Preparation of Fluorescent Molecularly Imprinted Polymers via Pickering Emulsion Interfaces and the Application for Visual Sensing Analysis of Listeria Monocytogenes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instruments
2.3. Bacterial Strains and Cultivation of Strains
2.4. Synthesis of CdTe QDs
2.5. Preparation of N-Acrylchitosan (NAC) and NAC-QD Complex
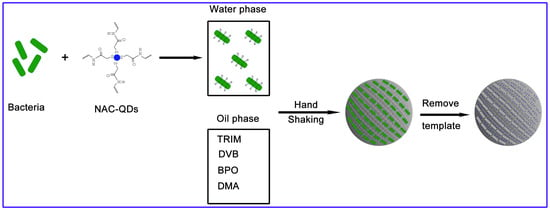
2.6. Synthesis of MIPs by Pickering Emulsion Polymerization
2.7. Analysis of Bacterial Binding Properties
2.8. Aplication to Real Samples
3. Results
3.1. Design and Preparation of MIPs
3.2. Characterization of Polymer Beads
3.3. Adsorption Performance of MIPs and NIPs
3.3.1. Bacterial Binding under Overloading Conditions
3.3.2. Kinetics Adsorption of MIPs
3.3.3. Adsorption Isotherm Analysis
3.3.4. Selectivity Study
3.4. Establishment of a Detection Method
3.5. Analysis in Real Samples
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, L.S.; Huang, R.; Liu, W.P.; Liu, H.X.; Zhou, X.M.; Xing, D. Rapid and visual detection of Listeria monocytogenes based on nanoparticle cluster catalyzed signal amplification. Biosens. Bioelectron. 2016, 86, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Bu, T.; Huang, Q.; Yan, L.Z.; Zhang, W.T.; Dou, L.N.; Huang, L.J.; Yang, Q.F.; Zhao, B.X.; Yang, B.W.; Li, T.; et al. Applicability of biological dye tracer in strip biosensor for ultrasensitive detection of pathogenic bacteria. Food Chem. 2019, 274, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Chiriaco, M.S.; Parlangeli, I.; Sirsi, F.; Poltronieri, P.; Primiceri, E. Impedance Sensing Platform for Detection of the Food Pathogen Listeria monocytogenes. Electronics 2018, 7, 11. [Google Scholar] [CrossRef]
- Zhao, X.; Lin, C.W.; Wang, J.; Oh, D.H. Advances in Rapid Detection Methods for Foodborne Pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alhogail, S.; Suaifan, G.; Zourob, M. Rapid colorimetric sensing platform for the detection of Listeria monocytogenes foodborne pathogen. Biosens. Bioelectron. 2016, 86, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Labrador, M.; Rota, M.C.; Perez-Arquillue, C.; Herrera, A.; Bayarri, S. Comparative evaluation of impedanciometry combined with chromogenic agars or RNA hybridization and real-time PCR methods for the detection of L. monocytogenes in dry-cured ham. Food Control 2018, 94, 108–115. [Google Scholar] [CrossRef]
- Lee, S.; Rakic-Martinez, M.; Graves, L.M.; Ward, T.J.; Siletzky, R.M.; Kathariou, S. Genetic Determinants for Cadmium and Arsenic Resistance among Listeria monocytogenes Serotype 4b Isolates from Sporadic Human Listeriosis Patients. Appl. Environ. Microbiol. 2013, 79, 2471–2476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef]
- Callejon, R.M.; Rodriguez-Naranjo, M.I.; Ubeda, C.; Hornedo-Ortega, R.; Garcia-Parrilla, M.C.; Troncoso, A.M. Reported Foodborne Outbreaks Due to Fresh Produce in the United States and European Union: Trends and Causes. Foodborne Pathog. Dis. 2015, 12, 32–38. [Google Scholar] [CrossRef]
- Song, S.X.; Wang, X.Y.; Xu, K.; Xia, G.M.; Yang, X.B. Visualized Detection of Vibrio parahaemolyticus in Food Samples Using Dual-Functional Aptamers and Cut-Assisted Rolling Circle Amplification. J. Agric. Food Chem. 2019, 67, 1244–1253. [Google Scholar] [CrossRef]
- Chen, J.; Park, B. Label-free screening of foodborne Salmonella using surface plasmon resonance imaging. Anal. Bioanal. Chem. 2018, 410, 5455–5464. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.J.; He, J.; Cao, X.H.; Huang, K.L.; Luo, Y.B.; Xu, W.T. Development of a double-antibody sandwich ELISA for rapid detection of Bacillus Cereus in food. Sci. Rep. 2016, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.Q.; Liang, T.B.; Zhan, Z.X.; Liu, R.; Li, F.; Xu, H.Y. Rapid and simultaneous quantification of viable Escherichia coli 0157:H7 and Salmonella spp. in milk through multiplex real-time PCR. J. Dairy Sci. 2017, 100, 8804–8813. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, S.D.; Barlen, B.; Kampfer, P.; Keusgen, M. Surface plasmon resonance (SPR) as a rapid tool for serotyping of Salmonella. Biosens. Bioelectron. 2010, 25, 967–971. [Google Scholar] [CrossRef]
- Liu, Y.S.; Zhao, C.; Song, X.L.; Xu, K.; Wang, J.; Li, J. Colorimetric immunoassay for rapid detection of Vibrio parahaemolyticus. Microchim. Acta 2017, 184, 4785–4792. [Google Scholar] [CrossRef]
- Kempe, H.; Pujolras, A.P.; Kempe, M. Molecularly Imprinted Polymer Nanocarriers for Sustained Release of Erythromycin. Pharm. Res. 2015, 32, 375–388. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, J.; Wang, J.; Wang, S. Development of water-compatible molecularly imprinted solid-phase extraction coupled with high performance liquid chromatography-tandem mass spectrometry for the detection of six sulfonamides in animal-derived foods. J. Chromatogr. A 2018, 1574, 9–17. [Google Scholar] [CrossRef]
- Han, S.; Su, L.Q.; Zhai, M.H.; Ma, L.; Liu, S.W.; Teng, Y. A molecularly imprinted composite based on graphene oxide for targeted drug delivery to tumor cells. J. Mater. Sci. 2019, 54, 3331–3341. [Google Scholar] [CrossRef]
- Wang, R.Y.; Pan, J.P.; Qin, M.; Guo, T.Y. Molecularly imprinted nanocapsule mimicking phosphotriesterase for the catalytic hydrolysis of organophosphorus pesticides. Eur. Polym. J. 2019, 110, 1–8. [Google Scholar] [CrossRef]
- Chen, L.X.; Xu, S.F.; Li, J.H. Recent advances in molecular imprinting technology: Current status, challenges and highlighted applications. Chem. Soc. Rev. 2011, 40, 2922–2942. [Google Scholar] [CrossRef]
- Shen, X.; Ye, L. Interfacial Molecular Imprinting in Nanoparticle-Stabilized Emulsions. Macromolecules 2011, 44, 5631–5637. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhou, T.; Ye, L. Molecular imprinting of protein in Pickering emulsion. Chem. Commun. 2012, 48, 8198–8200. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.T.; Bonde, J.S.; Kamra, T.; Bulow, L.; Leo, J.C.; Linke, D.; Ye, L. Bacterial Imprinting at Pickering Emulsion Interfaces. Angew. Chem. Int. Ed. 2014, 53, 10687–10690. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.M.; Qu, Q.; Cao, J.; Yan, D.; Liu, J.X.; Dai, X.H.; Yan, Y.S. Molecularly imprinted polymer foams with well-defined open-cell structure derived from Pickering HIPEs and their enhanced recognition of lambda-cyhalothrin. Chem. Eng. J. 2014, 253, 138–147. [Google Scholar] [CrossRef]
- Gan, M.Y.; Pan, J.M.; Zhang, Y.L.; Dai, X.H.; Yin, Y.J.; Qu, Q.; Yan, Y.S. Molecularly imprinted polymers derived from lignin-based Pickering emulsions and their selectively adsorption of lambda-cyhalothrin. Chem. Eng. J. 2014, 257, 317–327. [Google Scholar] [CrossRef]
- Zhu, W.J.; Ma, W.; Li, C.X.; Pan, J.M.; Dai, X.H. Well-designed multihollow magnetic imprinted microspheres based on cellulose nanocrystals (CNCs) stabilized Pickering double emulsion polymerization for selective adsorption of bifenthrin. Chem. Eng. J. 2015, 276, 249–260. [Google Scholar] [CrossRef]
- Liang, W.X.; Hu, H.W.; Guo, P.R.; Ma, Y.F.; Li, P.Y.; Zheng, W.R.; Zhang, M. Combining Pickering Emulsion Polymerization with Molecular Imprinting to Prepare Polymer Microspheres for Selective Solid-Phase Extraction of Malachite Green. Polymers 2017, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, Y.; Yang, J.J.; Sun, X.L.; Huang, C.N.; Zhang, X.D.; Chen, J.P. Preparation of dummy-imprinted polymers by Pickering emulsion polymerization for the selective determination of seven bisphenols from sediment samples. J. Sep. Sci. 2016, 39, 2188–2195. [Google Scholar] [CrossRef]
- Amjadi, M.; Jalili, R. Molecularly imprinted mesoporous silica embedded with carbon dots and semiconductor quantum dots as a ratiometric fluorescent sensor for diniconazole. Biosens. Bioelectron. 2017, 96, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Ding, H.; Yu, X.R.; Shi, X.Z.; Sun, A.L.; Li, D.X.; Zhao, J. Characterization and application of molecularly imprinted polymer-coated quantum dots for sensitive fluorescent determination of diethylstilbestrol in water samples. Talanta 2019, 197, 98–104. [Google Scholar] [CrossRef]
- Feng, J.W.; Tao, Y.; Shen, X.L.; Jin, H.; Zhou, T.T.; Zhou, Y.S.; Hu, L.Q.; Luo, D.; Mei, S.R.; Lee, Y.I. Highly sensitive and selective fluorescent sensor for tetrabromobisphenol-A in electronic waste samples using molecularly imprinted polymer coated quantum dots. Microchem. J. 2019, 144, 93–101. [Google Scholar] [CrossRef]
- Huang, S.Y.; Guo, M.L.; Tan, J.A.; Geng, Y.Y.; Wu, J.Y.; Tang, Y.W.; Su, C.C.; Lin, C.C.; Liang, Y. Novel Fluorescence Sensor Based on All-Inorganic Perovskite Quantum Dots Coated with Molecularly Imprinted Polymers for Highly Selective and Sensitive Detection of Omethoate. ACS Appl. Mater. Interfaces 2018, 10, 39056–39063. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.B.; Du, X.J.; Zang, Y.X.; Li, P.; Wang, S. SERS-Based Lateral Flow Strip Biosensor for Simultaneous Detection of Listeria monocytogenes and Salmonella enterica Serotype Enteritidis. J. Agric. Food Chem. 2017, 65, 10290–10299. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.L.; Dou, J.; Zhang, J.; Zhang, S.F. A simple and economical one-pot method to synthesize high-quality water soluble CdTe QDs. J. Mater. Chem. 2012, 22, 14573–14578. [Google Scholar] [CrossRef]
- Wongkongkatep, P.; Manopwisedjaroen, K.; Tiposoth, P.; Archakunakorn, S.; Pongtharangkul, T.; Suphantharika, M.; Honda, K.; Hamachi, I.; Wongkongkatep, J. Bacteria Interface Pickering Emulsions Stabilized by Self-assembled Bacteria-Chitosan Network. Langmuir 2012, 28, 5729–5736. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.M.; Yin, Y.J.; Gan, M.Y.; Meng, M.J.; Dai, X.H.; Wu, R.R.; Shi, W.D.; Yan, Y.S. Fabrication and evaluation of molecularly imprinted multi-hollow microspheres adsorbents with tunable inner pore structures derived from templating Pickering double emulsions. Chem. Eng. J. 2015, 266, 299–308. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, X.; Meng, M.J.; Liu, Z.C.; Ni, L.; Meng, X.G.; Qiu, J. RAFT-mediated microemulsion polymerization to synthesize a novel high-performance graphene oxide-based cadmium imprinted polymer. Chem. Eng. J. 2016, 302, 609–618. [Google Scholar] [CrossRef]
- Liang, X.Y.; Liu, F.; Wan, Y.Q.; Yin, X.Y.; Liu, W.T. Facile synthesis of molecularly imprinted polymers for selective extraction of tyrosine metabolites in human urine. J. Chromatogr. A 2019, 1587, 34–41. [Google Scholar] [CrossRef]
- Martinez, V.M.; Arbeloa, F.L.; Prieto, J.B.; Arbeloa, I.L. Characterization of rhodamine 6G aggregates intercalated in solid thin films of laponite clay. 2-Fluorescence spectroscopy. J. Phys. Chem. B 2005, 109, 7443–7450. [Google Scholar] [CrossRef]
- Kim, Y.S.; Lee, Y.; Kim, Y.; Kim, D.; Choi, H.S.; Park, J.C.; Nam, Y.S.; Jeon, D.Y. Synthesis of efficient near-infrared-emitting CuInS2/ZnS quantum dots by inhibiting cation-exchange for bio application. RSC Adv. 2017, 7, 10675–10682. [Google Scholar] [CrossRef] [Green Version]









© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Cui, Y.; Wang, J.; Wang, J. Preparation of Fluorescent Molecularly Imprinted Polymers via Pickering Emulsion Interfaces and the Application for Visual Sensing Analysis of Listeria Monocytogenes. Polymers 2019, 11, 984. https://doi.org/10.3390/polym11060984
Zhao X, Cui Y, Wang J, Wang J. Preparation of Fluorescent Molecularly Imprinted Polymers via Pickering Emulsion Interfaces and the Application for Visual Sensing Analysis of Listeria Monocytogenes. Polymers. 2019; 11(6):984. https://doi.org/10.3390/polym11060984
Chicago/Turabian StyleZhao, Xiaolei, Yan Cui, Junping Wang, and Junying Wang. 2019. "Preparation of Fluorescent Molecularly Imprinted Polymers via Pickering Emulsion Interfaces and the Application for Visual Sensing Analysis of Listeria Monocytogenes" Polymers 11, no. 6: 984. https://doi.org/10.3390/polym11060984