Insight into the Structure and Dynamics of Polymers by Neutron Scattering Combined with Atomistic Molecular Dynamics Simulations
Abstract
:1. Introduction
2. Grounds of the Strategy
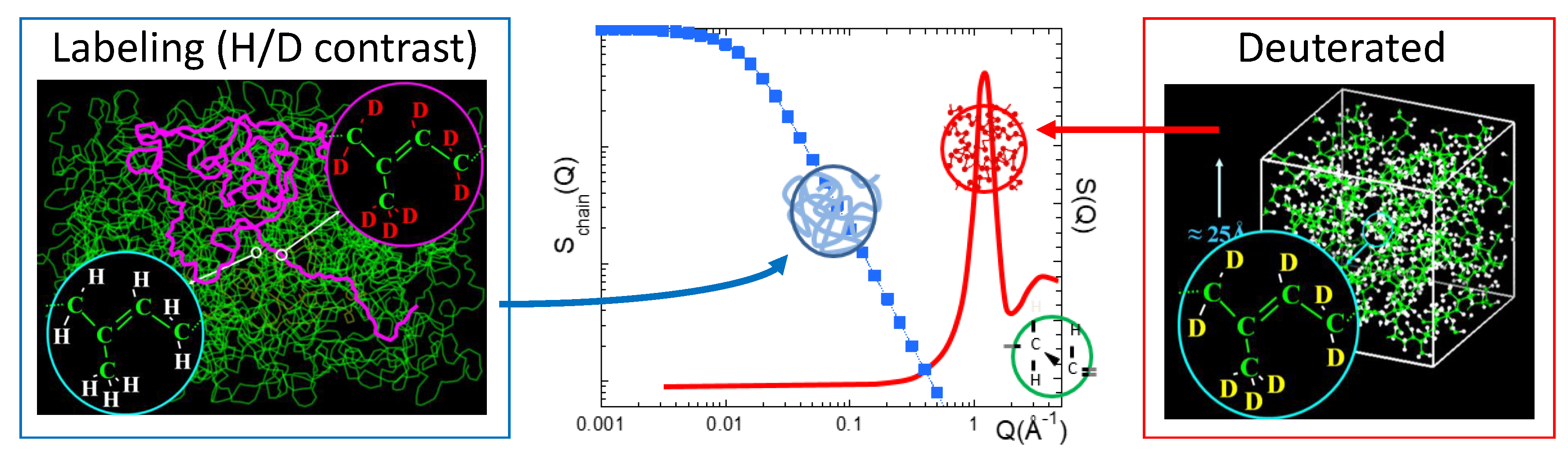
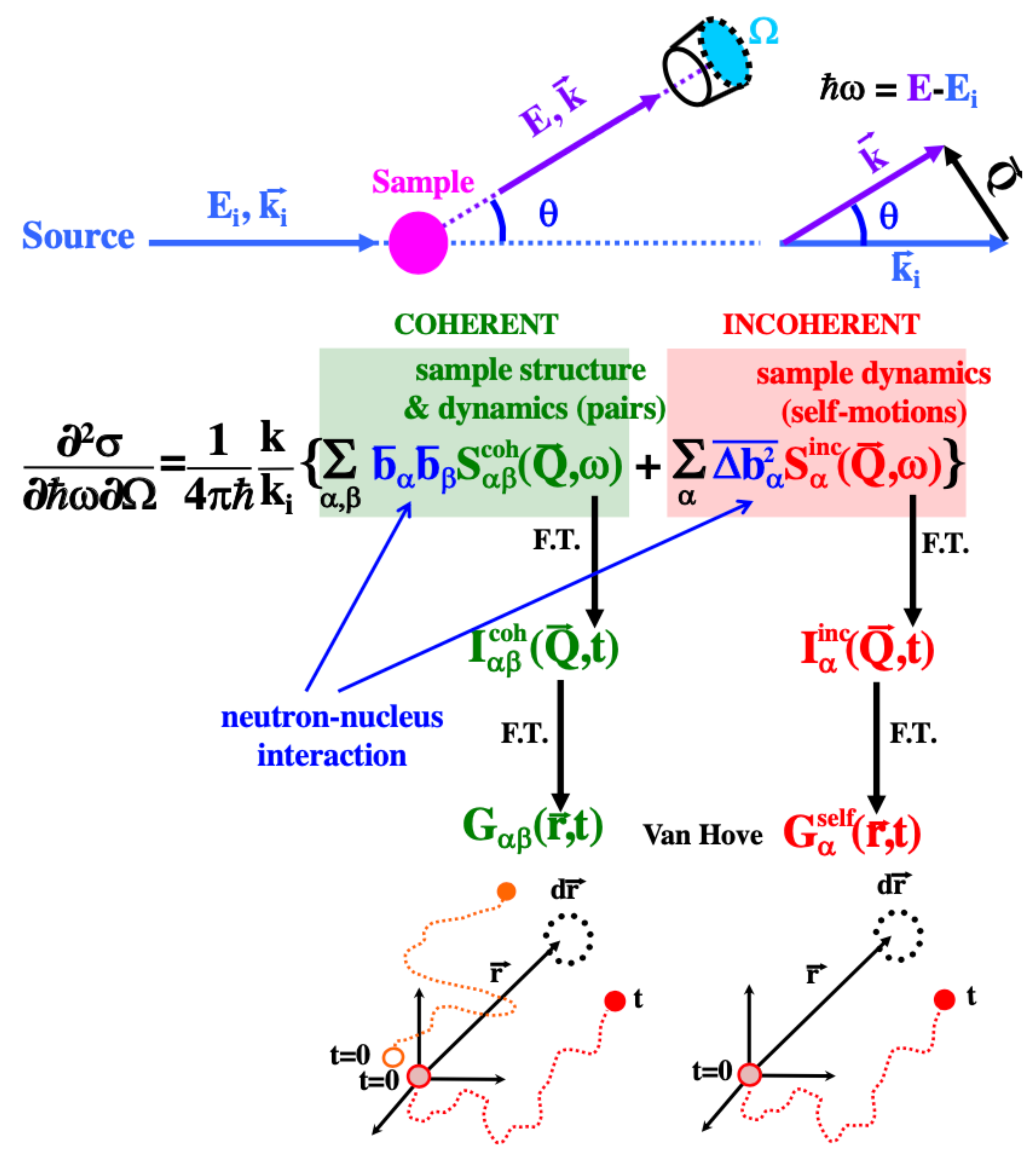
- Due to the large value of (and, thus, of ), in H-containing systems and the signal is dominated by the incoherent scattering from hydrogens. It thus reveals their self-motions
- If H is substituted by D, this incoherent contribution is drastically reduced. Then, we obtain differently weighted coherent contributions
- Since in a fully deuterated sample, the intensity scattered is mainly coherent. Given that , all pair correlations are almost equally weighted
- NS accesses correlation functions in the reciprocal space (), never in real space
- NSE does not distinguish the signals of different atoms, if they are of the same isotopic species (e.g., main-chain hydrogens vs side-group hydrogens)
- Self-motions of C and O are not accessible ( = = 0)
- With exception of the neutron spin echo (NSE) technique (F. Mezei, 1972 [18]), that directly accesses the intermediate scattering functions, experiments are performed in the frequency domain and the results are affected by the instrumental resolution through convolution
- Spectrometers cover relatively narrow dynamic windows and usually several instruments have to be combined
- Though polarization analysis (PA) allows separation of coherent and incoherent contributions, in the practice this is currently available only for diffraction experiments (structural information)
3. Results on ‘Simple’ Linear Homopolymers
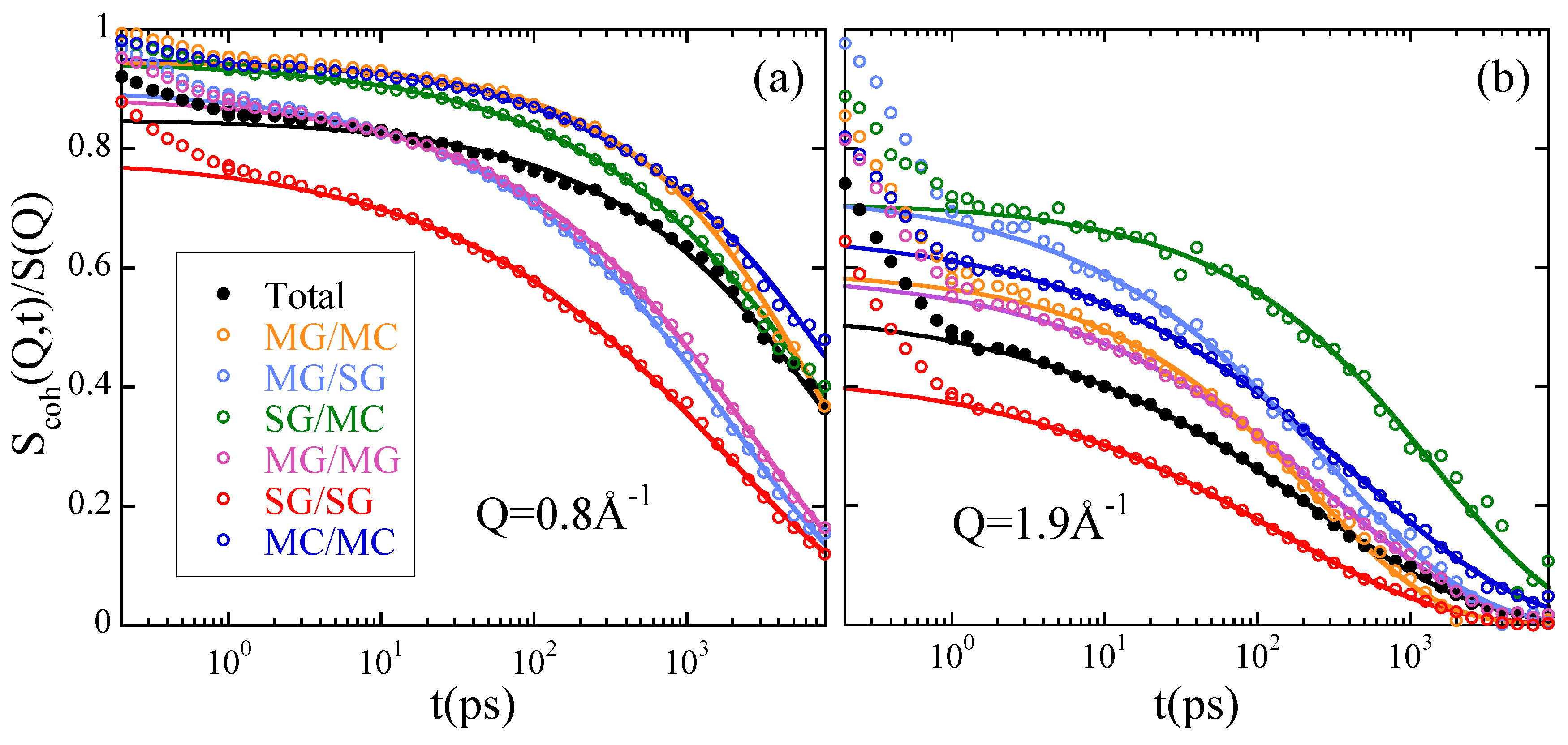
3.1. Structural Properties
3.2. Dynamical Behavior
3.2.1. Localized Motions
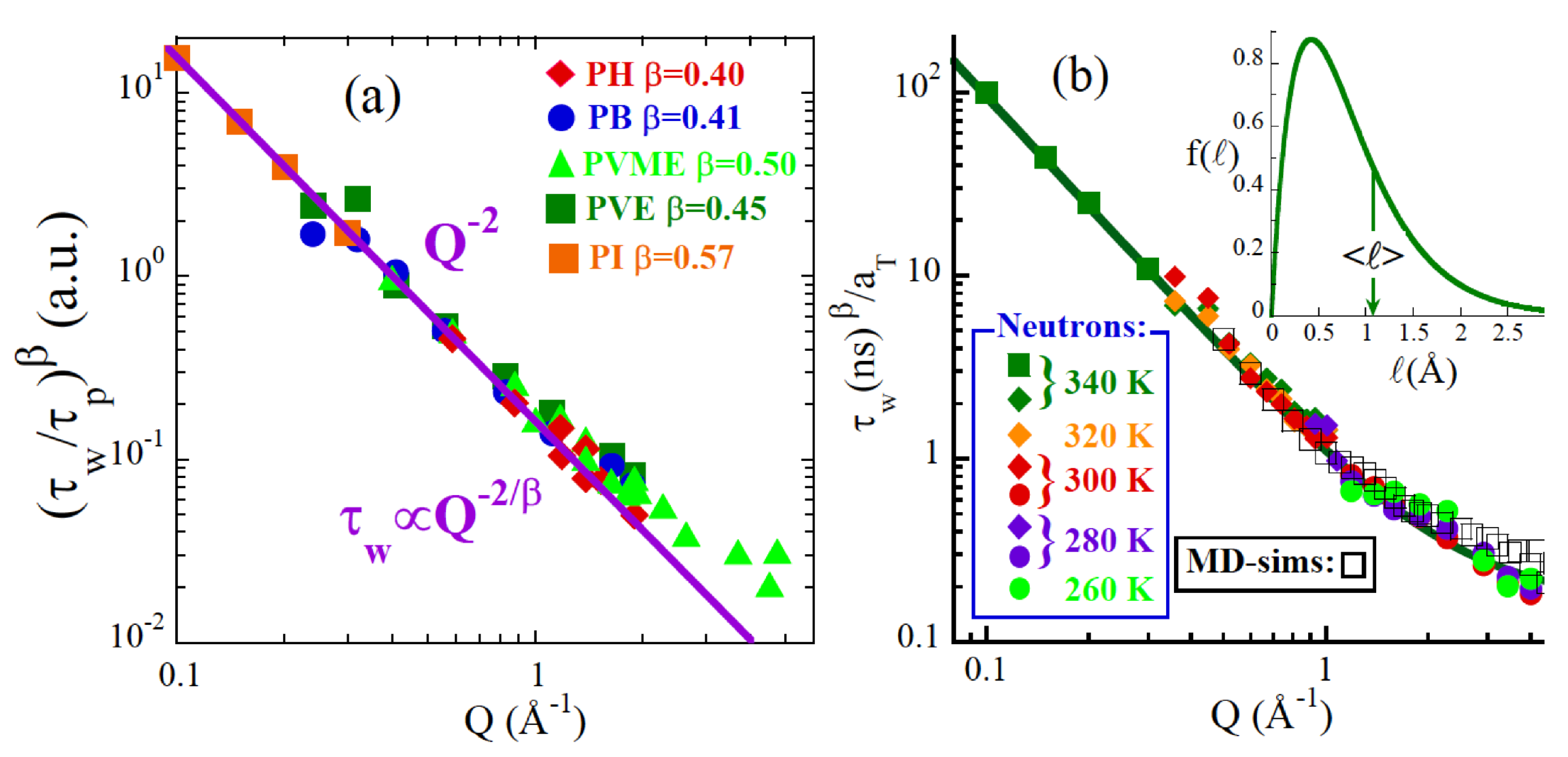
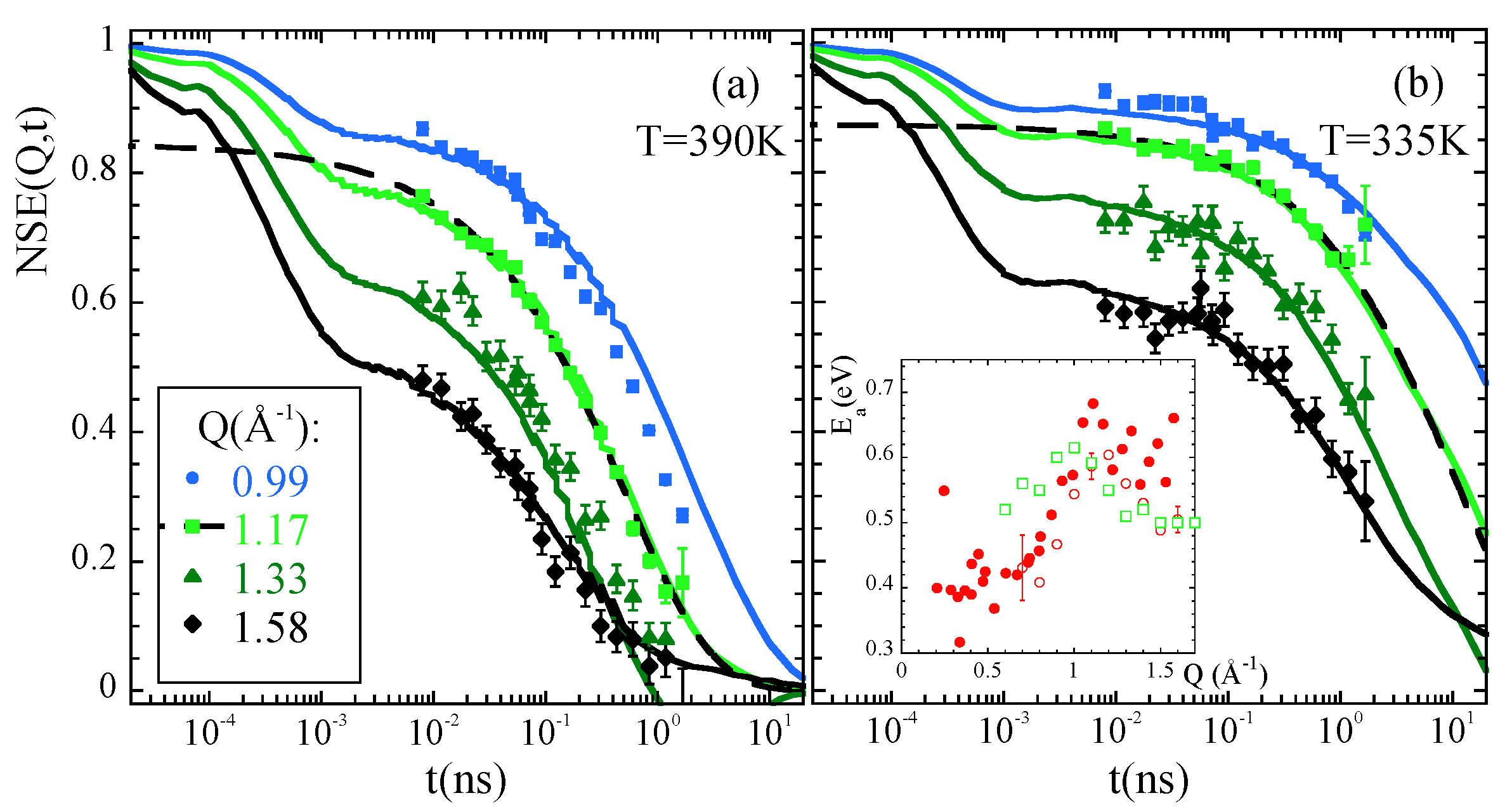
3.2.2. The Structural Relaxation
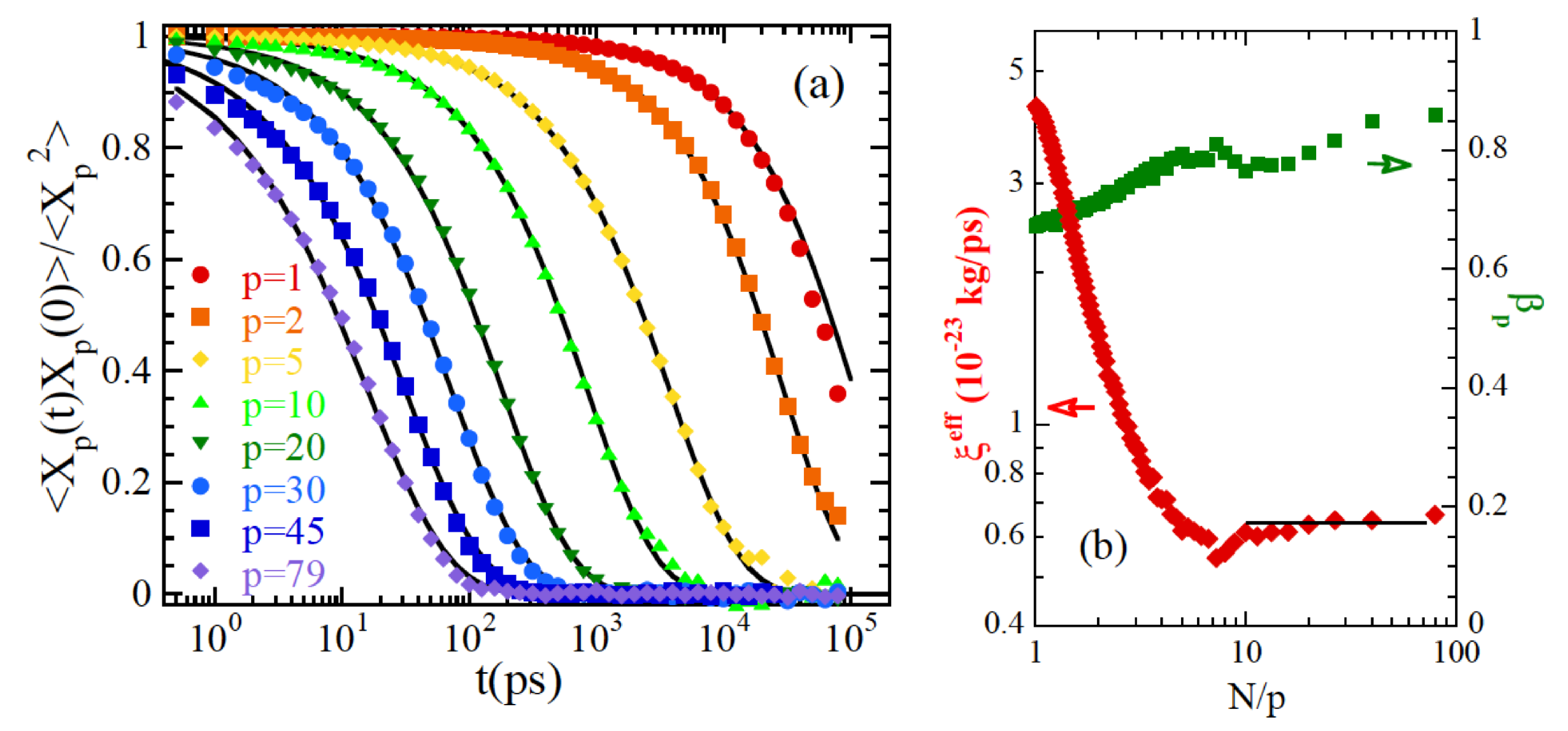
3.2.3. Chain Dynamics
4. Beyond ‘Simple’ Polymers and ‘Standard’ Experiments
4.1. Nanosegregation in More Complex Polymers
4.2. The Collective Dynamics at Intermediate Length Scales: An Unknown Territory
4.3. A Breakthrough Experiment on Water: A ‘Simple’ and Yet Enormously Complex System
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NS | Neutron scattering |
| MD | Molecular dynamics |
| NSE | Neutron spin echo |
| PA | Polarization analysis |
| PE | Polyethylene |
| AMBER | Assisted Model Building with Energy Refinement |
| COMPASS | Condensed-phase Optimized Molecular Potentials for Atomistic Simulation Studies |
| CHARMM | Chemistry at Harvard Macromolecular Mechanics |
| GROMOS | Groningen Molecular Simulation |
| OPLS | Optimized Potentials for Liquid Simulations |
| NAMD | Nanoscale Molecular Dynamics |
| GAFF | Generalized AMBER Force Field |
| GROMACS | Groningen Machine for Chemical Simulations |
| LAMMPS | Large-scale Atomic/Molecular Massively Parallel Simulator |
| OCTA | Open Computational Tool for Advanced material technology |
| PMMA | Poly(methyl metacrylate) |
| MC | Main chain |
| MG | Methyl group |
| SG | Side group |
| PI | Polyisoprene |
| PS | Polystyrene |
| PB | 1,4-Polybutadiene |
| PIB | Polyisobutylene |
| RRDM | Rotational rate distribution model |
| PVAc | Poly(vinyl acetate) |
| PPO | poly(propylene oxide) |
| KWW | Kohlrausch-Williams-Watts |
| PId3 | Polyisoprene with deuterated methyl groups |
| PVE | Poly(vinyl ethylene) |
| PVME | poly(vinyl methyl ether) |
| PEMA | Poly(ethyl methacrylate) |
| PTHF | Poly(tetrahydrofurane) |
| a-PP | atactic Polypropylene |
| PU | Polyurethene |
| PVC | Poly(vinyl chloride) |
| PEP | Poly(ethylene propylene) |
| MCT | Mode Coupling Theory |
| PPG | Poly(propylene glycol) |
| ARS | All rotational state model |
| PEO | Poly(ethylene oxide) |
| msd | mean squared displacement |
| PnMAs | Poly(n-alkyl methacrylates) |
| PAOs | Poly(alkylene oxides) |
| PVP | Poly(vinylpyrrolidone) |
| ILS | Intermediate length scales |
| NMR | Nuclear magnetic resonance |
| SANS | Small angle neutron scattering |
| SCNPs | Single-chain nano-particles |
References
- Flory, P. Principles of Polymer Chemistry; Cornell University Press: London, UK, 1953. [Google Scholar]
- Götze, W. Liquids, Freezing, Glass Transition; Elsevier: Amsterdam, The Netherlands, 1991; p. 287. [Google Scholar]
- Colmenero, J.; Mukhopadhyay, R.; Alegría, A.; Frick, B. Quantum rotational tunneling of methyl groups in polymers. Phys. Rev. Lett. 1998, 80, 2350–2353. [Google Scholar]
- Johari, G.; Goldstein, M. Viscous liquids and the glass transition. II. Secondary relaxations in glasses of rigid molecules. J. Chem. Phys. 1970, 53, 2372. [Google Scholar]
- Götze, M.; Mayr, M.R. Evolution of vibrational excitations in glassy systems. Phys. Rev. E 2000, 61, 587. [Google Scholar]
- Arbe, A.; Alvarez, F.; Colmenero, J. Neutron scattering and molecular dynamics simulations: Synergetic tools to unravel structure and dynamics in polymers. Soft Matter 2012, 8, 8257–8270. [Google Scholar]
- Lovesey, S.W. Theory of Neutron Scattering from Condensed Matter; Clarendon Press: Oxford, UK, 1984. [Google Scholar]
- Squires, G.L. Introduction to the Theory of Thermal Neutron Scattering; Dover Publication Inc.: New York, NY, USA, 1996. [Google Scholar]
- Squires, G.L. Introduction to Thermal Neutron Scattering; Cambridge University Press: Cambridge, UK, 1988. [Google Scholar]
- Kirste, R.G.; Kruse, W.A.; Schelten, J. Die Bestimmung des Trägheitsradius von Polymethylmethacrylat in Glaszustand durch Neutronenbeugung. Die Makromolekulare Chemie 1973, 162, 299–303. [Google Scholar]
- Benoit, H.; Decker, D.; Higgins, J.S.; Picot, C.; Cotton, J.P.; Farnoux, B.; Jannink, G.; Ober, R. Dimensions of a flexible polymer chain in the bulk and in solution. Nat. Phys. Sci. 1973, 245, 13. [Google Scholar]
- Rouse, P.E. A theory of the linear viscoelastic properties of dilute solutions of coiling polymers. J. Chem. Phys. 1953, 21, 1272. [Google Scholar]
- Richter, D.; Monkenbusch, M.; Arbe, A.; Colmenero, J. Neutron Spin Echo Investigations on Polymer Dynamics. In Advances in Polymer Science; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2005; Volume 174. [Google Scholar]
- Schleger, P.; Farago, B.; Lartigue, C.; Kollmar, A.; Richter, D. Clear evidence of reptation in polyethylene from neutron spin-echo spectroscopy. Phys. Rev. Lett. 1998, 81, 124. [Google Scholar]
- Doi, M.; Edwards, S.F. The Theory of Polymer Dynamics; Clarendon Press: Oxford, UK, 1986. [Google Scholar]
- De Gennes, P.G. Reptation of a polymer chain in the presence of fixed obstacles. J. Chem. Phys. 1971, 55, 572. [Google Scholar]
- Available online: http://neutronsources.org (accessed on 17 December 2020).
- Mezei, F. Neutron Spin Echo, Lecture Notes in Physics; Springer: Heidelberg, Germany, 1980; Volume 28. [Google Scholar]
- Arbe, A.; Nilsen, G.J.; Stewart, J.R.; Alvarez, F.; Sakai, V.G.; Colmenero, J. Coherent structural relaxation of water from meso- to intermolecular scales measured using neutron spectroscopy with polarization analysis. Phys. Rev. Res. 2020, 2, 022015. [Google Scholar] [CrossRef] [Green Version]
- Rahman, A. Correlations in the motion of atoms in liquid argon. Phys. Rev. 1964, 136, A405. [Google Scholar]
- Fetters, L.J.; Lohse, D.J.; Richter, D.; Witten, T.A.; Zirkel, A. Connection between polymer molecular weight, density, chain dimensions, and melt viscoelastic properties. Macromolecules 1994, 27, 4639. [Google Scholar]
- Theodorou, D.; Suter, U. Atomistic modeling of mechanical properties of polymeric glasses. Macromolecules 1986, 19, 139. [Google Scholar]
- Theodorou, D.; Suter, U. Local structure and the mechanism of response to elastic deformation in a glassy polymer. Macromolecules 1986, 19, 379. [Google Scholar]
- Karayiannis, N.; Mavrantzas, V.; Theodorou, D. A novel Monte Carlo scheme for the rapid equilibration of atomistic model polymer systems of precisely defined molecular architecture. Phys. Rev. Lett. 2002, 88, 105503. [Google Scholar] [PubMed]
- Karayiannis, N.C.; Mavrantzas, V.G. Hierarchical modeling of the dynamics of polymers with a nonlinear molecular architecture: Calculation of branch point friction and chain reptation time of H-shaped polyethylene melts from long molecular dynamics simulations. Macromolecules 2005, 38, 8583–8596. [Google Scholar]
- Cornell, W.; Cieplak, P.; Bayly, C.; Gould, I.; Merz, K.J.; Ferguson, D.; Spellmeyer, D.; Fox, T.; Caldwell, J.; Kollman, P. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995, 117, 5179. [Google Scholar]
- Sun, H. COMPASS: An ab initio force-field optimized for condensed-phase applications—Overview with details on alkane and benzene compounds. J. Phys. Chem. B 1998, 102, 7338. [Google Scholar]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586. [Google Scholar]
- Scott, W.R.P.; Hünenberger, P.H.; Tironi, I.G.; Mark, A.E.; Billeter, S.R.; Fennen, J.; Torda, A.E.; Huber, T.; Krüger, P.; van Gunsteren, W.F. The GROMOS biomolecular simulation program package. J. Phys. Chem. A 1999, 103, 3596. [Google Scholar]
- Jorgensen, W.; Tirado-Rives, J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 1988, 110, 1657. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Mayo, S.; Olafson, B.; Goddard, W. DREIDING: A generic force field for molecular simulations. J. Phys. Chem. 1990, 94, 8897. [Google Scholar] [CrossRef]
- Allen, M.; Tildesley, D. Computer Simulation of Liquids; Oxford Science Publications: Oxford, UK, 1987. [Google Scholar]
- Available online: www.accelrys.com (accessed on 17 December 2020).
- Available online: www.gromacs.org (accessed on 17 December 2020).
- Available online: www.ccp5.ac.uk (accessed on 17 December 2020).
- Large-Scale Atomic/Molecular Massively Parallel Simulator (LAMMPS) is a Molecular Dynamics Program from Sandia National Laboratories.
- Available online: www.ks.uiuc.edu/Research/namd (accessed on 17 December 2020).
- Available online: octa.jp (accessed on 17 December 2020).
- Tsolou, G.; Mavrantzas, V.; Theodorou, D. Detailed atomistic molecular dynamics simulation of cis-1,4-poly(butadiene). Macromolecules 2005, 38, 1478. [Google Scholar] [CrossRef]
- Stephanou, P.; Baig, C.; Tsolou, G.; Mavrantzas, V.; Kröger. Quantifying chain reptation in entangled polymer melts: Topological and dynamical mapping of atomistic simulation results onto the tube model. J. Chem. Phys. 2010, 132, 124904. [Google Scholar] [CrossRef] [Green Version]
- Binder, K. (Ed.) Monte Carlo and Molecular Dynamics Simulations in Polymer Science; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- Kotelyanski, M.; Theodorou, D. (Eds.) Simulation Methods for Polymers; Marcel Dekker: New York, NY, USA, 2004. [Google Scholar]
- Smith, G.D.; Paul, W.; Yoon, D.Y.; Zirkel, A.; Hendricks, J.; Richter, D.; Schober, H. Local dynamics in a long-chain alkane melt from molecular dynamics simulations and neutron scattering experiments. J. Chem. Phys. 1997, 107, 4751–4755. [Google Scholar] [CrossRef]
- Paul, W.; Smith, G.; Yoon, D.; Farago, B.; Rathgeber, S.; Zirkel, A.; Willner, L.; Richter, D. Chain motion in an unentangled polyethylene melt: A critical test of the Rouse model by molecular dynamics simulations and neutron spin echo spectroscopy. Phys. Rev. Lett. 1998, 80, 2346. [Google Scholar] [CrossRef]
- Eilhard, J.; Zirkel, A.; Tschop, W.; Hahn, O.; Kremer, K.; Scharpf, O.; Richter, D.; Buchenau, U. Spatial correlations in polycarbonates: Neutron scattering and simulation. J. Chem. Phys. 1999, 110, 1819. [Google Scholar] [CrossRef] [Green Version]
- Smith, G.; Paul, W.; Monkenbusch, M.; Richter, D. A comparison of neutron scattering studies and computer simulations of polymer melts. Chem. Phys. 2000, 261, 61. [Google Scholar] [CrossRef]
- Smith, G.; Paul, W.; Monkenbusch, M.; Richter, D. On the non-Gaussianity of chain motion in unentangled polymer melts. J. Chem. Phys. 2001, 114, 4285. [Google Scholar] [CrossRef] [Green Version]
- Triolo, A.; Arrighi, V.; Triolo, R.; Passerini, S.; Mastragostino, M.; Lechner, R.; Ferguson, R.; Borodin, O.; Smith, G. Dynamic heterogenity in polymer electrolytes. Comparison between QENS data and MD simulations. Physica B 2001, 301, 163. [Google Scholar] [CrossRef]
- Ahumada, O.; Theodorou, D.; Triolo, A.; Arrighi, V.; Karatasos, C.; Ryckaert, J. Segmental dynamics of atactic polypropylene as revealed by molecular simulations and quasielastic neutron scattering. Macromolecules 2002, 35, 7110. [Google Scholar] [CrossRef] [Green Version]
- Arialdi, G.; Karatasos, C.; Ryckaert, J.; Arrighi, V.; Saggio, F.; Triolo, A.; Desmedt, A.; Pieper, J.; Lechner, R. Local dynamics of polyethylene and its oligomers: A molecular dynamics interpretation of the incoherent dynamic structure factor. Macromolecules 2003, 36, 8864. [Google Scholar] [CrossRef]
- Paul, W.; Smith, G. Structure and dynamics of amorphous polymers: Computer simulations compared to experiment and theory. Rep. Prog. Phys. 2004, 67, 1117. [Google Scholar] [CrossRef]
- Tengroth, C.; Engberg, D.; Wahnström, G.; Börjesson, L.; Carlsson, P.; Ahlström, P.; Howells, W.S. The segmental and rotational dynamics of PPO, above the glass-transition, investigated by neutron scattering and molecular dynamics simulations. Soft Mater. 2005, 3, 1. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Depa, P.; García-Sakai, V.; Maranas, J.; Lynn, J.; Peral, I.; Copley, J. A comparison of united atom, explicit atom, and coarse-grained simulation models for poly(ethylene oxide). J. Chem. Phys. 2006, 124, 234901. [Google Scholar] [CrossRef]
- Chen, C.; Depa, P.; Maranas, J.; García-Sakai, V. Comparison of explicit atom, united atom, and coarse-grained simulations of poly(methyl methacrylate). J. Chem. Phys. 2008, 128, 124906. [Google Scholar] [CrossRef]
- Frick, B.; Richter, D.; Ritter, C. Structural changes near the glass transition – neutron diffraction on a simple polymer. Europhys. Lett. 1989, 9, 557. [Google Scholar] [CrossRef]
- Genix, A.C.; Arbe, A.; Alvarez, F.; Colmenero, J.; Schweika, W.; Richter, D. Local structure of syndiotactic poly(methyl methacrylate). A combined study by neutron diffraction with polarization analysis and atomistic molecular dynamics simulations. Macromolecules 2006, 39, 3947–3958. [Google Scholar] [CrossRef] [Green Version]
- Arbe, A.; Genix, A.C.; Colmenero, J.; Richter, D.; Fouquet, P. Anomalous relaxation of self-assembled alkyl nanodomains in high-order poly(n-alkyl methacrylates). Soft Matter 2008, 4, 1792–1795. [Google Scholar] [CrossRef]
- Sundarajan, P.; Flory, P. Configurational characteristics of poly (methyl methacrylate). J. Am. Chem. Soc. 1974, 96, 5025. [Google Scholar] [CrossRef]
- Alvarez, F.; Colmenero, J.; Zorn, R.; Willner, L.; Richter, D. Partial structure factors of polyisoprene: Neutron scattering and molecular dynamics simulation. Macromolecules 2003, 36, 238. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, F.; Arbe, A.; Colmenero, J.; Zorn, R.; Richter, D. Partial structure factors of a simulated polymer melt. Comput. Mater. Sci. 2002, 25, 596–606. [Google Scholar] [CrossRef]
- Colmenero, J.; Arbe, A.; Alvarez, F. Short-range order of glassy polymers by neutron diffraction with polarization analysis and MD-simulations. Comm. Powder Diffr. Int. Union Crystallogr. Newsl. 2004, 31, 35. [Google Scholar]
- Iradi, I.; Alvarez, F.; Colmenero, J.; Arbe, A. Structure factors in polystyrene: A neutron scattering and MD-simulation study. Physica B 2004, 350, 881. [Google Scholar] [CrossRef]
- Narros, A.; Arbe, A.; Alvarez, F.; Colmenero, J.; Zorn, R.; Schweika, W.; Richter, D. Partial Structure Factors in 1,4-Polybutadiene. A Combined Neutron Scattering and Molecular Dynamics Simulations Study. Macromolecules 2005, 38, 9847–9853. [Google Scholar] [CrossRef] [Green Version]
- Khairy, Y.; Alvarez, F.; Arbe, A.; Colmenero, J. Collective features in polyisobutylene. A study of the static and dynamic structure factor by molecular dynamics simulations. Macromolecules 2014, 47, 447–459. [Google Scholar] [CrossRef] [Green Version]
- Colmenero, J.; Moreno, A.J.; Alegría, A. Neutron scattering investigations on methyl group dynamics in polymers. Progr. Polym. Sci. 2005, 30, 1147–1184. [Google Scholar] [CrossRef] [Green Version]
- Chahid, A.; Alegría, A.; Colmenero, J. Methyl group dynamics in poly(vinyl methyl ether). A rotation rate distribution model. Macromolecules 1994, 27, 3282–3288. [Google Scholar] [CrossRef]
- Alvarez, F.; Alegría, A.; Colmenero, J.; Nicholson, T.M.; Davis, G. Origin of the distribution of potential barriers for methyl group dynamics in glassy polymers: A molecular dynamics simulation in polyisoprene. Macromolecules 2000, 33, 8077–8084. [Google Scholar] [CrossRef]
- Alvarez, F.; Arbe, A.; Colmenero, J. Methyl group dynamics above the glass transition temperature: A molecular dynamics simulation in polyisoprene. Chem. Phys. 2000, 261, 47–59. [Google Scholar] [CrossRef]
- Genix, A.C.; Arbe, A.; Alvarez, F.; Colmenero, J.; Farago, B.; Wischnewski, A.; Richter, D. Self- and collective dynamics of syndiotactic poly(methyl methacrylate). A combined study by quasielastic neutron scattering and atomistic molecular dynamics simulations. Macromolecules 2006, 39, 6260–6272. [Google Scholar] [CrossRef] [Green Version]
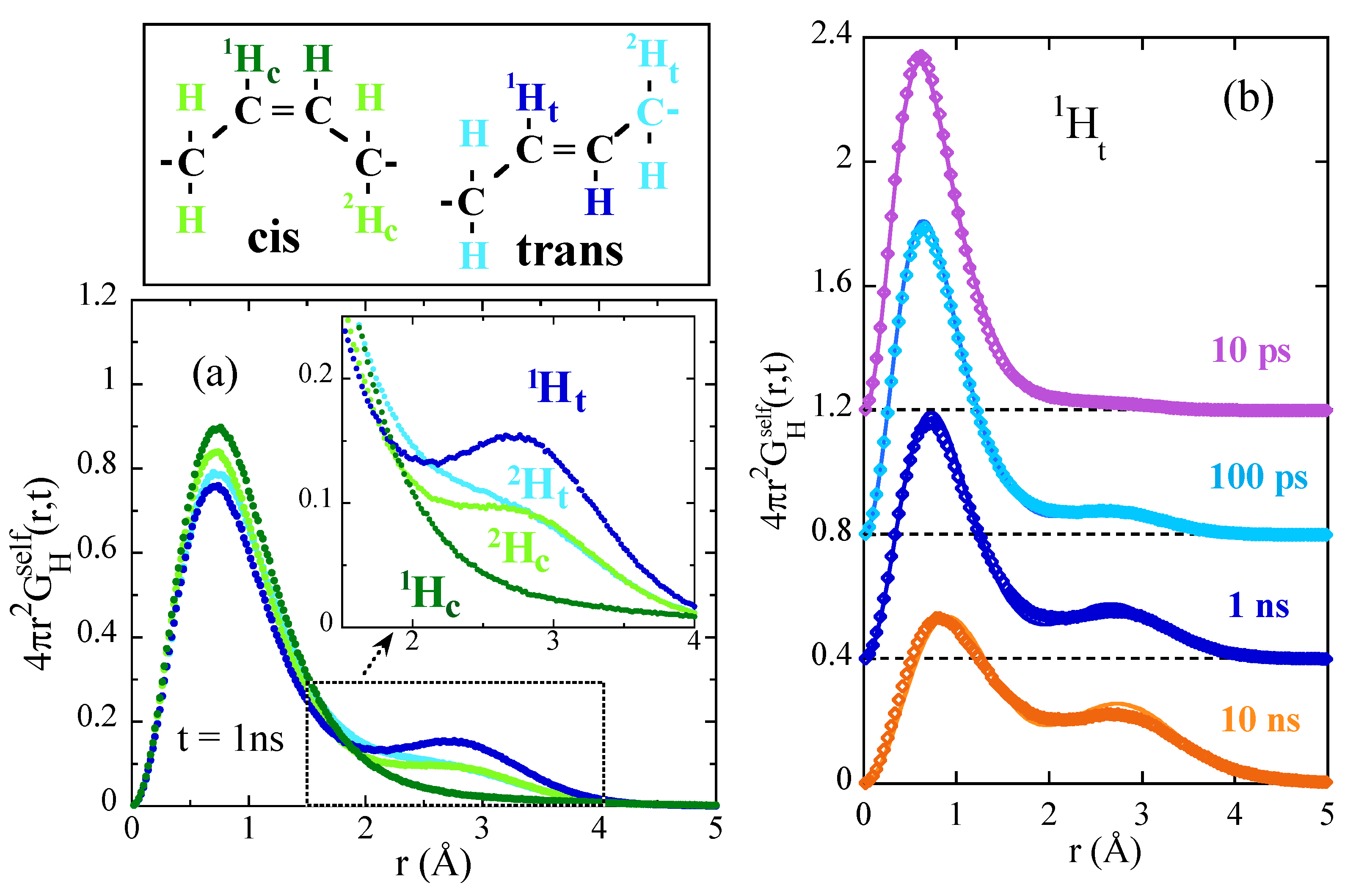
- Arbe, A.; Buchenau, U.; Willner, L.; Richter, D.; Farago, B.; Colmenero, J. Study of the dynamic structure factor in the β relaxation regime of polybutadiene. Phys. Rev. Lett. 1996, 76, 1872–1875. [Google Scholar] [CrossRef] [PubMed]
- Arbe, A.; Richter, D.; Colmenero, J.; Farago, B. Merging of the α and β relaxations in polybutadiene: A neutron spin echo and dielectric study. Phys. Rev. E 1996, 54, 3853–3869. [Google Scholar] [CrossRef]
- Aouadi, A.; Lebon, M.J.; Dreyfus, C.; Strube, B.; Steffen, W.; Patkowski, A.; Pick, R.M. A light-scattering stdy of 1,4 cis-trans polybutadiene. J. Phys. Cond. Mat. 1997, 9, 3803. [Google Scholar] [CrossRef]
- Ding, Y.; Novikov, V.; Sokolov, A. Unusual relaxation process in polybutadiene: Resolving the controversy. J. Polym. Sci. Part B Polym. Phys. 2004, 42, 994. [Google Scholar] [CrossRef]
- Colmenero, J.; Arbe, A.; Alvarez, F.; Narros, A.; Monkenbusch, M.; Richter, D. The decisive influence of local chain dynamics on the overall dynamic structure factor close to the glass transition. Europhys. Lett. 2005, 71, 262–268. [Google Scholar] [CrossRef]
- Narros, A.; Arbe, A.; Alvarez, F.; Colmenero, J.; Richter, D. Atomic motions in the αβ-merging region of 1,4-polybutadiene: A molecular dynamics simulation study. J. Chem. Phys. 2008, 128, 224905. [Google Scholar] [CrossRef]
- Colmenero, J.; Alegría, A.; Arbe, A.; Frick, B. Correlation between non-Debye behavior and Q-behavior of the α relaxation in glass-forming polymeric systems. Phys. Rev. Lett. 1992, 69, 478–481. [Google Scholar] [CrossRef]
- Arbe, A.; Colmenero, J.; Monkenbusch, M.; Richter, D. Dynamics of glass-forming polymers: “homogeneous” versus “heterogeneous” scenario. Phys. Rev. Lett. 1998, 81, 590–593. [Google Scholar] [CrossRef]
- Kob, W.; Andersen, H.C. Testing mode-coupling theory for a supercooled binary Lennard-Jones mixture I: The van Hove correlation function. Phys. Rev. E 1995, 51, 4626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kob, W.; Donati, C.; Plimpton, S.J.; Poole, P.H.; Glotzer, S.C. Dynamical heterogeneities in a supercooled Lennard-Jones liquid. Phys. Rev. Lett. 1997, 79, 2827. [Google Scholar] [CrossRef] [Green Version]
- Sciortino, F.; Gallo, P.; Tartaglia, P.; Chen, S.H. Supercooled water and the kinetic glass transition. Phys. Rev. E 1996, 54, 6331. [Google Scholar] [CrossRef] [PubMed]
- Mossa, S.; Leonardo, R.D.; Ruocco, G.; Sampoli, M. Molecular dynamics simulation of the fragile glass-former orthoterphenyl: A flexible molecule model. Phys. Rev. E 2000, 62, 612. [Google Scholar] [CrossRef] [Green Version]
- Caprion, D.; Matsui, J.; Schober, H.R. Dynamic heterogeneity of relaxations in glasses and liquids. Phys. Rev. Lett. 2000, 85, 4293. [Google Scholar] [CrossRef]
- Caprion, D.; Matsui, J.; Schober, H.R. Structure and relaxation in liquid and amorphous selenium. Phys. Rev. B 2000, 62, 3709. [Google Scholar] [CrossRef] [Green Version]
- Arbe, A.; Colmenero, J.; Alvarez, F.; Monkenbusch, M.; Richter, D.; Farago, B.; Frick, B. Non-Gaussian nature of the α relaxation of glass-forming polyisoprene. Phys. Rev. Lett. 2002, 89, 245701. [Google Scholar] [CrossRef]
- Arbe, A.; Colmenero, J.; Alvarez, F.; Monkenbusch, M.; Richter, D.; Farago, B.; Frick, B. Experimental evidence by neutron scattering of a crossover from Gaussian to non-Gaussian behavior in the α relaxation of polyisoprene. Phys. Rev. E 2003, 67, 051802. [Google Scholar] [CrossRef] [Green Version]
- Colmenero, J.; Alvarez, F.; Arbe, A. Self-motion and the α relaxation in a simulated glass-forming polymer: Crossover from Gaussian to non-Gaussian dynamic behavior. Phys. Rev. E 2002, 65, 041804. [Google Scholar] [CrossRef]
- Singwi, K.S.; Sjölander, A. Diffusive motions in water and cold neutron scattering. Phys. Rev. 1960, 119, 863. [Google Scholar] [CrossRef]
- Colmenero, J.; Arbe, A.; Alvarez, F.; Narros, A.; Richter, D.; Monkenbusch, M.; Farago, B. Hydrogen motions and the α-relaxation in glass-forming polymers: Molecular dynamics simulation and quasi-elastic neutron scattering results. PRAMANA J. Phys. 2004, 63, 25. [Google Scholar] [CrossRef]
- Narros, A.; Alvarez, F.; Arbe, A.; Colmenero, J.; Richter, D.; Farago, B. Hydrogen motions in the α-relaxation regime of poly(vinyl ethylene): A molecular dynamics simulation and neutron scattering study. J. Chem. Phys. 2004, 121, 3282–3294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capponi, S.; Arbe, A.; Cerveny, S.; Busselez, R.; Frick, B.; Embs, J.P.; Colmenero, J. Quasielastic neutron scattering study of hydrogen motions in an aqueous poly(vinyl methyl ether) solution. J. Chem. Phys. 2011, 134, 204906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genix, A.C.; Arbe, A.; Colmenero, J.; Wuttke, J.; Richter, D. Neutron scattering and X-ray Investigation of the structure and dynamics of poly(ethyl methacrylate). Macromolecules 2012, 45, 2522–2536. [Google Scholar] [CrossRef]
- Arbe, A.; Rubio, J.; Malo de Molina, P.; Maiz, J.; Pomposo, J.A.; Fouquet, P.; Prevost, S.; Juranyi, F.; Khaneft, M.; Colmenero, J. Melts of single-chain nanoparticles: A neutron scattering investigation. J. Appl. Phys. 2020, 127, 044305. [Google Scholar] [CrossRef]
- Götze, W. Recent tests of the mode-coupling theory for glassy dynamics. J. Phys. Cond. Matter 1999, 11, A1–A45. [Google Scholar] [CrossRef]
- Götze, W. Complex Dynamics of Glass-Forming Liquids. A Mode Coupling Theory; Oxford University Press: New York, NY, USA, 2009. [Google Scholar]
- Richter, D.; Frick, B.; Farago, B. Neutron-spin-echo investigation on the dynamics of polybutadiene near the glass transition. Phys. Rev. Lett. 1988, 61, 2465–2468. [Google Scholar] [CrossRef]
- Zorn, R.; Richter, D.; Farago, B.; Frick, B.; Kremer, F.; Kirst, U.; Fetters, L. Comparative study of the segmental relaxation in polyisoprene by quasi-elastic neutron scattering and dielectric spectroscopy. Physica B 1992, 180, 534–536. [Google Scholar] [CrossRef]
- Richter, D.; Arbe, A.; Colmenero, J.; Monkenbusch, M.; Farago, B.; Faust, R. Molecular motions in polyisobutylene: A neutron spin-echo and dielectric investigation. Macromolecules 1998, 31, 1133–1143. [Google Scholar] [CrossRef]
- Farago, B.; Arbe, A.; Colmenero, J.; Faust, R.; Buchenau, U.; Richter, D. Intermediate length scale dynamics of polyisobutylene. Phys. Rev. E 2002, 65, 051803. [Google Scholar] [CrossRef] [Green Version]
- Arrighi, V.; Pappas, C.; Triolo, A.; Pouget, S. Temperature dependence of local chain dynamics in atactic polypropylene: A neutron spin-echo study. Physica B 2001, 301, 157–162. [Google Scholar] [CrossRef]
- Faivre, A.; Levelut, C.; Durand, D.; Longeville, S.; Ehlers, G. Influence of the microstructure of PPO-based glass-formers on their dynamics as investigated by neutron spin echo. J. Non Cryst. Solids 2002, 307–310, 712–718. [Google Scholar] [CrossRef]
- Arbe, A.; Moral, A.; Alegría, A.; Colmenero, J.; Pyckhout-Hintzen, W.; Richter, D.; Farago, B.; Frick, B. Heterogeneous structure of poly(vinyl chloride) as the origin of anomalous dynamical behavior. J. Chem. Phys. 2002, 117, 1336–1350. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Aparicio, R.; Arbe, A.; Alvarez, F.; Colmenero, J.; Willner, L. Quasielastic neutron scattering and molecular dynamics simulation study on the structure factor of poly(ethylene-alt-propylene). Macromolecules 2009, 42, 8271–8285. [Google Scholar] [CrossRef]
- Arbe, A.; Rubio-Cervilla, J.; Alegría, A.; Moreno, A.J.; Pomposo, J.A.; Robles-Hernández, B.; Malo de Molina, P.; Fouquet, P.; Juranyi, F.; Colmenero, J. Mesoscale dynamics in melts of single-chain polymeric nanoparticles. Macromolecules 2019, 52, 6935–6942. [Google Scholar] [CrossRef]
- Colmenero, J.; Arbe, A.; Richter, D.; Farago, B.; Monkenbusch, M. Neutron spin echo spectroscopy. Basics, trends and applications; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2003; Volume 601. [Google Scholar]
- Colmenero, J.; Narros, A.; Alvarez, F.; Arbe, A.; Moreno, A.J. Atomic motions in the αβ-region of glass-forming polymers: Molecular versus mode coupling theory approach. J. Phys. Cond. Matter 2006, 19, 205127. [Google Scholar] [CrossRef] [Green Version]
- Capponi, S.; Arbe, A.; Alvarez, F.; Colmenero, J.; Frick, B.; Embs, J.P. Atomic motions in poly(vinyl methyl ether): A combined study by quasielastic neutron scattering and molecular dynamics simulations in the light of the mode coupling theory. J. Chem. Phys. 2009, 131, 204901. [Google Scholar] [CrossRef] [Green Version]
- Khairy, Y.; Alvarez, F.; Arbe, A.; Colmenero, J. Applicability of mode-coupling theory to polyisobutylene: A molecular dynamics simulation study. Phys. Rev. E 2013, 88, 042302. [Google Scholar] [CrossRef] [Green Version]
- Zorn, R.; Richter, D.; Frick, B.; Farago, B. Neutron scattering experiments on the glass transition of polymers. Physica A 1993, 201, 52–66. [Google Scholar] [CrossRef]
- Bergman, R.; Borjesson, L.; Torell, L.M.; Fontana, A. Dynamics around the liquid-glass transition in poly(propylene-glycol) investigated by wide-frequency-range light-scattering techniques. Phys. Rev. B 1997, 56, 11619. [Google Scholar] [CrossRef]
- Kisliuk, A.; Mathers, R.T.; Sokolov, A.P. Crossover in dynamics of polymeric liquids: Back to Tll? J. Polym. Sci. Part B Polym. Phys. 2000, 38, 2785–2790. [Google Scholar] [CrossRef]
- Bernabei, M.; Moreno, A.J.; Colmenero, J. Dynamic arrest in polymer melts: Competition between packing and intramolecular barriers. Phys. Rev. Lett. 2008, 101, 255701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, D.; Monkenbusch, M.; Allgaier, J.; Arbe, A.; Colmenero, J.; Farago, B.; Cheol Bae, Y.; Faust, R. From Rouse dynamics to local relaxation: A neutron spin echo study on polyisobutylene melts. J. Chem. Phys. 1999, 111, 6107. [Google Scholar] [CrossRef]
- Arbe, A.; Monkenbusch, M.; Stellbrink, J.; Richter, D.; Farago, B.; Almdal, K.; Faust, R. Origin of internal viscosity effects in flexible polymers: A comparative neutron spin-echo and light scattering study on poly(dimethylsiloxane) and polyisobutylene. Macromolecules 2001, 34, 1281–1290. [Google Scholar] [CrossRef] [Green Version]
- Richter, D.; Monkenbusch, M.; Willner, L.; Arbe, A.; Colmenero, J.; Farago, B. Direct observation of the crossover from α-relaxation to Rouse dynamics in a polymer melt. Europhys. Lett. 2004, 66, 239. [Google Scholar] [CrossRef]
- Smith, G.D.; Paul, W.; Monkenbusch, M.; Willner, L.; Richter, D.; Qiu, X.H.; Ediger, M.D. Molecular dynamics of a 1,4-polybutadiene melt. Comparison of experiment and simulation. Macromolecules 1999, 32, 8857. [Google Scholar] [CrossRef]
- Zamponi, M.; Wischnewski, A.; Monkenbusch, M.; Willner, L.; Richter, D.; Falus, P.; Farago, B.; Guenza, M.G. Cooperative dynamics in homopolymer melts: A comparison of theoretical predictions with neutron spin echo experiments. J. Phys. Chem. B 2008, 112, 16220. [Google Scholar] [CrossRef]
- Brodeck, M.; Alvarez, F.; Arbe, A.; Juranyi, F.; Unruh, T.; Holderer, O.; Colmenero, J.; Richter, D. Study of the dynamics of poly(ethylene oxide) by combining molecular dynamic simulations and neutron scattering experiments. J. Chem. Phys. 2009, 130, 094908. [Google Scholar] [CrossRef]
- Pérez-Aparicio, R.; Alvarez, F.; Arbe, A.; Willner, L.; Richter, D.; Falus, P.; Colmenero, J. Chain dynamics of unentangled poly(ethylene-alt-propylene) melts by means of neutron scattering and fully atomistic molecular dynamics simulations. Macromolecules 2011, 44, 3129–3139. [Google Scholar] [CrossRef] [Green Version]
- Brueckner, S. Configurational statistics of polymer chains accounting for all rotational states. General extension to finite chains and copolymers. Macromolecules 1981, 14, 449. [Google Scholar] [CrossRef]
- Pérez Aparicio, R.; Colmenero, J.; Alvarez, F.; Padding, J.T.; Briels, W.J. Chain dynamics of poly(ethylene-alt-propylene) melts by means of coarse-grained simulations based on atomistic molecular dynamics. J. Chem. Phys. 2010, 132, 024904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodeck, M.; Alvarez, F.; Colmenero, J.; Richter, D. Single chain dynamic structure factor of poly(ethylene oxide) in dynamically asymmetric blends with poly(methyl methacrylate). Neutron scattering and molecular dynamics simulations. Macromolecules 2011, 45, 536. [Google Scholar] [CrossRef]
- Heijboer, J. Physics of Non-Crystalline Solids; North-Holland: Amsterdam, The Netherlands, 1965; p. 231. [Google Scholar]
- McCrum, N.; Read, B.; Williams, G. Anelastic, Dielectric Effects in Polymeric Solids; Wiley: London, UK, 1967. [Google Scholar]
- Cowie, J. Relaxation processes in the glassy state: Molecular aspects. J. Macromol. Sci. Phys. B 1980, 18, 569. [Google Scholar] [CrossRef]
- Meier, G.; Fytas, G.; Dorfmüller, T. Photon correlation study of the relaxation processes in bulk poly(n-butyl methacrylate). Macromolecules 1984, 17, 957. [Google Scholar] [CrossRef]
- Giebel, L.; Meier, G.; Fytas, G.; Fischer, E.W. Dynamic light scattering from bulk poly(n-hexylmethacrylate) near and above the glass transition. J. Polym. Sci. B Polym. Phys. 1992, 30, 1291. [Google Scholar] [CrossRef]
- Garwe, F.; Schönhals, A.; Lockwenz, H.; Beiner, M.; Schröter, K.; Donth, E. Influence of cooperative α dynamics on local β relaxation during the development of the dynamic glass transition in poly(n-alkyl methacrylate)s. Macromolecules 1996, 29, 247. [Google Scholar] [CrossRef]
- Schröter, K.; Unger, R.; Reissig, S.; Garwe, F.; Kahle, S.; Beiner, M.; Donth, E. Dielectric spectroscopy in the αβ splitting region of glass transition in poly(ethyl methacrylate) and poly(n-butyl methacrylate): Different evaluation methods and experimental conditions. Macromolecules 1998, 31, 8966. [Google Scholar] [CrossRef]
- Floudas, G.; Stepanek, P. Structure and dynamics of poly(n-decyl methacrylate) below and above the glass transition. Macromolecules 1998, 31, 6951. [Google Scholar] [CrossRef]
- Beiner, M.; Schröter, K.; Hempel, E.; Reissig, S.; Donth, E. Multiple glass transition and nanophase separation in poly(n-alkyl methacrylate) homopolymers. Macromolecules 1999, 32, 6278. [Google Scholar] [CrossRef]
- Dudognon, E.; Bernès, A.; Lacabanne, C. Study by thermostimulated currents of dielectric relaxations through the glass transition in an amorphous polymer: poly(n-butyl methacrylate). Macromolecules 2001, 34, 3988. [Google Scholar] [CrossRef]
- Dudognon, E.; Bernès, A.; Lacabanne, C. Low-frequency chain dynamics of poly(n-hexyl methacrylate) by dielectric spectroscopies. Macromolecules 2002, 35, 5927. [Google Scholar] [CrossRef]
- Beiner, M. Relaxation in poly (alkyl methacrylate)s: Crossover region and nanophase separation. Macromol. Rapid Commun. 2001, 22, 869. [Google Scholar] [CrossRef]
- Beiner, M.; Kabisch, O.; Reichl, S.; Huth, H. Structural and dynamic nanoheterogeneities in higher poly(alkyl methacrylate)s. J. Non Cryst. Sol. 2002, 307-310, 658. [Google Scholar] [CrossRef]
- Hempel, E.; Beiner, M.; Huth, H.; Donth, E. Temperature modulated DSC for the multiple glass transition in poly (n-alkyl methacrylates). Thermochimica Acta 2002, 391, 219. [Google Scholar] [CrossRef]
- Pascui, O.; Beiner, M.; Reichert, D. Identification of slow dynamic processes in poly(n-hexyl methacrylate) by solid-state 1D-MAS exchange NMR. Macromolecules 2003, 36, 3992. [Google Scholar] [CrossRef]
- Beiner, M.; Huth, H. Nanophase separation and hindered glass transition in side-chain polymers. Nat. Mater. 2003, 2, 595. [Google Scholar] [CrossRef]
- Hiller, S.; Pascui, O.; Kabisch, O.; Reichert, D.; Beiner, M. Nanophase separation in side chain polymers: New evidence from structure and dynamics. New J. Phys. 2004, 6, 10. [Google Scholar] [CrossRef]
- Menissez, C.; Sixou, B.; David, L.; Vigier, G. Dielectric and mechanical relaxation behavior in poly(butyl methacrylate) isomers. J. Non Cryst. Sol. 2005, 351, 595. [Google Scholar] [CrossRef]
- Wind, M.; Graf, R.; Renker, S.; Spiess, H.; Steffen, W. Structure of amorphous poly-(ethylmethacrylate): A wide-angle X-ray scattering study. J. Chem. Phys. 2005, 122, 014906. [Google Scholar] [CrossRef]
- Beiner, M. CP832, Flow Dynamics, The Second International Conference on Flow Dynamics; American Institute of Physics: College Park, MD, USA, 2006; p. 134. [Google Scholar]
- Arbe, A.; Genix, A.C.; Arrese-Igor, S.; Colmenero, J.; Richter, D. Dynamics in poly(n-alkyl methacrylates): A neutron scattering, calorimetric, and dielectric study. Macromolecules 2010, 43, 3107. [Google Scholar] [CrossRef] [Green Version]
- Gaborieau, M.; Graf, R.; Kahle, S.; Pakula, T.; Spiess, H.W. Chain dynamics in poly(n-alkyl acrylates) by solid-state NMR, dielectric, and mechanical spectroscopies. Macromolecules 2007, 40, 6249. [Google Scholar] [CrossRef]
- Cowie, J.; Haq, Z.; McEwen, I.; Velickovic, J. Poly (alkyl itaconates): 8. Observations of dual glass transitions and crystallinity in the dialkyl ester series diheptyl to di-eicosyl. Polymer 1981, 22, 327. [Google Scholar] [CrossRef]
- Arrighi, V.; Triolo, A.; McEwen, I.J.; Holmes, P.; Triolo, R.; Amenitsch, H. Observation of local order in poly(di-n-alkyl itaconate)s. Macromolecules 2000, 33, 4989. [Google Scholar] [CrossRef]
- Genix, A.C.; Lauprêtre, F. Subglass and glass transitions of poly(di-n-alkylitaconate)s with various side-chain lengths: Dielectric relaxation investigation. Macromolecules 2005, 38, 2786. [Google Scholar] [CrossRef]
- Gerstl, C.; Schneider, G.J.; Pyckhout-Hintzen, W.; Allgaier, J.; Richter, D.; Alegría, A.; Colmenero, J. Segmental and normal mode relaxation of poly(alkylene oxide)s studied by dielectric spectroscopy and rheology. Macromolecules 2010, 43, 4968. [Google Scholar] [CrossRef]
- Gerstl, C.; Schneider, G.J.; Fuxman, A.; Zamponi, M.; Frick, B.; Seydel, T.; Koza, M.; Genix, A.C.; Allgaier, J.; Richter, D.; et al. Quasielastic neutron scattering study on the dynamics of poly(alkylene oxide)s. Macromolecules 2012, 45, 4394–4405. [Google Scholar] [CrossRef]
- Gerstl, C.; Brodeck, M.; Schneider, G.J.; Su, Y.; Allgaier, J.; Arbe, A.; Colmenero, J.; Richter, D. Short and intermediate range order in poly(alkylene oxide)s. A neutron diffraction and molecular dynamics simulations study. Macromolecules 2012, 45, 7293–7303. [Google Scholar] [CrossRef]
- Colmenero, J.; Brodeck, M.; Arbe, A.; Richter, D. Dynamics of poly(butylene oxide) well above the glass transition. A fully atomistic molecular dynamics simulation study. Macromolecules 2013, 46, 1678–1685. [Google Scholar] [CrossRef]
- Moreno, A.; Arbe, A.; Colmenero, J. Structure and dynamics of self-assembled comb copolymers: Comparison between simulations of a generic model and neutron scattering experiments. Macromolecules 2011, 44, 1695. [Google Scholar] [CrossRef] [Green Version]
- Busselez, R.; Arbe, A.; Alvarez, F.; Colmenero, J.; Frick, B. Study of the structure and dynamics of poly(vinyl pyrrolidone) by molecular dynamics simulations validated by quasielastic neutron scattering and X-ray diffraction experiments. J. Chem. Phys. 2011, 134, 054904. [Google Scholar] [CrossRef] [Green Version]
- Colmenero, J.; Arbe, A. Segmental dynamics in miscible polymer blends: Recent results and open questions. Soft Matter 2007, 3, 1474–1485. [Google Scholar] [CrossRef] [PubMed]
- Colmenero, J.; Alvarez, F.; Khairy, Y.; Arbe, A. Modeling the collective relaxation time of glass-forming polymers at intermediate length scales: Application to polyisobutylene. J. Chem. Phys. 2013, 139, 044906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colmenero, J.; Alvarez, F.; Arbe, A. Collective dynamics of glass-forming polymers at intermediate length scales—A synergetic combination of neutron scattering, atomistic simulations and theoretical modelling. EPJ Web Conf. 2015, 83, 01001. [Google Scholar] [CrossRef] [Green Version]
- De Gennes, P.G. Liquid dynamics and inelastic scattering of neutrons. Physica 1959, 25, 825. [Google Scholar] [CrossRef]
- Sköld, K. Small energy transfer scattering of cold neutrons from liquid argon. Phys. Rev. Lett. 1967, 19, 1023–1025. [Google Scholar] [CrossRef]
- Arbe, A.; Colmenero, J. Characterization of the “simple-liquid” state in a polymeric system: Coherent and incoherent scattering functions. Phys. Rev. E 2009, 80, 041805. [Google Scholar] [CrossRef]
- Tyagi, M.; Arbe, A.; Alvarez, F.; Colmenero, J.; González, M.A. Short-range order and collective dynamics of poly(vinyl acetate): A combined study by neutron scattering and molecular dynamics simulations. J. Chem. Phys. 2008, 129, 224903. [Google Scholar] [CrossRef]
- Novikov, V.N.; Schweizer, K.S.; Sokolov, A.P. Coherent neutron scattering and collective dynamics on mesoscale. J. Chem.Phys. 2013, 138, 164508. [Google Scholar] [CrossRef] [Green Version]
- Heil, W.; Andersen, K.H.; Cywinski, R.; Humblot, H.; Ritter, C.; Roberts, T.W.; Stewart, J.R. Large solid-angle neutron polarization analysis at thermal neutron wavelengths using a 3He spin-filter. Nucl. Inst. Meth. Phys. Res. A 2002, 485, 551–570. [Google Scholar] [CrossRef]
- Stewart, J.R.; Deen, P.P.; Andersen, K.H.; Schober, H.; Barthélémy, J.F.; Hillier, J.M.; Murani, A.P.; Hayes, T.; Lindenau, B. Disordered materials studied using neutron polarization analysis on the multi-detector spectrometer, D7. J. Appl. Cryst. 2009, 42, 69–84. [Google Scholar] [CrossRef]
- Burankova, T.; Hempelmann, R.; Wildes, A.R.; Embs, J.P. Collective ion diffusion and localized single particle dynamics in pyridinium-based ionic liquids. J. Phys. Chem. B 2014, 118, 14452–14460. [Google Scholar] [CrossRef] [PubMed]
- Casella, G.; Stewart, J.R.; Paternò, G.M.; García-Sakai, V.; Devonport, M.; Galsworthy, P.J.; Bewley, R.I.; Voneshen, D.J.; Raspino, D.; Nilsen, G.J. Polarization analysis on the LET cold neutron spectrometer using a 3He spin-filter: First results. J. Phys. Conf. Ser. 2019, 1316, 012007. [Google Scholar] [CrossRef]
- Chen, W.; Watson, S.; Qiu, Y.; Rodriguez-Rivera, J.A.; Faraone, A. Wide-angle polarization analysis on the multi-axis crystal spectrometer for the study of collective and single particle dynamics of methanol at its prepeak. Phys. B Cond. Matter 2019, 564, 166–171. [Google Scholar] [CrossRef]
- Arbe, A.; Malo de Molina, P.; Alvarez, F.; Frick, B.; Colmenero, J. Dielectric susceptibility of liquid water: Microscopic insights from coherent and incoherent neutron scattering. Phys. Rev. Lett. 2016, 117, 185501. [Google Scholar] [CrossRef] [PubMed]
- Arbe, A.; Colmenero, J.; Richter, D. Broadband Dielectric Spectroscopy; Kremer, F., Schönhals, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; p. 685. [Google Scholar]
- Qvist, J.; Schober, H.; Halle, B. Structural dynamics of supercooled water from quasielastic neutron scattering and molecular simulations. J. Chem. Phys. 2011, 134, 144508. [Google Scholar] [CrossRef]
- Arbe, A.; Moreno, A.J.; Allgaier, J.; Ivanova, O.; Fouquet, P.; Colmenero, J.; Richter, D. Role of dynamic asymmetry on the collective dynamics of comblike polymers: Insights from neutron spin-echo experiments and coarse-grained molecular dynamics simulations. Macromolecules 2016, 49, 4989–5000. [Google Scholar] [CrossRef]
- Moreno, A.J.; Lo Verso, F.; Sanchez-Sanchez, A.; Arbe, A.; Colmenero, J.; Pomposo, J.A. Advantages of orthogonal folding of single polymer chains to soft nanoparticles. Macromolecules 2013, 46, 9748–9759. [Google Scholar] [CrossRef]
- Pomposo, J.A.; Perez-Baena, I.; Lo Verso, F.; Moreno, A.J.; Arbe, A.; Colmenero, J. How far are single-chain polymer nanoparticles in solution from the globular state? ACS Macro Lett. 2014, 3, 767–772. [Google Scholar] [CrossRef]
- Moreno, A.J.; Lo Verso, F.; Arbe, A.; Pomposo, J.A.; Colmenero, J. Concentrated solutions of single-chain nanoparticles: A simple model for intrinsically disordered proteins under crowding conditions. J. Phys. Chem. Lett. 2016, 7, 838–844. [Google Scholar] [CrossRef] [Green Version]
- González-Burgos, M.; Arbe, A.; Moreno, A.J.; Pomposo, J.A.; Radulescu, A.; Colmenero, J. Crowding the environment of single-chain nanoparticles: A combined study by SANS and simulations. Macromolecules 2018, 51, 1573–1585. [Google Scholar] [CrossRef] [Green Version]
- Arbe, A.; Pomposo, J.; Moreno, A.; LoVerso, F.; González-Burgos, M.; Asenjo-Sanz, I.; Iturrospe, A.; Radulescu, A.; Ivanova, O.; Colmenero, J. Structure and dynamics of single-chain nano-particles in solution. Polymer 2016, 105, 532–544. [Google Scholar] [CrossRef]





| Isotope | /fm | /fm | /fm | /barns | /barns |
|---|---|---|---|---|---|
| H | −3.7406 | 13.992 | 638.78 | 1.7583 | 80.26 |
| H (D) | 6.6710 | 44.502 | 16.322 | 5.592 | 2.05 |
| C | 6.6511 | 44.237 | 0 | 5.559 | 0 |
| O | 5.8030 | 33.675 | 0 | 4.232 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arbe, A.; Alvarez, F.; Colmenero, J. Insight into the Structure and Dynamics of Polymers by Neutron Scattering Combined with Atomistic Molecular Dynamics Simulations. Polymers 2020, 12, 3067. https://doi.org/10.3390/polym12123067
Arbe A, Alvarez F, Colmenero J. Insight into the Structure and Dynamics of Polymers by Neutron Scattering Combined with Atomistic Molecular Dynamics Simulations. Polymers. 2020; 12(12):3067. https://doi.org/10.3390/polym12123067
Chicago/Turabian StyleArbe, Arantxa, Fernando Alvarez, and Juan Colmenero. 2020. "Insight into the Structure and Dynamics of Polymers by Neutron Scattering Combined with Atomistic Molecular Dynamics Simulations" Polymers 12, no. 12: 3067. https://doi.org/10.3390/polym12123067