of clathrin. Cell. Mol. Life Sci. 2006, 63, 1823–1832. [Google Scholar] [CrossRef] [PubMed]
- Stern, C.M.; Mermelstein, P.G. Caveolin regulation of neuronal intracellular signaling. Cell. Mol. Life Sci. 2010, 67, 3785–3795. [Google Scholar] [CrossRef]
- Gan, Q.; Wang, T.; Cochrane, C.; McCarron, P. Modulation of surface charge, particle size and morphological properties of chitosan–TPP nanoparticles intended for gene delivery. Colloids Surf. B Biointerfaces 2005, 44, 65–73. [Google Scholar] [CrossRef]
- Panyam, J.; Zhou, W.-Z.; Prabha, S.; Sahoo, S.K.; Labhasetwar, V. Rapid endo-lysosomal escape of poly(dl-lactide-co-glycolide) nanoparticles: Implications for drug and gene delivery. FASEB J. 2002, 16, 1217–1226. [Google Scholar] [CrossRef]
- Pagano, R.E.; Huang, L. Interaction of phospholipid vesicles with cultured mammalian cells. II. Studies of mechanism. J. Cell Biol. 1975, 67, 49–60. [Google Scholar] [CrossRef]
- Schindler, H. Exchange and interactions between lipid layers at the surface of a liposome solution. Biochim. Biophys. Acta (BBA) Biomembr. 1979, 555, 316–336. [Google Scholar] [CrossRef]










| Polymer | Mean Diameter by Number (nm) | Mean Diameter by Intensity (nm) | Mean PDI |
|---|---|---|---|
| mPEG-b-PCL (2000–12,000) | 58.8 ± 34.4 | 139.2 ± 0.7 | 0.16 ± 0.02 |
| mPEG-b-PLGA (5000–45,000) | 77.8 ± 14.8 | 640.3 ± 24.1 | 0.21 ± 0.04 |
| mPEG-b-PLGA (2000–11,500) | 235.9 ± 59.1 | 281.9 ± 7.8 | 0.20 ± 0.04 |
| mPEG-b-PLLA (5000–20,000) | 129.6 ± 52.0 | 316.5 ± 5.7 | 0.25 ± 0.06 |
| Polymer | Organic Solvent | Mean Diameter by Number (nm) | Mean Diameter by Intensity (nm) | Mean PDI |
|---|---|---|---|---|
| mPEG-b-PCL (2000–12,000) | Dichloromethane | 58.8 ± 34.4 | 139.2 ± 0.7 | 0.16 ± 0.02 |
| Chloroform | 209.0 ± 109.4 | 173.3 ± 0.8 | 0.23 ± 0.02 | |
| Ethyl acetate | 256.4 ± 106.7 | 228.1 ± 11.4 | 0.31 ± 0.02 | |
| DCM+acetone (1:1, v/v) | 87.7 ± 1.8 | 299.8 ± 42.1 | 0.38 ± 0.00 | |
| Tetrahydrofuran | 76.3 ± 29.6 | 179.4 ± 17.3 | 0.18 ± 0.02 | |
| Acetonitrile | 72.1 ± 4.2 | 136.8 ± 7.2 | 0.28 ± 0.01 |
| Polymer | Polymer Loading (mg/mL) | Mean Diameter by Number (nm) | Mean Diameter by Intensity (nm) | Mean PDI |
|---|---|---|---|---|
| mPEG-b-PCL (2000–12,000) | 1 | 69.3 ± 22.1 | 146.0 ± 2.7 | 0.20 ± 0.01 |
| 5 | 92.2 ± 3.5 | 132.0 ± 0.3 | 0.21 ± 0.00 | |
| 10 | 70.7 ± 13.8 | 128.3 ± 1.2 | 0.21 ± 0.02 |
| Polymer | Mean Diameter by Number (nm) | Mean Diameter by Intensity (nm) | Mean PDI | Mean Zeta Potential (mV) | Mean E.E. (%) | |
|---|---|---|---|---|---|---|
| mPEG-b-PCL | 59.0 ± 53.8 | 116.7 ± 10.1 | 0.13 ± 0.07 | −0.09 ± 0.49 | P3V8 | 22.90 ± 8.91 |
| QBP1 | 34.20 ± 3.70 | |||||
| Mix ligands PEG-b-PCL | 53.1 ± 16.8 | 201.3 ± 1.5 | 0.23 ± 0.02 | 0.02 ± 0.05 | P3V8 | 34.04 ± 4.88 |
| QBP1 | 46.26 ± 4.50 | |||||
| Polymer | pH of Water Phase | Mean Diameter by Number (nm) | Mean Diameter by Intensity (nm) | Mean PDI | Mean Zeta Potential (mV) | Mean E.E. (%) | |
|---|---|---|---|---|---|---|---|
| mPEG-b-PCL | 3 | 63.6 ± 17.2 | 245.2 ± 12.4 | 0.34 ± 0.01 | −52.1 ± 2.9 | P3V8 | 100.00 ± 0.00 |
| QBP1 | 98.06 ± 2.37 | ||||||
| 5 | 57.6 ± 20.5 | 269.9 ± 21.5 | 0.34 ± 0.02 | −44.1 ± 4.6 | P3V8 | 94.85 ± 0.41 | |
| QBP1 | 96.07 ± 0.26 | ||||||
| 7 | 87.3 ± 26.5 | 319.0 ± 60.8 | 0.33 ± 0.03 | −44.8 ± 7.9 | P3V8 | 94.33 ± 0.11 | |
| QBP1 | 88.48 ± 1.97 | ||||||
| Mix ligands PEG-b-PCL | 3 | 80.8 ± 16.5 | 246.2 ± 14.2 | 0.31 ± 0.01 | −38.7 ± 0.8 | P3V8 | 100.00 ± 0.00 |
| QBP1 | 99.36 ± 0.56 | ||||||
| 5 | 74.9 ± 37.3 | 284.1 ± 40.3 | 0.31 ± 0.03 | −47.6 ± 4.4 | P3V8 | 98.27 ± 0.33 | |
| QBP1 | 100.00 ± 0.00 | ||||||
| 7 | 63.5 ± 1.9 | 321.0 ± 107.3 | 0.32 ± 0.05 | −42.0 ± 2.4 | P3V8 | 94.68 ± 0.69 | |
| QBP1 | 84.89 ± 5.57 | ||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.R.; Feng, T.; Zhang, Q.; Chan, H.Y.E.; Chau, Y. Co-Encapsulation and Co-Delivery of Peptide Drugs via Polymeric Nanoparticles. Polymers 2019, 11, 288. https://doi.org/10.3390/polym11020288
Kim MR, Feng T, Zhang Q, Chan HYE, Chau Y. Co-Encapsulation and Co-Delivery of Peptide Drugs via Polymeric Nanoparticles. Polymers. 2019; 11(2):288. https://doi.org/10.3390/polym11020288
Chicago/Turabian StyleKim, Ma Rie, Teng Feng, Qian Zhang, Ho Yin Edwin Chan, and Ying Chau. 2019. "Co-Encapsulation and Co-Delivery of Peptide Drugs via Polymeric Nanoparticles" Polymers 11, no. 2: 288. https://doi.org/10.3390/polym11020288