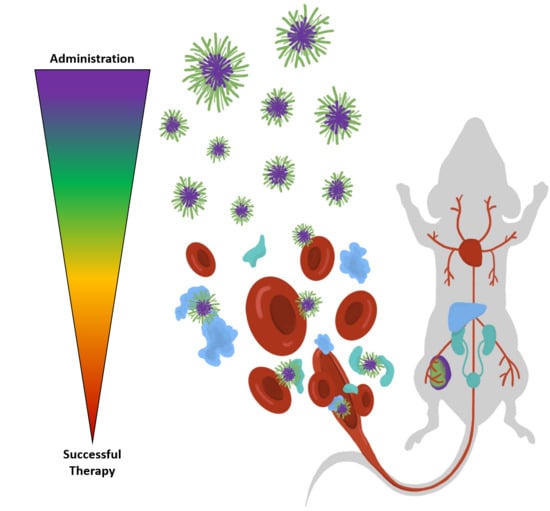
Engineered Polymeric Materials for Biological Applications: Overcoming Challenges of the Bio–Nano Interface
Abstract
:1. Introduction
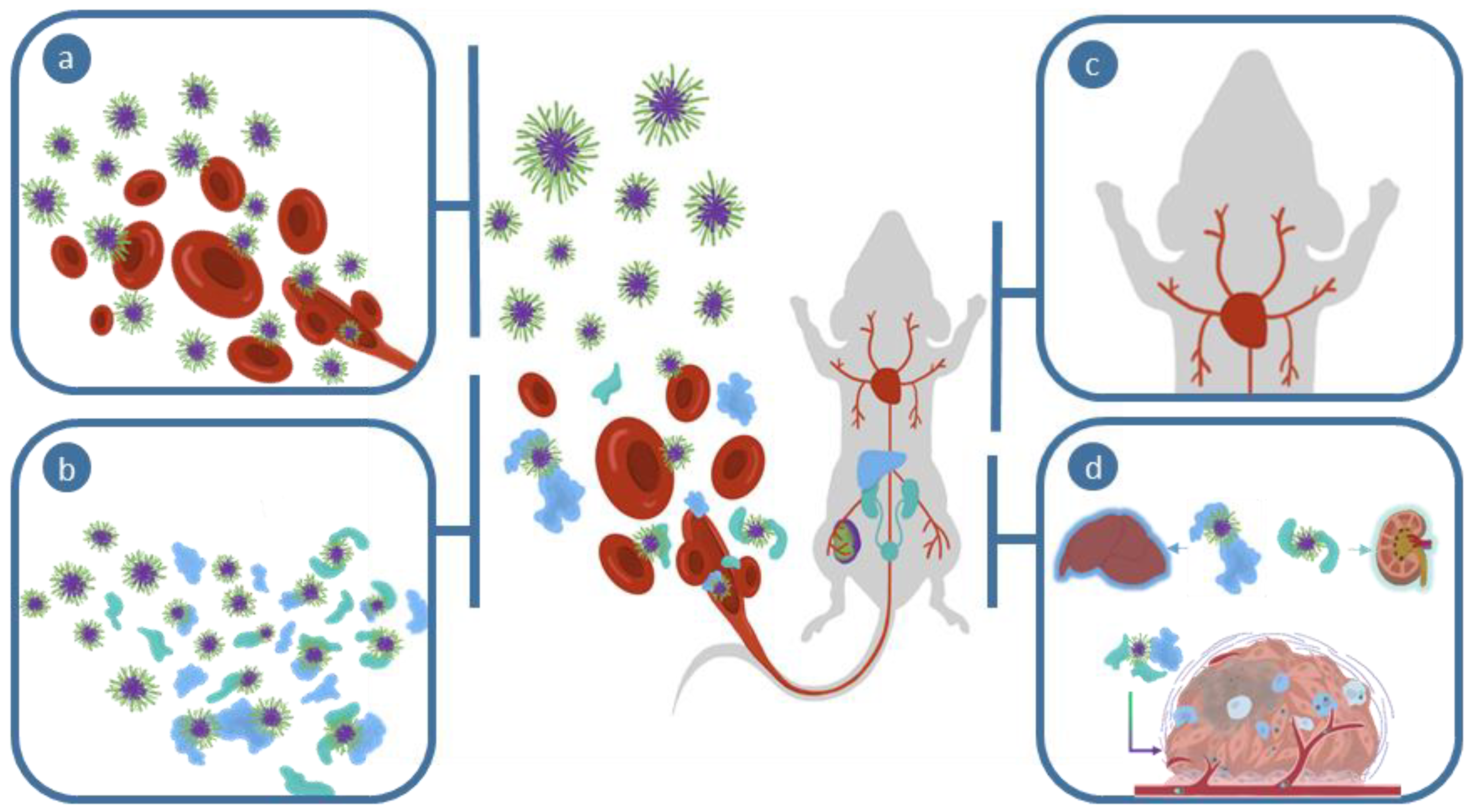
2. Interactions in the Blood Stream
2.1. Opsonization
2.1.1. The Protein Corona
2.1.2. Measuring and Monitoring Opsonization
2.2. Hemolysis
3. Biological Fate of Nanomedicines
3.1. Biodistribution
3.2. Clearance Pathways
4. Gathering at the Tumor Site
4.1. Extravasation
4.1.1. Impact of Vessel Architecture
4.1.2. Role of Endothelial Integrity
4.1.3. Emerging Tools for Imaging and Overcoming the Endothelial Barrier
4.2. Tumor Tissue Distribution
4.2.1. Tumor Stroma and Microenvironments
4.2.2. Spatial Regulation of Receptor Expression
4.2.3. Downstream Influence of Opsonization on Receptor Affinity
4.2.4. Current and Emerging Tools for Improving Penetration of the Tumor Mass
5. Cellular Level Interactions and Behaviors
5.1. Internalization
5.1.1. Nanoparticle Properties That Influence Cell Internalization
5.1.2. Tools to Understand Cellular Internalization and Limitations of In Vitro Models
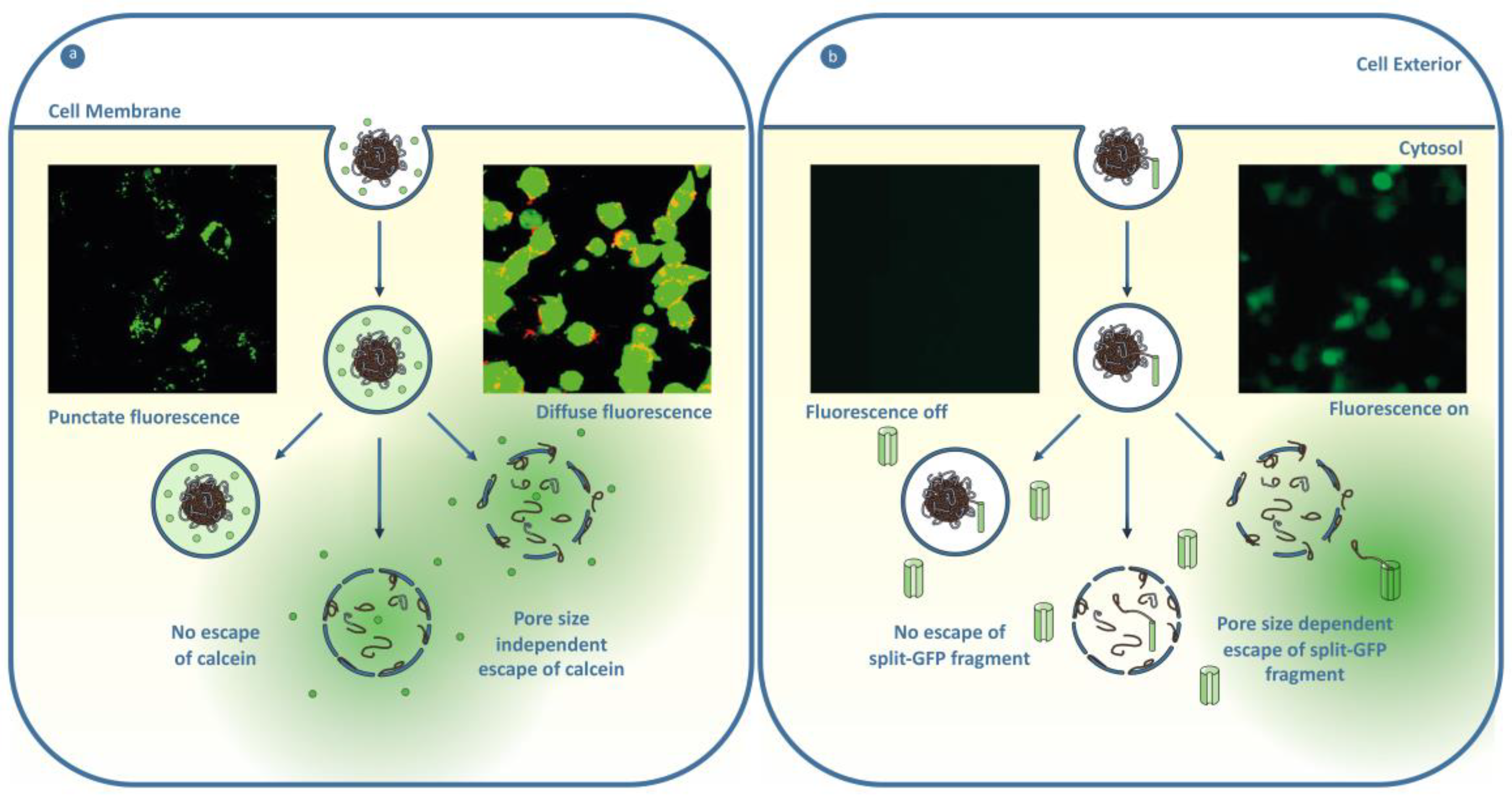
5.2. Endosomal Escape
5.2.1. Engineering Materials to Escape the Endosome
5.2.2. Tools to Understand Endosomal Escape
5.3. Trafficking to Subcellular Locations
5.3.1. Trafficking to the Nucleus
5.3.2. Trafficking to the Mitochondria
5.3.3. Emerging Tools for Quantification of Sub-Cellular Localization
6. Conclusions
Supplementary Files
Supplementary File 1Funding
Acknowledgments
Conflicts of Interest
References
- Bobo, D.; Robinson, K.J.; Islam, J.; Thurecht, K.J.; Corrie, S.R. Nanoparticle-Based Medicines: A Review of FDA-Approved Materials and Clinical Trials to Date. Pharm. Res. 2016, 33, 2373–2387. [Google Scholar] [CrossRef] [PubMed]
- Landesman-Milo, D.; Peer, D. Transforming Nanomedicines From Lab Scale Production to Novel Clinical Modality. Bioconjugate Chem. 2016, 27, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, S.; Tavares, A.J.; Dai, Q.; Ohta, S.; Audet, J.; Dvorak, H.F.; Chan, W.C.W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016, 1, 16014. [Google Scholar] [CrossRef]
- Gilleron, J.; Querbes, W.; Zeigerer, A.; Borodovsky, A.; Marsico, G.; Schubert, U.; Manygoats, K.; Seifert, S.; Andree, C.; Stoter, M.; et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013, 31, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Spicer, C.D.; Jumeaux, C.; Gupta, B.; Stevens, M.M. Peptide and protein nanoparticle conjugates: Versatile platforms for biomedical applications. Chem. Soc. Rev. 2018, 47, 3574–3620. [Google Scholar] [CrossRef] [PubMed]
- Giner-Casares, J.J.; Henriksen-Lacey, M.; Coronado-Puchau, M.; Liz-Marzan, L.M. Inorganic nanoparticles for biomedicine: Where materials scientists meet medical research. Mater. Today 2016, 19, 19–28. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; McNeil, S.E. Immunological Properties of Engineered Nanomaterials: An Introduction. Front. Nanobiomed. Res. 2013, 1, 1–23. [Google Scholar]
- Owens, D.E., 3rd; Peppas, N.A. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int. J. Pharm. 2006, 307, 93–102. [Google Scholar] [CrossRef]
- Yang, S.H.; Heo, D.; Park, J.; Na, S.; Suh, J.S.; Haam, S.; Park, S.W.; Huh, Y.M.; Yang, J. Role of surface charge in cytotoxicity of charged manganese ferrite nanoparticles towards macrophages. Nanotechnology 2012, 23, 505702. [Google Scholar] [CrossRef]
- Schottler, S.; Becker, G.; Winzen, S.; Steinbach, T.; Mohr, K.; Landfester, K.; Mailander, V.; Wurm, F.R. Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers. Nat. Nanotechnol. 2016, 11, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Pederzoli, F.; Tosi, G.; Vandelli, M.A.; Belletti, D.; Forni, F.; Ruozi, B. Protein corona and nanoparticles: How can we investigate on? Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017, 9, e1467. [Google Scholar] [CrossRef]
- Maiolo, D.; Del Pino, P.; Metrangolo, P.; Parak, W.J.; Baldelli Bombelli, F. Nanomedicine delivery: Does protein corona route to the target or off road? Nanomedicine 2015, 10, 3231–3247. [Google Scholar] [CrossRef]
- Schottler, S.; Landfester, K.; Mailander, V. Controlling the Stealth Effect of Nanocarriers through Understanding the Protein Corona. Angew. Chem. Int. Ed. Engl. 2016, 55, 8806–8815. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.; Morsbach, S.; Landfester, K. Possibilities and Limitations of Different Separation Techniques for the Analysis of the Protein Corona. Angew. Chem. Int. Ed. Engl. 2019, 58. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Wang, G.; Griffin, J.I.; Brenneman, B.; Banda, N.K.; Holers, V.M.; Backos, D.S.; Wu, L.; Moghimi, S.M.; Simberg, D. Complement proteins bind to nanoparticle protein corona and undergo dynamic exchange in vivo. Nat. Nanotechnol. 2017, 12, 387–393. [Google Scholar] [CrossRef]
- Lazarovits, J.; Chen, Y.Y.; Sykes, E.A.; Chan, W.C. Nanoparticle-blood interactions: The implications on solid tumour targeting. Chem. Commun. 2015, 51, 2756–2767. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Chen, C. The Crown and the Scepter: Roles of the Protein Corona in Nanomedicine. Adv. Mater. 2018, 0, e1805740. [Google Scholar] [CrossRef] [PubMed]
- Papi, M.; Caputo, D.; Palmieri, V.; Coppola, R.; Palchetti, S.; Bugli, F.; Martini, C.; Digiacomo, L.; Pozzi, D.; Caracciolo, G. Clinically approved PEGylated nanoparticles are covered by a protein corona that boosts the uptake by cancer cells. Nanoscale 2017, 9, 10327–10334. [Google Scholar] [CrossRef]
- Tavakol, M.; Montazeri, A.; Naghdabadi, R.; Hajipour, M.J.; Zanganeh, S.; Caracciolo, G.; Mahmoudi, M. Disease-related metabolites affect protein-nanoparticle interactions. Nanoscale 2018, 10, 7108–7115. [Google Scholar] [CrossRef] [PubMed]
- Palchetti, S.; Pozzi, D.; Capriotti, A.L.; Barbera, G.; Chiozzi, R.Z.; Digiacomo, L.; Peruzzi, G.; Caracciolo, G.; Lagana, A. Influence of dynamic flow environment on nanoparticle-protein corona: From protein patterns to uptake in cancer cells. Colloids Surf. B 2017, 153, 263–271. [Google Scholar] [CrossRef]
- Vasti, C.; Bonnet, L.V.; Galiano, M.R.; Rojas, R.; Giacomelli, C.E. Relevance of protein-protein interactions on the biological identity of nanoparticles. Colloids Surf. B 2018, 166, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gustafsson, O.J.R.; Pilkington, E.H.; Kakinen, A.; Javed, I.; Faridi, A.; Davis, T.P.; Ke, P.C. Nanoparticle–proteome in vitro and in vivo. J. Mater. Chem. B 2018, 6, 6026–6041. [Google Scholar] [CrossRef]
- Bertrand, N.; Grenier, P.; Mahmoudi, M.; Lima, E.M.; Appel, E.A.; Dormont, F.; Lim, J.M.; Karnik, R.; Langer, R.; Farokhzad, O.C. Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics. Nat. Commun. 2017, 8, 777. [Google Scholar] [CrossRef]
- Miceli, E.; Kar, M.; Calderon, M. Interactions of organic nanoparticles with proteins in physiological conditions. J. Mater. Chem. B 2017, 5, 4393–4405. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, T.; Yu, W.; Ruan, S.; He, Q.; Gao, H. Ligand Size and Conformation Affect the Behavior of Nanoparticles Coated with in Vitro and in Vivo Protein Corona. ACS Appl. Mater. Interfaces 2018, 10, 9094–9103. [Google Scholar] [CrossRef] [PubMed]
- Burnand, D.; Milosevic, A.; Balog, S.; Spuch-Calvar, M.; Rothen-Rutishauser, B.; Dengjel, J.; Kinnear, C.; Moore, T.L.; Petri-Fink, A. Beyond Global Charge: Role of Amine Bulkiness and Protein Fingerprint on Nanoparticle-Cell Interaction. Small 2018, 14, e1802088. [Google Scholar] [CrossRef]
- Jayaram, D.T.; Pustulka, S.M.; Mannino, R.G.; Lam, W.A.; Payne, C.K. Protein Corona in Response to Flow: Effect on Protein Concentration and Structure. Biophys. J. 2018, 115, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Kelly, P.M.; Mahon, E.; Stockmann, H.; Rudd, P.M.; Caruso, F.; Dawson, K.A.; Yan, Y.; Monopoli, M.P. The “sweet” side of the protein corona: Effects of glycosylation on nanoparticle-cell interactions. ACS Nano 2015, 9, 2157–2166. [Google Scholar] [CrossRef]
- Garcia-Alvarez, R.; Hadjidemetriou, M.; Sanchez-Iglesias, A.; Liz-Marzan, L.M.; Kostarelos, K. In vivo formation of protein corona on gold nanoparticles. The effect of their size and shape. Nanoscale 2018, 10, 1256–1264. [Google Scholar] [CrossRef] [PubMed]
- Bigdeli, A.; Palchetti, S.; Pozzi, D.; Hormozi-Nezhad, M.R.; Baldelli Bombelli, F.; Caracciolo, G.; Mahmoudi, M. Exploring Cellular Interactions of Liposomes Using Protein Corona Fingerprints and Physicochemical Properties. ACS Nano 2016, 10, 3723–3737. [Google Scholar] [CrossRef]
- Corbo, C.; Molinaro, R.; Tabatabaei, M.; Farokhzad, O.C.; Mahmoudi, M. Personalized protein corona on nanoparticles and its clinical implications. Biomater. Sci. 2017, 5, 378–387. [Google Scholar] [CrossRef]
- Curran, A.M.; Fogarty Draper, C.; Scott-Boyer, M.P.; Valsesia, A.; Roche, H.M.; Ryan, M.F.; Gibney, M.J.; Kutmon, M.; Evelo, C.T.; Coort, S.L.; et al. Sexual Dimorphism, Age, and Fat Mass Are Key Phenotypic Drivers of Proteomic Signatures. J. Proteome Res. 2017, 16, 4122–4133. [Google Scholar] [CrossRef]
- Geyer, P.E.; Kulak, N.A.; Pichler, G.; Holdt, L.M.; Teupser, D.; Mann, M. Plasma Proteome Profiling to Assess Human Health and Disease. Cell Syst. 2016, 2, 185–195. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Buil, A.; Collins, B.C.; Gillet, L.C.; Blum, L.C.; Cheng, L.Y.; Vitek, O.; Mouritsen, J.; Lachance, G.; Spector, T.D.; et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol. Syst. Biol. 2015, 11, 786. [Google Scholar] [CrossRef]
- Geyer, P.E.; Wewer Albrechtsen, N.J.; Tyanova, S.; Grassl, N.; Iepsen, E.W.; Lundgren, J.; Madsbad, S.; Holst, J.J.; Torekov, S.S.; Mann, M. Proteomics reveals the effects of sustained weight loss on the human plasma proteome. Mol. Syst. Biol. 2016, 12, 901. [Google Scholar] [CrossRef]
- Hennig, R.; Cajic, S.; Borowiak, M.; Hoffmann, M.; Kottler, R.; Reichl, U.; Rapp, E. Towards personalized diagnostics via longitudinal study of the human plasma N-glycome. Biochim. Biophys. Acta 2016, 1860, 1728–1738. [Google Scholar] [CrossRef] [Green Version]
- Carrillo-Carrion, C.; Carril, M.; Parak, W.J. Techniques for the experimental investigation of the protein corona. Curr. Opin. Biotechnol. 2017, 46, 106–113. [Google Scholar] [CrossRef] [Green Version]
- Duan, Y.; Liu, Y.; Coreas, R.; Zhong, W. Mapping Molecular Structure of Protein Locating on Nanoparticles with Limited Proteolysis. Anal. Chem. 2019, 91, 4204–4212. [Google Scholar] [CrossRef]
- Simon, J.; Müller, L.K.; Kokkinopoulou, M.; Lieberwirth, I.; Morsbach, S.; Landfester, K.; Mailänder, V. Exploiting the biomolecular corona: Pre-coating of nanoparticles enables controlled cellular interactions. Nanoscale 2018, 10, 10731–10739. [Google Scholar] [CrossRef]
- Prozeller, D.; Pereira, J.; Simon, J.; Mailander, V.; Morsbach, S.; Landfester, K. Prevention of Dominant IgG Adsorption on Nanocarriers in IgG-Enriched Blood Plasma by Clusterin Precoating. Adv. Sci. 2019, 6, 1802199. [Google Scholar] [CrossRef]
- Pan, Z.; Fang, D.; Song, N.; Song, Y.; Ding, M.; Li, J.; Luo, F.; Tan, H.; Fu, Q. Surface Distribution and Biophysicochemical Properties of Polymeric Micelles Bearing Gemini Cationic and Hydrophilic Groups. ACS Appl. Mater. Interfaces 2017, 9, 2138–2149. [Google Scholar] [CrossRef]
- Atkinson, S.P.; Andreu, Z.; Vicent, M.J. Polymer Therapeutics: Biomarkers and New Approaches for Personalized Cancer Treatment. J. Pers. Med. 2018, 8, 6. [Google Scholar] [CrossRef]
- Björnmalm, M.; Thurecht, K.J.; Michael, M.; Scott, A.M.; Caruso, F. Bridging Bio–Nano Science and Cancer Nanomedicine. ACS Nano 2017, 11, 9594–9613. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; Aggarwal, P.; Hall, J.B.; McNeil, S.E. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol. Pharmaceutics 2008, 5, 487–495. [Google Scholar] [CrossRef]
- Pavan, C.; Rabolli, V.; Tomatis, M.; Fubini, B.; Lison, D. Why does the hemolytic activity of silica predict its pro-inflammatory activity? Part. Fibre Toxicol. 2014, 11, 1. [Google Scholar] [CrossRef]
- Cohen, J.J. Apoptosis: The physiologic pathway of cell death. Hosp. Pract. (Off. Ed.) 1993, 28, 35–43. [Google Scholar] [CrossRef]
- Schaer, D.J.; Alayash, A.I.; Buehler, P.W. Gating the radical hemoglobin to macrophages: The anti-inflammatory role of CD163, a scavenger receptor. Antioxid Redox Sign 2007, 9, 991–999. [Google Scholar] [CrossRef]
- Schroit, A.J.; Madsen, J.W.; Tanaka, Y. In vivo recognition and clearance of red blood cells containing phosphatidylserine in their plasma membranes. J. Biol. Chem. 1985, 260, 5131–5138. [Google Scholar]
- Cheng, F.-Y.; Su, C.-H.; Yang, Y.-S.; Yeh, C.-S.; Tsai, C.-Y.; Wu, C.-L.; Wu, M.-T.; Shieh, D.-B. Characterization of aqueous dispersions of Fe 3 O 4 nanoparticles and their biomedical applications. Biomaterials 2005, 26, 729–738. [Google Scholar] [CrossRef]
- Goodman, C.M.; McCusker, C.D.; Yilmaz, T.; Rotello, V.M. Toxicity of gold nanoparticles functionalized with cationic and anionic side chains. Bioconjugate Chem. 2004, 15, 897–900. [Google Scholar] [CrossRef]
- Kim, D.; El-Shall, H.; Dennis, D.; Morey, T. Interaction of PLGA nanoparticles with human blood constituents. Colloids Surf. B 2005, 40, 83–91. [Google Scholar] [CrossRef]
- Schubert, M.A.; Muller-Goymann, C.C. Characterisation of surface-modified solid lipid nanoparticles (SLN): Influence of lecithin and nonionic emulsifier. Eur. J. Pharm. Biopharm. 2005, 61, 77–86. [Google Scholar] [CrossRef]
- Dutta, T.; Agashe, H.B.; Garg, M.; Balasubramanium, P.; Kabra, M.; Jain, N.K. Poly (propyleneimine) dendrimer based nanocontainers for targeting of efavirenz to human monocytes/macrophages in vitro: Research Paper. J. Drug Target. 2007, 15, 89–98. [Google Scholar] [CrossRef]
- Kainthan, R.K.; Gnanamani, M.; Ganguli, M.; Ghosh, T.; Brooks, D.E.; Maiti, S.; Kizhakkedathu, J.N. Blood compatibility of novel water soluble hyperbranched polyglycerol-based multivalent cationic polymers and their interaction with DNA. Biomaterials 2006, 27, 5377–5390. [Google Scholar] [CrossRef]
- Chen, L.; Simpson, J.D.; Fuchs, A.V.; Rolfe, B.E.; Thurecht, K.J. Effects of Surface Charge of Hyperbranched Polymers on Cytotoxicity, Dynamic Cellular Uptake and Localization, Hemotoxicity, and Pharmacokinetics in Mice. Mol. Pharmaceutics 2017, 14, 4485–4497. [Google Scholar] [CrossRef]
- Brownlie, A.; Uchegbu, I.F.; Schatzlein, A.G. PEI-based vesicle-polymer hybrid gene delivery system with improved biocompatibility. Int. J. Pharm. 2004, 274, 41–52. [Google Scholar] [CrossRef]
- Jin, Y.G.; Tong, L.; Ai, P.; Li, M.; Hou, X.P. Self-assembled drug delivery systems - 1. Properties and in vitro/in vivo behavior of acyclovir self-assembled nanoparticles (SAN). Int. J. Pharm. 2006, 309, 199–207. [Google Scholar] [CrossRef]
- Chouly, C.; Bordenave, L.; Bareille, R.; Guerin, V.; Baquey, A.; Pouliquen, D.; Baquey, C.; Jallet, P. In vitro study of the hemocompatibility of superparamagnetic contrast agent for magnetic resonance imaging. Clin. Mater. 1994, 15, 293–301. [Google Scholar] [CrossRef]
- Duguid, J.G.; Li, C.; Shi, M.; Logan, M.J.; Alila, H.; Rolland, A.; Tomlinson, E.; Sparrow, J.T.; Smith, L.C. A physicochemical approach for predicting the effectiveness of peptide-based gene delivery systems for use in plasmid-based gene therapy. Biophys. J. 1998, 74, 2802–2814. [Google Scholar] [CrossRef]
- Guowei, D.; Adriane, K.; Chen, X.; Jie, C.; Yinfeng, L. PVP magnetic nanospheres: Biocompatibility, in vitro and in vivo bleomycin release. Int. J. Pharm. 2007, 328, 78–85. [Google Scholar] [CrossRef]
- Koziara, J.M.; Oh, J.J.; Akers, W.S.; Ferraris, S.P.; Mumper, R.J. Blood compatibility of cetyl alcohol/polysorbate-based nanoparticles. Pharm. Res. 2005, 22, 1821–1828. [Google Scholar] [CrossRef]
- Lim, S.J.; Lee, M.K.; Kim, C.K. Altered chemical and biological activities of all-trans retinoic acid incorporated in solid lipid nanoparticle powders. J. Control. Release 2004, 100, 53–61. [Google Scholar] [CrossRef]
- Nimesh, S.; Goyal, A.; Pawar, V.; Jayaraman, S.; Kumar, P.; Chandra, R.; Singh, Y.; Gupta, K.C. Polyethylenimine nanoparticles as efficient transfecting agents for mammalian cells. J. Control. Release 2006, 110, 457–468. [Google Scholar] [CrossRef]
- Hall, J.B.; Dobrovolskaia, M.A.; Patri, A.K.; McNeil, S.E. Characterization of nanoparticles for therapeutics. Nanomedicine 2007, 2, 789–803. [Google Scholar] [CrossRef]
- Faria, M.; Bjornmalm, M.; Thurecht, K.J.; Kent, S.J.; Parton, R.G.; Kavallaris, M.; Johnston, A.P.R.; Gooding, J.J.; Corrie, S.R.; Boyd, B.J.; et al. Minimum information reporting in bio-nano experimental literature. Nat. Nanotechnol. 2018, 13, 777–785. [Google Scholar] [CrossRef]
- Brenner, J.S.; Pan, D.C.; Myerson, J.W.; Marcos-Contreras, O.A.; Villa, C.H.; Patel, P.; Hekierski, H.; Chatterjee, S.; Tao, J.Q.; Parhiz, H.; et al. Red blood cell-hitchhiking boosts delivery of nanocarriers to chosen organs by orders of magnitude. Nat. Commun. 2018, 9, 2684. [Google Scholar] [CrossRef]
- Moss, D.M.; Siccardi, M. Optimizing nanomedicine pharmacokinetics using physiologically based pharmacokinetics modelling. Br. J. Pharmacol. 2014, 171, 3963–3979. [Google Scholar] [CrossRef]
- Dogra, P.; Butner, J.D.; Chuang, Y.L.; Caserta, S.; Goel, S.; Brinker, C.J.; Cristini, V.; Wang, Z. Mathematical modeling in cancer nanomedicine: A review. Biomed. Microdevices 2019, 21, 40. [Google Scholar] [CrossRef]
- Almeida, J.P.; Chen, A.L.; Foster, A.; Drezek, R. In vivo biodistribution of nanoparticles. Nanomedicine 2011, 6, 815–835. [Google Scholar] [CrossRef]
- Gaspar, R.; Duncan, R. Polymeric carriers: Preclinical safety and the regulatory implications for design and development of polymer therapeutics. Adv. Drug Deliv. Rev. 2009, 61, 1220–1231. [Google Scholar] [CrossRef]
- Baumann, A.; Tuerck, D.; Prabhu, S.; Dickmann, L.; Sims, J. Pharmacokinetics, metabolism and distribution of PEGs and PEGylated proteins: Quo vadis? Drug Discov. Today 2014, 19, 1623–1631. [Google Scholar] [CrossRef]
- Hoshyar, N.; Gray, S.; Han, H.; Bao, G. The effect of nanoparticle size on in vivo pharmacokinetics and cellular interaction. Nanomedicine 2016, 11, 673–692. [Google Scholar] [CrossRef]
- Haute, D.V.; Berlin, J.M. Challenges in realizing selectivity for nanoparticle biodistribution and clearance: Lessons from gold nanoparticles. Ther. Deliv. 2017, 8, 763–774. [Google Scholar] [CrossRef]
- Thurecht, K.J.; Blakey, I.; Peng, H.; Squires, O.; Hsu, S.; Alexander, C.; Whittaker, A.K. Functional hyperbranched polymers: Toward targeted in vivo 19F magnetic resonance imaging using designed macromolecules. J. Am. Chem. Soc. 2010, 132, 5336–5337. [Google Scholar] [CrossRef]
- Choi, H.S.; Liu, W.; Misra, P.; Tanaka, E.; Zimmer, J.P.; Itty Ipe, B.; Bawendi, M.G.; Frangioni, J.V. Renal clearance of quantum dots. Nat. Biotechnol. 2007, 25, 1165–1170. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.C.; MacKay, J.A.; Frechet, J.M.; Szoka, F.C. Designing dendrimers for biological applications. Nat. Biotechnol. 2005, 23, 1517–1526. [Google Scholar] [CrossRef]
- Rolfe, B.E.; Blakey, I.; Squires, O.; Peng, H.; Boase, N.R.; Alexander, C.; Parsons, P.G.; Boyle, G.M.; Whittaker, A.K.; Thurecht, K.J. Multimodal polymer nanoparticles with combined 19F magnetic resonance and optical detection for tunable, targeted, multimodal imaging in vivo. J. Am. Chem. Soc. 2014, 136, 2413–2419. [Google Scholar] [CrossRef]
- Drobník, J.; Rypáček, F. Soluble synthetic polymers in biological systems. Proceedings of Polymers in Medicine, Berlin/Heidelberg, Germany, 1984; pp. 1–50. [Google Scholar]
- Alexis, F.; Pridgen, E.; Molnar, L.K.; Farokhzad, O.C. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharmaceutics 2008, 5, 505–515. [Google Scholar] [CrossRef]
- Park, J.-K.; Utsumi, T.; Seo, Y.-E.; Deng, Y.; Satoh, A.; Saltzman, W.M.; Iwakiri, Y. Cellular distribution of injected PLGA-nanoparticles in the liver. Nanomedicine 2016, 12, 1365–1374. [Google Scholar] [CrossRef] [Green Version]
- Wong, A.D.; Ye, M.; Ulmschneider, M.B.; Searson, P.C. Quantitative Analysis of the Enhanced Permeation and Retention (EPR) Effect. PLoS ONE 2015, 10, e0123461. [Google Scholar] [CrossRef]
- Cui, J.; Ju, Y.; Houston, Z.H.; Glass, J.J.; Fletcher, N.L.; Alcantara, S.; Dai, Q.; Howard, C.B.; Mahler, S.M.; Wheatley, A.K.; et al. Modulating Targeting of Poly(ethylene glycol) Particles to Tumor Cells Using Bispecific Antibodies. Adv. Healthcare Mater. 2019, 8, e1801607. [Google Scholar] [CrossRef]
- Bailly, C.; Bodet-Milin, C.; Rousseau, C.; Faivre-Chauvet, A.; Kraeber-Bodere, F.; Barbet, J. Pretargeting for imaging and therapy in oncological nuclear medicine. EJNMMI Radiopharm. Chem. 2017, 2, 6. [Google Scholar] [CrossRef] [Green Version]
- Tienken, L.; Drude, N.; Schau, I.; Winz, O.H.; Temme, A.; Weinhold, E.; Mottaghy, F.M.; Morgenroth, A. Evaluation of a Pretargeting Strategy for Molecular Imaging of the Prostate Stem Cell Antigen with a Single Chain Antibody. Sci. Rep. 2018, 8, 3755. [Google Scholar] [CrossRef]
- Membreno, R.; Keinanen, O.M.; Cook, B.E.; Tully, K.M.; Fung, K.C.; Lewis, J.S.; Zeglis, B.M. Toward the Optimization of Click-Mediated Pretargeted Radioimmunotherapy. Mol. Pharmaceutics 2019, 16, 2259–2263. [Google Scholar] [CrossRef]
- Versteegen, R.M.; Ten Hoeve, W.; Rossin, R.; de Geus, M.A.R.; Janssen, H.M.; Robillard, M.S. Click-to-Release from trans-Cyclooctenes: Mechanistic Insights and Expansion of Scope from Established Carbamate to Remarkable Ether Cleavage. Angew. Chem. Int. Ed. Engl. 2018, 57, 10494–10499. [Google Scholar] [CrossRef]
- Ji, X.; Pan, Z.; Yu, B.; De La Cruz, L.K.; Zheng, Y.; Ke, B.; Wang, B. Click and release: Bioorthogonal approaches to “on-demand” activation of prodrugs. Chem. Soc. Rev. 2019, 48, 1077–1094. [Google Scholar] [CrossRef]
- Dewhirst, M.W.; Secomb, T.W. Transport of drugs from blood vessels to tumour tissue. Nat. Rev. Cancer 2017, 17, 738–750. [Google Scholar] [CrossRef]
- Klein, D. The Tumor Vascular Endothelium as Decision Maker in Cancer Therapy. Front. Oncol. 2018, 8, 367. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Nichols, J.W.; Toh, K.; Nomoto, T.; Cabral, H.; Miura, Y.; Christie, R.J.; Yamada, N.; Ogura, T.; Kano, M.R.; et al. Vascular bursts enhance permeability of tumour blood vessels and improve nanoparticle delivery. Nat. Nanotechnol. 2016, 11, 533–538. [Google Scholar] [CrossRef]
- Lee, S.S.; Bindokas, V.P.; Kron, S.J. Multiplex Three-Dimensional Mapping of Macromolecular Drug Distribution in the Tumor Microenvironment. Mol. Cancer Ther. 2019, 18, 213–226. [Google Scholar] [CrossRef]
- Suzuki, K.; Miura, Y.; Mochida, Y.; Miyazaki, T.; Toh, K.; Anraku, Y.; Melo, V.; Liu, X.; Ishii, T.; Nagano, O.; et al. Glucose transporter 1-mediated vascular translocation of nanomedicines enhances accumulation and efficacy in solid tumors. J. Control. Release 2019, 301, 28–41. [Google Scholar] [CrossRef]
- Sulheim, E.; Kim, J.; van Wamel, A.; Kim, E.; Snipstad, S.; Vidic, I.; Grimstad, I.H.; Wideroe, M.; Torp, S.H.; Lundgren, S.; et al. Multi-modal characterization of vasculature and nanoparticle accumulation in five tumor xenograft models. J. Control. Release 2018, 279, 292–305. [Google Scholar] [CrossRef]
- Golombek, S.K.; May, J.-N.; Theek, B.; Appold, L.; Drude, N.; Kiessling, F.; Lammers, T. Tumor targeting via EPR: Strategies to enhance patient responses. Adv. Drug Deliv. Rev. 2018. [Google Scholar] [CrossRef]
- Kutova, M.O.; Guryev, L.E.; Sokolova, A.E.; Alzeibak, R.; Balalaeva, V.I. Targeted Delivery to Tumors: Multidirectional Strategies to Improve Treatment Efficiency. Cancers 2019, 11, 68. [Google Scholar] [CrossRef]
- Quiros-Gonzalez, I.; Tomaszewski, M.R.; Aitken, S.J.; Ansel-Bollepalli, L.; McDuffus, L.A.; Gill, M.; Hacker, L.; Brunker, J.; Bohndiek, S.E. Optoacoustics delineates murine breast cancer models displaying angiogenesis and vascular mimicry. Br. J. Cancer 2018, 118, 1098–1106. [Google Scholar] [CrossRef]
- Atukorale, P.U.; Covarrubias, G.; Bauer, L.; Karathanasis, E. Vascular targeting of nanoparticles for molecular imaging of diseased endothelium. Adv. Drug Deliv. Rev. 2017, 113, 141–156. [Google Scholar] [CrossRef]
- Youn, Y.S.; Bae, Y.H. Perspectives on the past, present, and future of cancer nanomedicine. Adv. Drug Deliv. Rev. 2018, 130, 3–11. [Google Scholar] [CrossRef]
- Lee, S.H.; Hu, W.; Matulay, J.T.; Silva, M.V.; Owczarek, T.B.; Kim, K.; Chua, C.W.; Barlow, L.J.; Kandoth, C.; Williams, A.B.; et al. Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell 2018, 173, 515–528.e517. [Google Scholar] [CrossRef]
- Fletcher, N.L.; Houston, Z.H.; Simpson, J.D.; Veedu, R.N.; Thurecht, K.J. Designed multifunctional polymeric nanomedicines: Long-term biodistribution and tumour accumulation of aptamer-targeted nanomaterials. Chem. Commun. 2018, 54, 11538–11541. [Google Scholar] [CrossRef]
- Du, S.; Xiong, H.; Xu, C.; Lu, Y.; Yao, J. Attempts to strengthen and simplify the tumor vascular normalization strategy using tumor vessel normalization promoting nanomedicines. Biomater. Sci. 2019, 7, 1147–1160. [Google Scholar] [CrossRef]
- Frenzel, T.; Hoffmann, B.; Schmitz, R.; Bethge, A.; Schumacher, U.; Wedemann, G. Radiotherapy and chemotherapy change vessel tree geometry and metastatic spread in a small cell lung cancer xenograft mouse tumor model. PLoS ONE 2017, 12, e0187144. [Google Scholar] [CrossRef]
- Jiang, W.; Huang, Y.H.; An, Y.; Kim, B.Y.S. Remodeling Tumor Vasculature to Enhance Delivery of Intermediate-Sized Nanoparticles. ACS Nano 2015, 9, 8689–8696. [Google Scholar] [CrossRef]
- Li, H.J.; Du, J.Z.; Du, X.J.; Xu, C.F.; Sun, C.Y.; Wang, H.X.; Cao, Z.T.; Yang, X.Z.; Zhu, Y.H.; Nie, S.; et al. Stimuli-responsive clustered nanoparticles for improved tumor penetration and therapeutic efficacy. Proc. Natl. Acad. Sci. USA 2016, 113, 4164–4169. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.L.; Hu, S.H. Functional Nanoparticles for Tumor Penetration of Therapeutics. Pharmaceutics 2018, 10, 193. [Google Scholar] [CrossRef]
- Cabral, H.; Miyata, K.; Osada, K.; Kataoka, K. Block Copolymer Micelles in Nanomedicine Applications. Chem. Rev. 2018, 118, 6844–6892. [Google Scholar] [CrossRef] [Green Version]
- El-Sawy, H.S.; Al-Abd, A.M.; Ahmed, T.A.; El-Say, K.M.; Torchilin, V.P. Stimuli-responsive nano-architecture drug-delivery systems to solid tumor micromilieu: past, present, and future perspectives. ACS Nano 2018, 12, 10636–10664. [Google Scholar] [CrossRef]
- Peng, F.; Setyawati, M.I.; Tee, J.K.; Ding, X.; Wang, J.; Nga, M.E.; Ho, H.K.; Leong, D.T. Nanoparticles promote in vivo breast cancer cell intravasation and extravasation by inducing endothelial leakiness. Nat. Nanotechnol. 2019, 14, 279–286. [Google Scholar] [CrossRef]
- Boase, N.R.B.; Blakey, I.; Thurecht, K.J. Molecular imaging with polymers. Polym. Chem.-Uk 2012, 3, 1384–1389. [Google Scholar] [CrossRef]
- Fuchs, A.V.; Gemmell, A.C.; Thurecht, K.J. Utilising polymers to understand diseases: Advanced molecular imaging agents. Polym. Chem.-Uk 2015, 6, 868–880. [Google Scholar] [CrossRef]
- Fuchs, A.V.; Bapat, A.P.; Cowin, G.J.; Thurecht, K.J. Switchable 19F MRI polymer theranostics: Towards in situ quantifiable drug release. Polym. Chem.-Uk 2017, 8, 5157–5166. [Google Scholar] [CrossRef]
- Upponi, J.R.; Jerajani, K.; Nagesha, D.K.; Kulkarni, P.; Sridhar, S.; Ferris, C.; Torchilin, V.P. Polymeric micelles: Theranostic co-delivery system for poorly water-soluble drugs and contrast agents. Biomaterials 2018, 170, 26–36. [Google Scholar] [CrossRef]
- Pan, Y.; Chen, W.; Yang, J.; Zheng, J.; Yang, M.; Yi, C. Facile Synthesis of Gadolinium Chelate-Conjugated Polymer Nanoparticles for Fluorescence/Magnetic Resonance Dual-Modal Imaging. Anal. Chem. 2018, 90, 1992–2000. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, W.; Zhu, G.; Xie, J.; Chen, X. Rethinking cancer nanotheranostics. Nat. Rev. Mater. 2017, 2, 17024. [Google Scholar] [CrossRef]
- Pautu, V.; Mellinger, A.; Resnier, P.; Lepeltier, E.; Martin, L.; Boussemart, L.; Letournel, F.; Passirani, C.; Clere, N. Melanoma tumour vasculature heterogeneity: From mice models to human. J. Cancer Res. Clin. Oncol. 2019, 145, 589–597. [Google Scholar] [CrossRef]
- Sindhwani, S.; Syed, A.M.; Wilhelm, S.; Chan, W.C. Exploring Passive Clearing for 3D Optical Imaging of Nanoparticles in Intact Tissues. Bioconjugate Chem. 2017, 28, 253–259. [Google Scholar] [CrossRef]
- Tang, Y.; Soroush, F.; Sheffield, J.B.; Wang, B.; Prabhakarpandian, B.; Kiani, M.F. A Biomimetic Microfluidic Tumor Microenvironment Platform Mimicking the EPR Effect for Rapid Screening of Drug Delivery Systems. Sci. Rep. 2017, 7, 9359. [Google Scholar] [CrossRef]
- Wang, H.-F.; Ran, R.; Liu, Y.; Hui, Y.; Zeng, B.; Chen, D.; Weitz, D.A.; Zhao, C.-X. Tumor-Vasculature-on-a-Chip for Investigating Nanoparticle Extravasation and Tumor Accumulation. ACS Nano 2018, 12, 11600–11609. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, H.; Fontana, F.; Hirvonen, J.T.; Santos, H.A. Current developments and applications of microfluidic technology toward clinical translation of nanomedicines. Adv. Drug Deliv. Rev. 2018, 128, 54–83. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Fletcher, N.L.; Liu, T.; Gemmell, A.C.; Houston, Z.H.; Blakey, I.; Thurecht, K.J. In vivo therapeutic evaluation of polymeric nanomedicines: Effect of different targeting peptides on therapeutic efficacy against breast cancer. Nanotheranostics 2018, 2, 360–370. [Google Scholar] [CrossRef]
- Beziere, N.; Lozano, N.; Nunes, A.; Salichs, J.; Queiros, D.; Kostarelos, K.; Ntziachristos, V. Dynamic imaging of PEGylated indocyanine green (ICG) liposomes within the tumor microenvironment using multi-spectral optoacoustic tomography (MSOT). Biomaterials 2015, 37, 415–424. [Google Scholar] [CrossRef]
- Laramie, M.D.; Smith, M.K.; Marmarchi, F.; McNally, L.R.; Henary, M. Small Molecule Optoacoustic Contrast Agents: An Unexplored Avenue for Enhancing In Vivo Imaging. Molecules 2018, 23, 2766. [Google Scholar] [CrossRef]
- Martin, J.D.; Seano, G.; Jain, R.K. Normalizing Function of Tumor Vessels: Progress, Opportunities, and Challenges. Annu. Rev. Physiol. 2019, 81, 505–534. [Google Scholar] [CrossRef]
- Ho, Y.J.; Wang, T.C.; Fan, C.H.; Yeh, C.K. Spatially Uniform Tumor Treatment and Drug Penetration by Regulating Ultrasound with Microbubbles. ACS Appl. Mater. Interfaces 2018, 10, 17784–17791. [Google Scholar] [CrossRef]
- Mathivet, T.; Bouleti, C.; Van Woensel, M.; Stanchi, F.; Verschuere, T.; Phng, L.K.; Dejaegher, J.; Balcer, M.; Matsumoto, K.; Georgieva, P.B.; et al. Dynamic stroma reorganization drives blood vessel dysmorphia during glioma growth. EMBO Mol. Med. 2017, 9, 1629–1645. [Google Scholar] [CrossRef]
- Yang, S.; Gao, H. Nanoparticles for modulating tumor microenvironment to improve drug delivery and tumor therapy. Pharmacol. Res. 2017, 126, 97–108. [Google Scholar] [CrossRef]
- Lucas, A.T.; White, T.F.; Deal, A.M.; Herity, L.B.; Song, G.; Santos, C.M.; Zamboni, W.C. Profiling the relationship between tumor-associated macrophages and pharmacokinetics of liposomal agents in preclinical murine models. Nanomedicine 2017, 13, 471–482. [Google Scholar] [CrossRef]
- Turley, S.J.; Cremasco, V.; Astarita, J.L. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat. Rev. Immunol. 2015, 15, 669–682. [Google Scholar] [CrossRef]
- Chen, Z.J.; Liu, W.P.; Wang, X.; Liu, Y.; Li, X.H. Sequential Drug Release to Modulate Collagen Synthesis and Promote Micelle Penetration in Tumors. ACS Biomater. Sci. Eng. 2019, 5, 1343–1353. [Google Scholar] [CrossRef]
- Jahanban-Esfahlan, R.; de la Guardia, M.; Ahmadi, D.; Yousefi, B. Modulating tumor hypoxia by nanomedicine for effective cancer therapy. J. Cell Physiol. 2018, 233, 2019–2031. [Google Scholar] [CrossRef]
- Stapleton, S.; Dunne, M.; Milosevic, M.; Tran, C.W.; Gold, M.J.; Vedadi, A.; McKee, T.D.; Ohashi, P.S.; Allen, C.; Jaffray, D.A. Radiation and Heat Improve the Delivery and Efficacy of Nanotherapeutics by Modulating Intratumoral Fluid Dynamics. ACS Nano 2018, 12, 7583–7600. [Google Scholar] [CrossRef]
- Moore, T.L.; Hauser, D.; Gruber, T.; Rothen-Rutishauser, B.; Lattuada, M.; Petri-Fink, A.; Lyck, R. Cellular Shuttles: Monocytes/Macrophages Exhibit Transendothelial Transport of Nanoparticles under Physiological Flow. ACS Appl. Mater. Interfaces 2017, 9, 18501–18511. [Google Scholar] [CrossRef]
- Sun, Q.; Zhou, Z.; Qiu, N.; Shen, Y. Rational Design of Cancer Nanomedicine: Nanoproperty Integration and Synchronization. Adv. Mater. 2017, 29, 1606628. [Google Scholar] [CrossRef]
- Zhang, L.; Hao, P.Y.; Yang, D.J.; Feng, S.; Peng, B.; Appelhans, D.; Zhang, T.H.; Zan, X.J. Designing nanoparticles with improved tumor penetration: Surface properties from the molecular architecture viewpoint. J. Mater. Chem. B 2019, 7, 953–964. [Google Scholar] [CrossRef]
- Mu, J.; Lin, J.; Huang, P.; Chen, X. Development of endogenous enzyme-responsive nanomaterials for theranostics. Chem. Soc. Rev. 2018, 47, 5554–5573. [Google Scholar] [CrossRef]
- Li, H.-J.; Du, J.-Z.; Liu, J.; Du, X.-J.; Shen, S.; Zhu, Y.-H.; Wang, X.; Ye, X.; Nie, S.; Wang, J. Smart Superstructures with Ultrahigh pH-Sensitivity for Targeting Acidic Tumor Microenvironment: Instantaneous Size Switching and Improved Tumor Penetration. ACS Nano 2016, 10, 6753–6761. [Google Scholar] [CrossRef]
- Zhao, Y.; Houston, Z.H.; Simpson, J.D.; Chen, L.; Fletcher, N.L.; Fuchs, A.V.; Blakey, I.; Thurecht, K.J. Using Peptide Aptamer Targeted Polymers as a Model Nanomedicine for Investigating Drug Distribution in Cancer Nanotheranostics. Mol. Pharmaceutics 2017, 14, 3539–3549. [Google Scholar] [CrossRef]
- Kong, L.; Campbell, F.; Kros, A. DePEGylation strategies to increase cancer nanomedicine efficacy. Nanoscale Horiz. 2019, 4, 378–387. [Google Scholar] [CrossRef]
- Zhao, G.; Long, L.; Zhang, L.; Peng, M.; Cui, T.; Wen, X.; Zhou, X.; Sun, L.; Che, L. Smart pH-sensitive nanoassemblies with cleavable PEGylation for tumor targeted drug delivery. Sci. Rep. 2017, 7, 3383. [Google Scholar] [CrossRef]
- Hannun, Y.A. Apoptosis and the dilemma of cancer chemotherapy. Blood 1997, 89, 1845–1853. [Google Scholar]
- Raghavan, D.; Koczwara, B.; Javle, M. Evolving strategies of cytotoxic chemotherapy for advanced prostate cancer. Eur. J. Cancer 1997, 33, 566–574. [Google Scholar] [CrossRef]
- Sen, S.; D’Incalci, M. Apoptosis. Biochemical events and relevance to cancer chemotherapy. FEBS Lett. 1992, 307, 122–127. [Google Scholar] [CrossRef]
- Ma, X.; Gong, N.; Zhong, L.; Sun, J.; Liang, X.J. Future of nanotherapeutics: Targeting the cellular sub-organelles. Biomaterials 2016, 97, 10–21. [Google Scholar] [CrossRef]
- Maity, A.R.; Stepensky, D. Limited Efficiency of Drug Delivery to Specific Intracellular Organelles Using Subcellularly "Targeted" Drug Delivery Systems. Mol. Pharmaceutics 2016, 13, 1–7. [Google Scholar] [CrossRef]
- Azevedo, C.; Macedo, M.H.; Sarmento, B. Strategies for the enhanced intracellular delivery of nanomaterials. Drug Discov. Today 2018, 23, 944–959. [Google Scholar] [CrossRef]
- Misra, R.; Sahoo, S.K. Intracellular trafficking of nuclear localization signal conjugated nanoparticles for cancer therapy. Eur. J. Pharm. Sci. 2010, 39, 152–163. [Google Scholar] [CrossRef]
- Pan, L.; He, Q.; Liu, J.; Chen, Y.; Ma, M.; Zhang, L.; Shi, J. Nuclear-targeted drug delivery of TAT peptide-conjugated monodisperse mesoporous silica nanoparticles. J. Am. Chem. Soc. 2012, 134, 5722–5725. [Google Scholar] [CrossRef]
- Maity, A.R.; Stepensky, D. Delivery of drugs to intracellular organelles using drug delivery systems: Analysis of research trends and targeting efficiencies. Int. J. Pharm. 2015, 496, 268–274. [Google Scholar] [CrossRef]
- Singh, M.S.; Tammam, S.N.; Shetab Boushehri, M.A.; Lamprecht, A. MDR in cancer: Addressing the underlying cellular alterations with the use of nanocarriers. Pharmacol. Res. 2017, 126, 2–30. [Google Scholar] [CrossRef]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef]
- Canton, I.; Battaglia, G. Endocytosis at the nanoscale. Chem. Soc. Rev. 2012, 41, 2718–2739. [Google Scholar] [CrossRef]
- Johnston, A.P.R. Life Under the Microscope: Quantifying Live Cell Interactions to Improve Nanoscale Drug Delivery. ACS Sens. 2017, 2, 4–9. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.C.; Stenzel, M.H. Entry of nanoparticles into cells: The importance of nanoparticle properties. Polym. Chem.-Uk 2018, 9, 259–272. [Google Scholar] [CrossRef]
- Steichen, S.D.; Caldorera-Moore, M.; Peppas, N.A. A review of current nanoparticle and targeting moieties for the delivery of cancer therapeutics. Eur. J. Pharm. Sci. 2013, 48, 416–427. [Google Scholar] [CrossRef] [Green Version]
- Huhn, D.; Kantner, K.; Geidel, C.; Brandholt, S.; De Cock, I.; Soenen, S.J.H.; Gil, P.R.; Montenegro, J.M.; Braeckmans, K.; Mullen, K.; et al. Polymer-Coated Nanoparticles Interacting with Proteins and Cells: Focusing on the Sign of the Net Charge. ACS Nano 2013, 7, 3253–3263. [Google Scholar] [CrossRef]
- Mann, S.K.; Czuba, E.; Selby, L.I.; Such, G.K.; Johnston, A.P. Quantifying Nanoparticle Internalization Using a High Throughput Internalization Assay. Pharm. Res. 2016, 33, 2421–2432. [Google Scholar] [CrossRef]
- Song, D.; Cui, J.; Ju, Y.; Faria, M.; Sun, H.; Howard, C.B.; Thurecht, K.J.; Caruso, F. Cellular Targeting of Bispecific Antibody-Functionalized Poly(ethylene glycol) Capsules: Do Shape and Size Matter? ACS Appl. Mater. Interfaces 2019. [Google Scholar] [CrossRef]
- Zhang, Z.; Shen, W.; Ling, J.; Yan, Y.; Hu, J.; Cheng, Y. The fluorination effect of fluoroamphiphiles in cytosolic protein delivery. Nat. Commun. 2018, 9, 1377. [Google Scholar] [CrossRef]
- Walkey, C.D.; Olsen, J.B.; Guo, H.; Emili, A.; Chan, W.C. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc. 2012, 134, 2139–2147. [Google Scholar] [CrossRef]
- Li, S.D.; Huang, L. Stealth nanoparticles: High density but sheddable PEG is a key for tumor targeting. J. Control. Release 2010, 145, 178–181. [Google Scholar] [CrossRef] [Green Version]
- Dai, Q.; Bertleff-Zieschang, N.; Braunger, J.A.; Björnmalm, M.; Cortez-Jugo, C.; Caruso, F. Particle targeting in complex biological media. Adv. Healthc. Mater. 2018, 7. [Google Scholar] [CrossRef]
- Jin, Q.; Deng, Y.Y.; Chen, X.H.; Ji, J. Rational Design of Cancer Nanomedicine for Simultaneous Stealth Surface and Enhanced Cellular Uptake. ACS Nano 2019, 13, 954–977. [Google Scholar] [CrossRef]
- Li, H.; Yu, S.S.; Miteva, M.; Nelson, C.E.; Werfel, T.; Giorgio, T.D.; Duvall, C.L. Matrix Metalloproteinase Responsive, Proximity-activated Polymeric Nanoparticles for siRNA Delivery. Adv. Funct. Mater. 2013, 23, 3040–3052. [Google Scholar] [CrossRef]
- Gordon, M.R.; Zhao, B.; Anson, F.; Fernandez, A.; Singh, K.; Homyak, C.; Canakci, M.; Vachet, R.W.; Thayumanavan, S. Matrix Metalloproteinase-9-Responsive Nanogels for Proximal Surface Conversion and Activated Cellular Uptake. Biomacromolecules 2018, 19, 860–871. [Google Scholar] [CrossRef]
- Selby, L.I.; Cortez-Jugo, C.M.; Such, G.K.; Johnston, A.P.R. Nanoescapology: Progress toward understanding the endosomal escape of polymeric nanoparticles. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017, 9, e1452. [Google Scholar] [CrossRef]
- Wang, T.; Bai, J.; Jiang, X.; Nienhaus, G.U. Cellular uptake of nanoparticles by membrane penetration: A study combining confocal microscopy with FTIR spectroelectrochemistry. ACS Nano 2012, 6, 1251–1259. [Google Scholar] [CrossRef]
- Sakaguchi, N.; Kojima, C.; Harada, A.; Koiwai, K.; Kono, K. The correlation between fusion capability and transfection activity in hybrid complexes of lipoplexes and pH-sensitive liposomes. Biomaterials 2008, 29, 4029–4036. [Google Scholar] [CrossRef]
- Kauffman, W.B.; Fuselier, T.; He, J.; Wimley, W.C. Mechanism matters: A taxonomy of cell penetrating peptides. Trends Biochem. Sci. 2015, 40, 749–764. [Google Scholar] [CrossRef]
- Mout, R.; Ray, M.; Tay, T.; Sasaki, K.; Yesilbag Tonga, G.; Rotello, V.M. General Strategy for Direct Cytosolic Protein Delivery via Protein-Nanoparticle Co-engineering. ACS Nano 2017, 11, 6416–6421. [Google Scholar] [CrossRef]
- Postupalenko, V.; Desplancq, D.; Orlov, I.; Arntz, Y.; Spehner, D.; Mely, Y.; Klaholz, B.P.; Schultz, P.; Weiss, E.; Zuber, G. Protein Delivery System Containing a Nickel-Immobilized Polymer for Multimerization of Affinity-Purified His-Tagged Proteins Enhances Cytosolic Transfer. Angew. Chem. Int. Ed. Engl. 2015, 54, 10583–10586. [Google Scholar] [CrossRef]
- Van Sluis, R.; Bhujwalla, Z.M.; Raghunand, N.; Ballesteros, P.; Alvarez, J.; Cerdán, S.; Galons, J.P.; Gillies, R.J. In vivo imaging of extracellular pH using 1H MRSI. Magn. Reson. Med. 1999, 41, 743–750. [Google Scholar] [CrossRef]
- Kono, K.; Torikoshi, Y.; Mitsutomi, M.; Itoh, T.; Emi, N.; Yanagie, H.; Takagishi, T. Novel gene delivery systems: Complexes of fusigenic polymer-modified liposomes and lipoplexes. Gene Ther. 2001, 8, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Brandenberger, C.; Clift, M.J.; Vanhecke, D.; Muhlfeld, C.; Stone, V.; Gehr, P.; Rothen-Rutishauser, B. Intracellular imaging of nanoparticles: Is it an elemental mistake to believe what you see? Part. Fibre Toxicol. 2010, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Reifarth, M.; Hoeppener, S.; Schubert, U.S. Uptake and Intracellular Fate of Engineered Nanoparticles in Mammalian Cells: Capabilities and Limitations of Transmission Electron Microscopy-Polymer-Based Nanoparticles. Adv. Mater. 2018, 30. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Rahman, M.A.; Zhao, Z.; Weiss, K.; Zhang, C.; Chen, Z.; Hurwitz, S.J.; Chen, Z.G.; Shin, D.M.; Ke, Y. Visualization of the Cellular Uptake and Trafficking of DNA Origami Nanostructures in Cancer Cells. J. Am. Chem. Soc. 2018, 140, 2478–2484. [Google Scholar] [CrossRef] [PubMed]
- Ariotti, N.; Hall, T.E.; Rae, J.; Ferguson, C.; McMahon, K.A.; Martel, N.; Webb, R.E.; Webb, R.I.; Teasdale, R.D.; Parton, R.G. Modular Detection of GFP-Labeled Proteins for Rapid Screening by Electron Microscopy in Cells and Organisms. Dev. Cell 2015, 35, 513–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, S.S.; Martell, J.D.; Kamer, K.J.; Deerinck, T.J.; Ellisman, M.H.; Mootha, V.K.; Ting, A.Y. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 2015, 12, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Hermann, R.; Walther, P.; Muller, M. Immunogold labeling in scanning electron microscopy. Histochem. Cell Biol. 1996, 106, 31–39. [Google Scholar] [CrossRef]
- Gottstein, C.; Wu, G.; Wong, B.J.; Zasadzinski, J.A. Precise quantification of nanoparticle internalization. ACS Nano 2013, 7, 4933–4945. [Google Scholar] [CrossRef]
- Selby, L.I.; Aurelio, L.; Yuen, D.; Graham, B.; Johnston, A.P.R. Quantifying Cellular Internalization with a Fluorescent Click Sensor. ACS Sens. 2018, 3, 1182–1189. [Google Scholar] [CrossRef]
- Liu, H.; Johnston, A.P. A programmable sensor to probe the internalization of proteins and nanoparticles in live cells. Angew. Chem. Int. Ed. Engl. 2013, 52, 5744–5748. [Google Scholar] [CrossRef]
- Dai, Q.; Wilhelm, S.; Ding, D.; Syed, A.M.; Sindhwani, S.; Zhang, Y.; Chen, Y.Y.; MacMillan, P.; Chan, W.C.W. Quantifying the Ligand-Coated Nanoparticle Delivery to Cancer Cells in Solid Tumors. ACS Nano 2018, 12, 8423–8435. [Google Scholar] [CrossRef] [PubMed]
- Tietjen, G.T.; Bracaglia, L.G.; Saltzman, W.M.; Pober, J.S. Focus on Fundamentals: Achieving Effective Nanoparticle Targeting. Trends Mol. Med. 2018, 24, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.C.; Zhang, Q.; Xia, Y. The effect of sedimentation and diffusion on cellular uptake of gold nanoparticles. Nat. Nanotechnol. 2011, 6, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Such, G.K.; Yan, Y.; Johnston, A.P.; Gunawan, S.T.; Caruso, F. Interfacing materials science and biology for drug carrier design. Adv. Mater. 2015, 27, 2278–2297. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A.; Selby, L.I.; Johnston, A.P.R.; Such, G.K. The Endosomal Escape of Nanoparticles: Toward More Efficient Cellular Delivery. Bioconjugate Chem. 2019, 30, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Litwin, T.; Nagaraja, A.R.; Kwong, B.; Katz, J.; Watson, N.; Irvine, D.J. Cytosolic delivery of membrane-impermeable molecules in dendritic cells using pH-Responsive core-shell nanoparticles. Nano Lett. 2007, 7, 3056–3064. [Google Scholar] [CrossRef]
- Tran, K.K.; Zhan, X.; Shen, H. Polymer blend particles with defined compositions for targeting antigen to both class I and II antigen presentation pathways. Adv. Healthc. Mater. 2014, 3, 690–702. [Google Scholar] [CrossRef]
- Su, X.; Yang, N.; Wittrup, K.D.; Irvine, D.J. Synergistic antitumor activity from two-stage delivery of targeted toxins and endosome-disrupting nanoparticles. Biomacromolecules 2013, 14, 1093–1102. [Google Scholar] [CrossRef]
- Kongkatigumjorn, N.; Cortez-Jugo, C.; Czuba, E.; Wong, A.S.M.; Hodgetts, R.Y.; Johnston, A.P.R.; Such, G.K. Probing Endosomal Escape Using pHlexi Nanoparticles. Macromol. Biosci. 2017, 17, 1600248. [Google Scholar] [CrossRef]
- Wong, A.S.; Mann, S.K.; Czuba, E.; Sahut, A.; Liu, H.; Suekama, T.C.; Bickerton, T.; Johnston, A.P.; Such, G.K. Self-assembling dual component nanoparticles with endosomal escape capability. Soft Matter 2015, 11, 2993–3002. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Luo, L.; Wang, Y.; Wu, Q.; Dai, H.B.; Li, J.S.; Durkan, C.; Wang, N.; Wang, G.X. Endogenous pH-responsive nanoparticles with programmable size changes for targeted tumor therapy and imaging applications. Theranostics 2018, 8, 3038–3058. [Google Scholar] [CrossRef] [PubMed]
- Selbo, P.K.; Weyergang, A.; Hogset, A.; Norum, O.J.; Berstad, M.B.; Vikdal, M.; Berg, K. Photochemical internalization provides time- and space-controlled endolysosomal escape of therapeutic molecules. J. Control. Release 2010, 148, 2–12. [Google Scholar] [CrossRef]
- Mora-Espi, I.; Barrios, L.; Ibanez, E.; Soriano, J.; Nogues, C. Membrane reorganization after photochemical internalization to release transferrin-biofunctionalized polystyrene microparticles. Sci. Rep. 2018, 8, 17617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Høgset, A. Photochemical internalisation in drug and gene delivery. Adv. Drug Deliv. Rev. 2004, 56, 95–115. [Google Scholar] [CrossRef]
- Nishiyama, N.; Iriyama, A.; Jang, W.D.; Miyata, K.; Itaka, K.; Inoue, Y.; Takahashi, H.; Yanagi, Y.; Tamaki, Y.; Koyama, H.; et al. Light-induced gene transfer from packaged DNA enveloped in a dendrimeric photosensitizer. Nat. Mater. 2005, 4, 934–941. [Google Scholar] [CrossRef] [PubMed]
- Febvay, S.; Marini, D.M.; Belcher, A.M.; Clapham, D.E. Targeted cytosolic delivery of cell-impermeable compounds by nanoparticle-mediated, light-triggered endosome disruption. Nano Lett. 2010, 10, 2211–2219. [Google Scholar] [CrossRef] [PubMed]
- Nischan, N.; Herce, H.D.; Natale, F.; Bohlke, N.; Budisa, N.; Cardoso, M.C.; Hackenberger, C.P. Covalent attachment of cyclic TAT peptides to GFP results in protein delivery into live cells with immediate bioavailability. Angew. Chem. Int. Ed. Engl. 2015, 54, 1950–1953. [Google Scholar] [CrossRef] [PubMed]
- Lonn, P.; Kacsinta, A.D.; Cui, X.S.; Hamil, A.S.; Kaulich, M.; Gogoi, K.; Dowdy, S.F. Enhancing Endosomal Escape for Intracellular Delivery of Macromolecular Biologic Therapeutics. Sci. Rep. 2016, 6, 32301. [Google Scholar] [CrossRef]
- Mix, K.A.; Lomax, J.E.; Raines, R.T. Cytosolic Delivery of Proteins by Bioreversible Esterification. J. Am. Chem. Soc. 2017, 139, 14396–14398. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, P.; He, D.; Rodl, W.; Preiss, T.; Radler, J.O.; Wagner, E.; Lachelt, U. pH-Reversible Cationic RNase A Conjugates for Enhanced Cellular Delivery and Tumor Cell Killing. Biomacromolecules 2016, 17, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Wissner, R.F.; Steinauer, A.; Knox, S.L.; Thompson, A.D.; Schepartz, A. Fluorescence Correlation Spectroscopy Reveals Efficient Cytosolic Delivery of Protein Cargo by Cell-Permeant Miniature Proteins. ACS Cent. Sci. 2018, 4, 1379–1393. [Google Scholar] [CrossRef] [PubMed]
- Bale, S.S.; Kwon, S.J.; Shah, D.A.; Kane, R.S.; Dordick, J.S. A GFP complementation system for monitoring and directing nanomaterial mediated protein delivery to human cellular organelles. Biotechnol. Bioeng. 2010, 107, 1040–1047. [Google Scholar] [CrossRef] [PubMed]
- Deprey, K.; Becker, L.; Kritzer, J.; Pluckthun, A. Trapped! A Critical Evaluation of Methods for Measuring Total Cellular Uptake versus Cytosolic Localization. Bioconjugate Chem. 2019, 30, 1006–1027. [Google Scholar] [CrossRef] [PubMed]
- Hinde, E.; Thammasiraphop, K.; Duong, H.T.; Yeow, J.; Karagoz, B.; Boyer, C.; Gooding, J.J.; Gaus, K. Pair correlation microscopy reveals the role of nanoparticle shape in intracellular transport and site of drug release. Nat. Nanotechnol. 2017, 12, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Milech, N.; Longville, B.A.; Cunningham, P.T.; Scobie, M.N.; Bogdawa, H.M.; Winslow, S.; Anastasas, M.; Connor, T.; Ong, F.; Stone, S.R.; et al. GFP-complementation assay to detect functional CPP and protein delivery into living cells. Sci. Rep. 2015, 5, 18329. [Google Scholar] [CrossRef] [PubMed]
- Holub, J.M.; Larochelle, J.R.; Appelbaum, J.S.; Schepartz, A. Improved assays for determining the cytosolic access of peptides, proteins, and their mimetics. Biochemistry 2013, 52, 9036–9046. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.; Adjobo-Hermans, M.J.; Wallbrecher, R.; Verdurmen, W.P.; Bovee-Geurts, P.H.; van Oostrum, J.; Milletti, F.; Enderle, T.; Brock, R. Detecting Cytosolic Peptide Delivery with the GFP Complementation Assay in the Low Micromolar Range. Angew. Chem. Int. Ed. Engl. 2015, 54, 15105–15108. [Google Scholar] [CrossRef]
- Pollard, H.; Remy, J.S.; Loussouarn, G.; Demolombe, S.; Behr, J.P.; Escande, D. Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. J. Biol. Chem. 1998, 273, 7507–7511. [Google Scholar] [CrossRef] [PubMed]
- Pouton, C.W.; Wagstaff, K.M.; Roth, D.M.; Moseley, G.W.; Jans, D.A. Targeted delivery to the nucleus. Adv. Drug Deliv. Rev. 2007, 59, 698–717. [Google Scholar] [CrossRef]
- Ray, M.; Tang, R.; Jiang, Z.; Rotello, V.M. Quantitative tracking of protein trafficking to the nucleus using cytosolic protein delivery by nanoparticle-stabilized nanocapsules. Bioconjugate Chem. 2015, 26, 1004–1007. [Google Scholar] [CrossRef]
- Nitin, N.; LaConte, L.; Rhee, W.J.; Bao, G. Tat peptide is capable of importing large nanoparticles across nuclear membrane in digitonin permeabilized cells. Ann. Biomed. Eng. 2009, 37, 2018–2027. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.D.; Ross, N.L.; Sullivan, M.O. Requirements for the nuclear entry of polyplexes and nanoparticles during mitosis. J. Gene Med. 2012, 14, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Ray, M.; Lee, Y.W.; Hardie, J.; Mout, R.; Yesilbag Tonga, G.; Farkas, M.E.; Rotello, V.M. CRISPRed Macrophages for Cell-Based Cancer Immunotherapy. Bioconjugate Chem. 2018, 29, 445–450. [Google Scholar] [CrossRef] [PubMed]
- van der Aa, M.A.; Koning, G.A.; d’Oliveira, C.; Oosting, R.S.; Wilschut, K.J.; Hennink, W.E.; Crommelin, D.J. An NLS peptide covalently linked to linear DNA does not enhance transfection efficiency of cationic polymer based gene delivery systems. J. Gene Med. 2005, 7, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Tammam, S.N.; Azzazy, H.M.; Breitinger, H.G.; Lamprecht, A. Chitosan Nanoparticles for Nuclear Targeting: The Effect of Nanoparticle Size and Nuclear Localization Sequence Density. Mol. Pharmaceutics 2015, 12, 4277–4289. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xian, L.; Xing, H.; Yu, J.; Yang, Z.; Yang, T.; Yang, L.; Ding, P. Factors influencing the nuclear targeting ability of nuclear localization signals. J. Drug Target. 2016, 24, 927–933. [Google Scholar] [CrossRef] [PubMed]
- Grandinetti, G.; Reineke, T.M. Exploring the mechanism of plasmid DNA nuclear internalization with polymer-based vehicles. Mol. Pharmaceutics 2012, 9, 2256–2267. [Google Scholar] [CrossRef] [PubMed]
- Grandinetti, G.; Smith, A.E.; Reineke, T.M. Membrane and nuclear permeabilization by polymeric pDNA vehicles: Efficient method for gene delivery or mechanism of cytotoxicity? Mol. Pharmaceutics 2012, 9, 523–538. [Google Scholar] [CrossRef]
- Sakhrani, N.M.; Padh, H. Organelle targeting: Third level of drug targeting. Drug Des. Dev. Ther. 2013, 7, 585–599. [Google Scholar] [CrossRef]
- Mahmoud, A.; de Jongh, P.; Briere, S.; Chen, M.Z.; Nowell, C.J.; Johnston, A.P.R.; Davis, T.P.; Haddleton, D.M.; Kempe, K. Carboxylated Cy5-labeled Comb Polymers Passively Diffuse the Cell Membrane and Target Mitochondria. ACS Appl. Mater. Interfaces 2019. [Google Scholar] [CrossRef]
- Milane, L.; Trivedi, M.; Singh, A.; Talekar, M.; Amiji, M. Mitochondrial biology, targets, and drug delivery. J. Control. Release 2015, 207, 40–58. [Google Scholar] [CrossRef] [PubMed]
- Battigelli, A.; Russier, J.; Venturelli, E.; Fabbro, C.; Petronilli, V.; Bernardi, P.; Da Ros, T.; Prato, M.; Bianco, A. Peptide-based carbon nanotubes for mitochondrial targeting. Nanoscale 2013, 5, 9110–9117. [Google Scholar] [CrossRef] [PubMed]
- Agemy, L.; Friedmann-Morvinski, D.; Kotamraju, V.R.; Roth, L.; Sugahara, K.N.; Girard, O.M.; Mattrey, R.F.; Verma, I.M.; Ruoslahti, E. Targeted nanoparticle enhanced proapoptotic peptide as potential therapy for glioblastoma. Proc. Natl. Acad. Sci. USA 2011, 108, 17450–17455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshino, A.; Fujioka, K.; Oku, T.; Nakamura, S.; Suga, M.; Yamaguchi, Y.; Suzuki, K.; Yasuhara, M.; Yamamoto, K. Quantum dots targeted to the assigned organelle in living cells. Microbiol. Immunol. 2004, 48, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Stepensky, D. Quantitative aspects of intracellularly-targeted drug delivery. Pharm. Res. 2010, 27, 2776–2780. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Ardoy, A.; Lostale-Seijo, I.; Montenegro, J. Where in the Cell Is our Cargo? Methods Currently Used To Study Intracellular Cytosolic Localisation. Chembiochem 2019, 20, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Sneh-Edri, H.; Stepensky, D. ‘IntraCell’ plugin for assessment of intracellular localization of nano-delivery systems and their targeting to the individual organelles. Biochem. Biophys. Res. Commun. 2011, 405, 228–233. [Google Scholar] [CrossRef]
- Efeoglu, E.; Keating, M.; McIntyre, J.; Casey, A.; Byrne, H.J. Determination of nanoparticle localisation within subcellular organelles in vitro using Raman spectroscopy. Anal. Methods 2015, 7, 10000–10017. [Google Scholar] [CrossRef] [Green Version]
- Dorney, J.; Bonnier, F.; Garcia, A.; Casey, A.; Chambers, G.; Byrne, H.J. Identifying and localizing intracellular nanoparticles using Raman spectroscopy. Analyst 2012, 137, 1111–1119. [Google Scholar] [CrossRef] [Green Version]
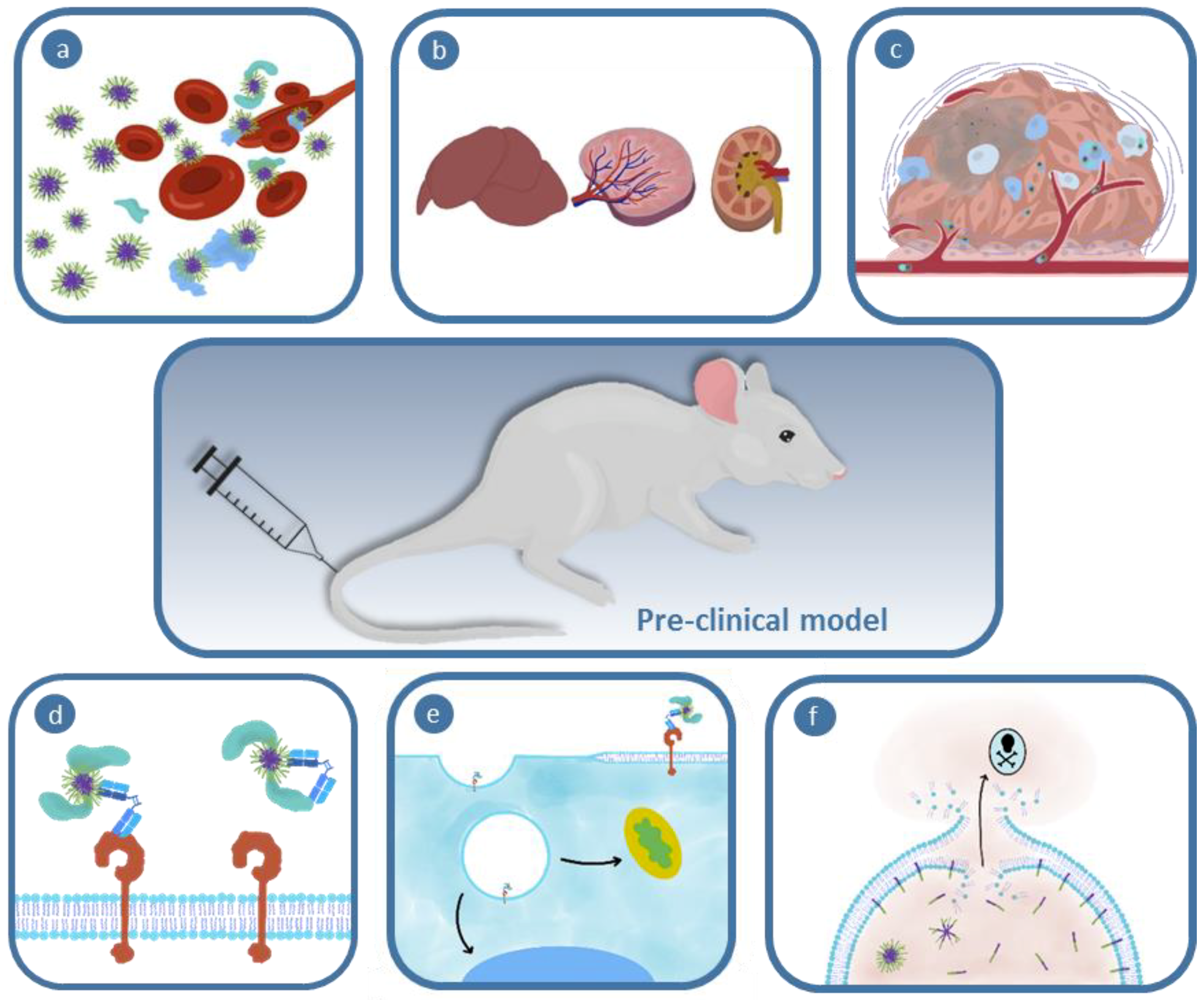


| Biological Barrier | Tools to Overcome | Current Challenges | Referred Section |
|---|---|---|---|
| Interactions in the Blood Stream | Tuning Nanoparticle Physicochemical Properties Pre-Incubation/First pass Artificial Hard Corona | Protein corona has not been extensively profiled for soft nanoparticles The plasma protein content between patients varies | Section 2.1.1 |
| Section 2.1.2 | |||
| Biodistribution | Nanoparticle Biophysical Properties Incorporation of Targeting Ligands Pre-Targeting Method Hitchhike onto Red Blood Cell | Defined sized cutoff for soft nanoparticle clearance remains challenging | Section 3.1 and Section 3.2 |
| Stealth property will be voided | Section 3.2 | ||
| Section 2.2 | |||
| Gathering at Tumor Site | Nanoparticle Size Nanoparticle Hardness Vascular Normalization and Remodeling | Section 4.1.2 | |
| Potential to induce metastasis | Section 4.1.3 | ||
| Tumor Tissue Distribution | Nanoparticle Physicochemical Properties Charge-Switching Nanoparticles Size-Switching Nanoparticles Sheddable PEG Corona Macrophage Shuttling | Few studies report on the intratumoral distribution | Section 4.2.4 |
| Prevents use of environmentally-responsive polymers | |||
| Receptor Affinity | Influenced by degree of opsonization | Section 4.2.3 | |
| Internalization | Nanoparticle Physicochemical Properties | Factors influencing nanoparticle internalization are not extensively investigated | Section 5.1.1 |
| Polymer Composition (e.g., Fluorous Substitution) Detachable Particle Corona Charge-Switching Particle Nanoparticle–Cell Membrane Fusion | |||
| Endosomal Escape | pH-Responsive Materials that Membrane Interact or Swell Modifying the Therapeutic with Cell-Penetrating Peptides Modifying the Therapeutic Physicochemical Properties Incorporation of a Photosensitizer | Internalization must be faster than material activation at tumor microenvironment pH | Section 5.2.1 |
| Requires encapsulation for selective delivery | |||
| Limitations with depth of penetration of light and toxicity | |||
| Subcellular Trafficking | (Nucleus) <9 nm Diameter of Nanoparticle or Therapeutic Conjugation of a Nuclear Localization Signal (Mitochondria) Incorporation of Mitochondrotropic Polymers Conjugation of a Mitochondria Targeting Signal | Factors influencing nanoparticle subcellular trafficking are not extensively investigated | Section 5.3.1 |
| Section 5.3.2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simpson, J.D.; Smith, S.A.; Thurecht, K.J.; Such, G. Engineered Polymeric Materials for Biological Applications: Overcoming Challenges of the Bio–Nano Interface. Polymers 2019, 11, 1441. https://doi.org/10.3390/polym11091441
Simpson JD, Smith SA, Thurecht KJ, Such G. Engineered Polymeric Materials for Biological Applications: Overcoming Challenges of the Bio–Nano Interface. Polymers. 2019; 11(9):1441. https://doi.org/10.3390/polym11091441
Chicago/Turabian StyleSimpson, Joshua D, Samuel A Smith, Kristofer J. Thurecht, and Georgina Such. 2019. "Engineered Polymeric Materials for Biological Applications: Overcoming Challenges of the Bio–Nano Interface" Polymers 11, no. 9: 1441. https://doi.org/10.3390/polym11091441