Increased Cytotoxic Efficacy of Protocatechuic Acid in A549 Human Lung Cancer Delivered via Hydrophobically Modified-Chitosan Nanoparticles As an Anticancer Modality
Abstract
:1. Introduction
2. Materials and Methods
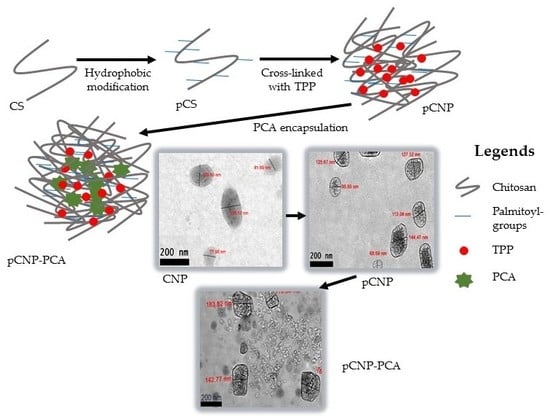
2.1. Formation of Chitosan Nanoparticles (CNP)
2.2. Hydrophobic Modification of Chitosan Nanoparticles (pCNP)
2.3. Synthesis of Protocatechuic Acid-Encapsulated Nanoparticles (CNP-PCA and pCNP-PCA)
2.4. Physicochemical Characterization of Nanoparticles
2.5. Determination of Free Amine Groups Using Trinitrobenzene Sulfonic Acid Assay (TNBS)
Determination of PCA Encapsulation Efficiency (%EE) in Nanoparticle Samples:
2.6. Assessment of In Vitro Vellular Efficacy of Nanoparticle Mediated PCA Uptake in A549 Lung Cancer Cell Line
3. Results and Discussions
3.1. The Colume of Cross-Linker Governing the Size and PDI of Nanoparticles
3.2. The Formation of Nanoparticles Utilized Free Amine Group of CS/pCS Polymer
3.3. Formation of PCA-Encapsulated Nanoparticles
3.4. Morphological Analysis of Nanoparticles
3.5. Functional Group Annotation of Nanoparticle Samples Using FTIR Spectroscopy
3.6. Assessment of In Vitro Cytotoxicity of CNP and pCNP in A549 Lung Cancer Cells
3.7. Assessment Of PCA Efficacy and Anticancer Properties Using Nanoparticle-Mediated In Vitro Cellular Delivery Systems
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Society, A.C. American Cancer Society: Facts & Figures 2018; American Cancer Society: Atlanta, GA, USA, 2018. [Google Scholar]
- Nakano, T.; Shimizu, K.; Kawashima, O.; Kamiyoshihara, M.; Kakegawa, S.; Sugano, M.; Ibe, T.; Nagashima, T.; Kaira, K.; Sunaga, N.; et al. Establishment of a Human Lung Cancer Cell Line with High Metastatic Potential to Multiple Organs: Gene Expression Associated with Metastatic Potential in Human Lung Cancer. Oncol. Rep. 2012, 28, 1727–1735. [Google Scholar] [CrossRef] [PubMed]
- Schirrmacher, V. From Chemotherapy to Biological Therapy: A Review of Novel Concepts to Reduce the Side Effects of Systemic Cancer Treatment (Review). Int. J. Oncol. 2019, 54, 407–419. [Google Scholar] [CrossRef]
- Chanvorachote, P.; Chamni, S.; ninsontia, C.; Phiboonchaiyanan, P.P. Potential Anti-Metastasis Natural Compounds for Lung Cancer. Anticancer Res. 2016, 36, 5707–5718. [Google Scholar] [CrossRef] [Green Version]
- Abotaleb, M.; Liskova, A.; Kubatka, P.; Büsselberg, D. Therapeutic Potential of Plant Phenolic Acids in the Treatment of Cancer. Biomolecules 2020, 10, 221. [Google Scholar] [CrossRef] [Green Version]
- Mitra, S.; Dash, R. Natural Products for the Management and Prevention of Breast Cancer. Evid.-Based Complement. Altern. Med. 2018, 2018, 8324696. [Google Scholar] [CrossRef]
- Demain, A.L.; Vaishnav, P. Natural Products for Cancer Chemotherapy. Microb. Biotechnol. 2011, 4, 687–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.-M. Green Tea and Prevention of Esophageal and Lung Cancers. Mol. Nutr. Food Res. 2011, 55, 886–904. [Google Scholar] [CrossRef]
- Fritz, H.; Seely, D.; Kennedy, D.A.; Fernandes, R.; Cooley, K.; Fergusson, D. Green Tea and Lung Cancer: A Systematic Review. Integr. Cancer Ther. 2013, 12, 7–24. [Google Scholar] [CrossRef]
- Mehta, H.J.; Patel, V.; Sadikot, R.T. Curcumin and Lung Cancer—A Review. Target. Oncol. 2014, 9, 295–310. [Google Scholar] [CrossRef]
- Tsai, J.-R.; Liu, P.-L.; Chen, Y.-H.; Chou, S.-H.; Cheng, Y.-J.; Hwang, J.-J.; Chong, I.-W. Curcumin Inhibits Non-Small Cell Lung Cancer Cells Metastasis through the Adiponectin/NF-κb/MMPs Signaling Pathway. PLoS ONE 2015, 10, e0144462. [Google Scholar] [CrossRef] [Green Version]
- Ulasli, S.S.; Celik, S.; Gunay, E.; Ozdemir, M.; Ozyurek, A.; Koyuncu, T.; Unlu, M. Anticancer Effects of Thymoquinone, Caffeic Acid Phenethyl Ester and Resveratrol on A549 Non-Small Cell Lung Cancer Cells Exposed to Benzo (a) Pyrene. Asian Pac. J. Cancer Prev. 2013, 14, 6159–6164. [Google Scholar] [CrossRef] [Green Version]
- Rajendra Prasad, N.; Karthikeyan, A.; Karthikeyan, S.; Venkata Reddy, B. Inhibitory Effect of Caffeic Acid on Cancer Cell Proliferation by Oxidative Mechanism in Human HT-1080 Fibrosarcoma Cell Line. Mol. Cell. Biochem. 2011, 349, 11–19. [Google Scholar] [CrossRef]
- Liu, R.H. Potential Synergy of Phytochemicals in Cancer Prevention: Mechanism of Action. J. Nutr. 2004, 134, 3479S–3485S. [Google Scholar] [CrossRef] [Green Version]
- Kakkar, S.; Bais, S. A Review on Protocatechuic Acid and Its Pharmacological Potential. ISRN Pharmacol. 2014, 2014, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, C.-Y.; Yin, M.-C. Antibacterial Effects of Roselle Calyx Extracts and Protocatechuic Acid in Ground Beef and Apple Juice. Foodborne Pathog. Dis. 2009, 6, 201–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xican, L.; Wang, X.; Chen, D.; Chen, S. Antioxidant Activity and Mechanism of Protocatechuic Acid in Vitro. Funct. Foods Health Dis. 2011, 1, 232–244. [Google Scholar]
- Tanaka, T.; Tanaka, T.; Tanaka, M. Potential Cancer Chemopreventive Activity of Protocatechuic Acid. J. Exp. Clin. Med. 2011, 3, 27–33. [Google Scholar] [CrossRef]
- Scazzocchio, B.; Varì, R.; Filesi, C.; D’Archivio, M.; Santangelo, C.; Giovannini, C.; Iacovelli, A.; Silecchia, G.; Volti, G.L.; Galvano, F.; et al. Cyanidin-3-O-β-Glucoside and Protocatechuic Acid Exert Insulin-like Effects by Upregulating PPARγ Activity in Human Omental Adipocytes. Diabetes 2011, 60, 2234–2244. [Google Scholar] [CrossRef] [Green Version]
- Shi, G.F.; An, L.J.; Jiang, B.; Guan, S.; Bao, Y.M. Alpinia Protocatechuic Acid Protects against Oxidative Damage in Vitro and Reduces Oxidative Stress in Vivo. Neurosci. Lett. 2006, 403, 206–210. [Google Scholar] [CrossRef]
- Lende, A.B.; Kshirsagar, A.D.; Deshpande, A.D.; Muley, M.M.; Patil, R.R.; Bafna, P.A.; Naik, S.R. Anti-Inflammatory and Analgesic Activity of Protocatechuic Acid in Rats and Mice. Inflammopharmacology 2011, 19, 255–263. [Google Scholar] [CrossRef]
- Yin, M.C.; Lin, C.C.; Wu, H.C.; Tsao, S.M.; Hsu, C.K. Apoptotic Effects of Protocatechuic Acid in Human Breast, Lung, Liver, Cervix, and Prostate Cancer Cells: Potential Mechanisms of Action. J. Agric. Food Chem. 2009, 57, 6468–6473. [Google Scholar] [CrossRef]
- Hu, J.; Lin, S.; Huang, J.J.; Cheung, P.C.K. Mechanistic Study of the in Vitro and in Vivo Inhibitory Effects of Protocatechuic Acid and Syringic Acid on VEGF-Induced Angiogenesis. J. Agric. Food Chem. 2018, 66, 6742–6751. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.M.; Hsia, T.C.; Yin, M.C. Protocatechuic Acid Inhibits Lung Cancer Cells by Modulating FAK, MAPK, and NF- B Pathways. Nutr. Cancer 2014, 66, 1331–1341. [Google Scholar] [CrossRef]
- Savjani, K.T.; Gajjar, A.K.; Savjani, J.K. Drug Solubility: Importance and Enhancement Techniques. ISRN Pharm. 2012, 2012, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Lu, L.; Wang, S.; Wu, J.; Shi, J.; Yan, T.; Xie, C.; Li, Q.; Hu, M.; Liu, Z. Oral Absorption Basics. In Developing Solid Oral Dosage Forms; Elsevier: Amsterdam, The Netherlands, 2017; pp. 297–329. [Google Scholar] [CrossRef]
- Sadhukha, T.; Prabha, S. Encapsulation in Nanoparticles Improves Anti-Cancer Efficacy of Carboplatin. AAPS PharmSciTech 2014, 15, 1029–1038. [Google Scholar] [CrossRef]
- Tiwari, G.; Tiwari, R.; Bannerjee, S.; Bhati, L.; Pandey, S.; Pandey, P.; Sriwastawa, B. Drug Delivery Systems: An Updated Review. Int. J. Pharm. Investig. 2012, 2, 2. [Google Scholar] [CrossRef] [Green Version]
- Benelmekki, M. An Introduction to Nanoparticles and Nanotechnology. Des. Hybrid Nanopart. 2014, 10, 5951–5959. [Google Scholar] [CrossRef] [Green Version]
- Nagarajan, R. Nanoparticles: Building Blocks for Nanotechnology; ACS: Washington, DC, USA, 2008; pp. 2–14. [Google Scholar] [CrossRef] [Green Version]
- Nasrollahzadeh, M.; Sajadi, S.M.; Sajjadi, M.; Issaabadi, Z. Applications of Nanotechnology in Daily Life, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2019; Volume 28. [Google Scholar] [CrossRef]
- Giustini, A.J.; Petryk, A.A.; Cassim, S.M.; Tate, J.A.; Baker, I.; Hoopes, P.J. Magnetic Nanoparticle Hyperthermia In Cancer Treatment. Nano Life 2010, 1, 17–32. [Google Scholar] [CrossRef]
- Wu, C.-H.; Lan, C.H.; Wu, K.L.; Wu, Y.M.; Jane, W.N.; Hsiao, M.; Wu, H.C. Hepatocellular Carcinoma-Targeted Nanoparticles for Cancer Therapy. Int. J. Oncol. 2018, 52, 389–401. [Google Scholar] [CrossRef] [Green Version]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, Applications and Toxicities. Arab. J. Chem. 2017, 12, 908–931. [Google Scholar] [CrossRef]
- Riccardi, C.; Musumeci, D.; Trifuoggi, M.; Irace, C.; Paduano, L.; Montesarchio, D. Anticancer ruthenium(III) Complexes and Ru(III)-Containing Nanoformulations: An Update on the Mechanism of Action and Biological Activity. Pharmaceuticals 2019, 12, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrand, A.M.; Rahman, M.F.; Hussain, S.M.; Schlager, J.J.; Smith, D.A.; Syed, A.F. Metal-Based Nanoparticles and Their Toxicity Assessment. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010, 2, 544–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Sun, P.; Bao, Y.; Liu, J.; An, L. Cytotoxicity of Single-Walled Carbon Nanotubes on PC12 Cells. Toxicol. Vitr. 2011, 25, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Pacurari, M.; Schwegler-Berry, D.; Friend, S.; Leonard, S.S.; Mercer, R.R.; Vallyathan, V.; Castranova, V. Raw Single-Walled Carbon Nanotube-Induced Cytotoxic Effects in Human Bronchial Epithelial Cells: Comparison to Asbestos. Toxicol. Environ. Chem. 2011, 93, 1045–1072. [Google Scholar] [CrossRef]
- Schins, R.P.F. Mechanisms of Genotoxicity of Particles and Fibers. Inhal. Toxicol. 2002, 14, 57–78. [Google Scholar] [CrossRef]
- Panyam, J.; Labhasetwar, V. Biodegradable Nanoparticles for Drug and Gene Delivery to Cells and Tissue. Adv. Drug Deliv. Rev. 2003, 55, 329–347. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Syeda, J.T.M.; Wasan, K.M.; Wasan, E.K. An Overview of Chitosan Nanoparticles and Its Application in Non-Parenteral Drug Delivery. Pharmaceutics 2017, 9, 53. [Google Scholar] [CrossRef] [Green Version]
- El-Shabouri, M.H. Positively Charged Nanoparticles for Improving the Oral Bioavailability of Cyclosporin-A. Int. J. Pharm. 2002, 249, 101–108. [Google Scholar] [CrossRef]
- Min, K.H.; Park, K.; Kim, Y.S.; Bae, S.M.; Lee, S.; Jo, H.G.; Park, R.W.; Kim, I.S.; Jeong, S.Y.; Kim, K.; et al. Hydrophobically Modified Glycol Chitosan Nanoparticles-Encapsulated Camptothecin Enhance the Drug Stability and Tumor Targeting in Cancer Therapy. J. Control. Release 2008, 127, 208–218. [Google Scholar] [CrossRef]
- Masarudin, M.J.; Cutts, S.M.; Evison, B.J.; Pigram, P.J. Factors Determining the Stability, Size Distribution, and Cellular Accumulation of Small, Monodisperse Chitosan Nanoparticles as Candidate Vectors for Anticancer Drug Delivery: Application to the Passive Encapsulation of [14 C]—Doxorubicin. Nanotechnol. Sci. Appl. 2015, 8, 67–80. [Google Scholar] [CrossRef] [Green Version]
- Shu, X.; Zhu, K. The Influence of Multivalent Phosphate Structure on the Properties of Ionically Cross-Linked Chitosan Films for Controlled Drug Release. Eur. J. Pharm. Biopharm. 2002, 54, 235–243. [Google Scholar] [CrossRef]
- Kavi Rajan, R.; Hussein, M.Z.; Fakurazi, S.; Yusoff, K.; Masarudin, M.J. Increased ROS Scavenging and Antioxidant Efficiency of Chlorogenic Acid Compound Delivered via a Chitosan Nanoparticulate System for Efficient In Vitro Visualization and Accumulation in Human Renal Adenocarcinoma Cells. Int. J. Mol. Sci. 2019, 20, 4667. [Google Scholar] [CrossRef] [Green Version]
- Naskar, S.; Koutsu, K.; Sharma, S. Chitosan-Based Nanoparticles as Drug Delivery Systems: A Review on Two Decades of Research. J. Drug Target. 2019, 27, 379–393. [Google Scholar] [CrossRef]
- Bangun, H.; Tandiono, S.; Arianto, A. Preparation and Evaluation of Chitosan-Tripolyphosphate Nanoparticles Suspension as an Antibacterial Agent. J. Appl. Pharm. Sci. 2018, 8, 147–156. [Google Scholar] [CrossRef] [Green Version]
- Koukaras, E.N.; Papadimitriou, S.A.; Bikiaris, D.N.; Froudakis, G.E. Insight on the Formation of Chitosan Nanoparticles through Ionotropic Gelation with Tripolyphosphate. Mol. Pharm. 2012, 9, 2856–2862. [Google Scholar] [CrossRef]
- Farhangi, M.; Kobarfard, F.; Mahboubi, A.; Vatanara, A.; Mortazavi, S.A. Preparation of an Optimized Ciprofloxacin-Loaded Chitosan Nanomicelle with Enhanced Antibacterial Activity. Drug Dev. Ind. Pharm. 2018, 44, 1273–1284. [Google Scholar] [CrossRef] [PubMed]
- Snyder, S.L.; Sobocinski, P.Z. An Improved 2,4,6-Trinitrobenzenesulfonic Acid Method for the Determination of Amines. Anal. Biochem. 1975, 64, 284–288. [Google Scholar] [CrossRef]
- Kuen, C.; Fakurazi, S.; Othman, S.; Masarudin, M. Increased Loading, Efficacy and Sustained Release of Silibinin, a Poorly Soluble Drug Using Hydrophobically-Modified Chitosan Nanoparticles for Enhanced Delivery of Anticancer Drug Delivery Systems. Nanomaterials 2017, 7, 379. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Lapitsky, Y. Monovalent Salt Enhances Colloidal Stability during the Formation of Chitosan/tripolyphosphate Microgels. Langmuir 2011, 27, 10392–10399. [Google Scholar] [CrossRef]
- Esquivel, R.; Juárez, J.; Almada, M.; Ibarra, J.; Valdez, M.A. Synthesis and Characterization of New Thiolated Chitosan Nanoparticles Obtained by Ionic Gelation Method. Int. J. Polym. Sci. 2015, 2015, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.S.; Liu, L.R.; Jiang, Q.; Zhang, Q.Q. Self-Aggregated Nanoparticles of Cholesterol-Modified Chitosan Conjugate as a Novel Carrier of Epirubicin. Eur. Polym. J. 2007, 43, 43–51. [Google Scholar] [CrossRef]
- Kim, Y.H.; Gihm, S.H.; Park, C.R.; Lee, K.Y.; Kim, T.W.; Kwon, I.C.; Chung, H.; Jeong, S.Y. Structural Characteristics of Size-Controlled Self-Aggregates of Deoxycholic Acid-Modified Chitosan and Their Application as a DNA Delivery Carrier. Bioconjug. Chem. 2001, 12, 932–938. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.Q.; Ren, G.F.; Yuan, H.; Du, Y.Z.; Zeng, S. Shell Cross-Linked Stearic Acid Grafted Chitosan Oligosaccharide Self-Aggregated Micelles for Controlled Release of Paclitaxel. Colloids Surf. B Biointerfaces 2006, 50, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Opanasopit, P.; Ngawhirunpat, T.; Rojanarata, T.; Choochottiros, C.; Chirachanchai, S. Camptothecin-Incorporating N-Phthaloylchitosan-G-mPEG Self-Assembly Micellar System: Effect of Degree of Deacetylation. Colloids Surf. B Biointerfaces 2007, 60, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Raja, M.A.; Arif, M.; Feng, C.; Zeenat, S.; Liu, C.G. Synthesis and Evaluation of pH-Sensitive, Self-Assembled Chitosan-Based Nanoparticles as Efficient Doxorubicin Carriers. J. Biomater. Appl. 2017, 31, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, X.G.; Li, Y.Y.; Liu, C.S. Self-Assembled Nanoparticles Based on Hydrophobically Modified Chitosan as Carriers for Doxorubicin. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 258–265. [Google Scholar] [CrossRef]
- Ortona, O.; D’Errico, G.; Mangiapia, G.; Ciccarelli, D. The Aggregative Behavior of Hydrophobically Modified Chitosans with High Substitution Degree in Aqueous Solution. Carbohydr. Polym. 2008, 74, 16–22. [Google Scholar] [CrossRef]
- Ways, T.M.M.; Lau, W.M.; Khutoryanskiy, V.V. Chitosan and Its Derivatives for Application in Mucoadhesive Drug Delivery Systems. Polymers (Basel) 2018, 10, 267. [Google Scholar] [CrossRef] [Green Version]
- Ong, S.; Ming, L.; Lee, K.; Yuen, K. Influence of the Encapsulation Efficiency and Size of Liposome on the Oral Bioavailability of Griseofulvin-Loaded Liposomes. Pharmaceutics 2016, 8, 25. [Google Scholar] [CrossRef]
- Othman, N.; Masarudin, M.; Kuen, C.; Dasuan, N.; Abdullah, L.; Md. Jamil, S. Synthesis and Optimization of Chitosan Nanoparticles Loaded with L-Ascorbic Acid and Thymoquinone. Nanomaterials 2018, 8, 920. [Google Scholar] [CrossRef] [Green Version]
- Roberts, S.A.; Agrawal, N. Enhancing the Drug Encapsulation Efficiency of Liposomes for Therapeutic Delivery. 2017 IEEE Healthc. Innov. Point Care Technol. HI-POCT 2017, 2017, 136–139. [Google Scholar] [CrossRef]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maruyama, C.R.; Guilger, M.; Pascoli, M.; Bileshy-José, N.; Abhilash, P.C.; Fraceto, L.F.; De Lima, R. Nanoparticles Based on Chitosan as Carriers for the Combined Herbicides Imazapic and Imazapyr. Sci. Rep. 2016, 6, 19768. [Google Scholar] [CrossRef] [PubMed]
- Kruse, A.; Dinjus, E. Hot Compressed Water as Reaction Medium and Reactant. Properties and Synthesis Reactions. J. Supercrit. Fluids 2007, 39, 362–380. [Google Scholar] [CrossRef]
- Malm, A.V.; Corbett, J.C.W. Improved Dynamic Light Scattering Using an Adaptive and Statistically Driven Time Resolved Treatment of Correlation Data. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Jonassen, H.; Kjøniksen, A.L.; Hiorth, M. Effects of Ionic Strength on the Size and Compactness of Chitosan Nanoparticles. Colloid Polym. Sci. 2012, 290, 919–929. [Google Scholar] [CrossRef]
- Mayeen, A.; Shaji, L.K.; Nair, A.K.; Kalarikkal, N. Morphological Characterization of Nanomaterials; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar] [CrossRef]
- Barhoum, A.; Luisa García-Betancourt, M. Physicochemical Characterization of Nanomaterials: Size, Morphology, Optical, Magnetic, and Electrical Properties; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar] [CrossRef]
- Dogan, Ü.; Çiftçi, H.; Cetin, D.; Suludere, Z.; Tamer, U. Nanoparticle Embedded Chitosan Film for Agglomeration Free TEM Images. Microsc. Res. Tech. 2017, 80, 163–166. [Google Scholar] [CrossRef]
- Coates, J. Interpretation of Infrared Spectra, A Practical Approach; Meyer, R.A., Ed.; John Wiley & Sons: Chichester, UK, 2000. [Google Scholar]
- N., M.D.; Eskandari, R.; Zolfagharian, H.; Mohammad, M. Preparation and in Vitro Characterization of Chitosan Nanoparticles Containing Mesobuthus Eupeus Scorpion Venom as an Antigen Delivery System. J. Venom. Anim. Toxins Trop. Dis. 2012, 18, 44–52. [Google Scholar]
- Martins, A.F.; de Oliveira, D.M.; Pereira, A.G.B.; Rubira, A.F.; Muniz, E.C. Chitosan/TPP Microparticles Obtained by Microemulsion Method Applied in Controlled Release of Heparin. Int. J. Biol. Macromol. 2012, 51, 1127–1133. [Google Scholar] [CrossRef] [Green Version]
- Queiroz, M.F.; Melo, K.R.T.; Sabry, D.A.; Sassaki, G.L.; Rocha, H.A.O. Does the Use of Chitosan Contribute to Oxalate Kidney Stone Formation? Mar. Drugs 2015, 13, 141–158. [Google Scholar] [CrossRef]
- Bi, X.; Zhang, H.; Dou, L. Layered Double Hydroxide-Based Nanocarriers for Drug Delivery. Pharmaceutics 2014, 6, 298–332. [Google Scholar] [CrossRef] [PubMed]
- Barahuie, F.; Hussein, M.Z.; Gani, S.A.; Fakurazi, S.; Zainal, Z. Synthesis of Protocatechuic Acid-Zinc/aluminium-Layered Double Hydroxide Nanocomposite as an Anticancer Nanodelivery System. J. Solid State Chem. 2015, 221, 21–31. [Google Scholar] [CrossRef]
- Sani, M.; Mohd, U.; Hussein, Z.; Umar, A.; Sharida, K.; Mas, F.; Masarudin, J. Synthesis and Characterization of Protocatechuic Acid—Loaded Gadolinium—Layered Double Hydroxide and Gold Nanocomposite for Theranostic Application. Appl. Nanosci. 2018, 8, 5. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Liu, Y.; Wu, C.; Qiu, Y.; Xu, X.; Lv, H.; Bai, A.; Liu, X. Development of Drug-Loaded Chitosan Hollow Nanoparticles for Delivery of Paclitaxel to Human Lung Cancer A549 Cells. Drug Dev. Ind. Pharm. 2017, 43, 1304–1313. [Google Scholar] [CrossRef]
- Uppal, S.; Kaur, K.; Kumar, R.; Kaur, N.D.; Shukla, G.; Mehta, S.K. Chitosan Nanoparticles as a Biocompatible and Efficient Nanowagon for Benzyl Isothiocyanate. Int. J. Biol. Macromol. 2018, 115, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Cheung, R.; Ng, T.; Wong, J.; Chan, W. Chitosan: An Update on Potential Biomedical and Pharmaceutical Applications. Mar. Drugs 2015, 13, 5156–5186. [Google Scholar] [CrossRef]
- Kong, B.; Seog, J.H.; Lee, S.B. Experimental Considerations on the Cytotoxicity of Nanoparticles Choice of Cell Types. Nanomedicine 2015, 6, 1–12. [Google Scholar] [CrossRef]
- Loutfy, S.A.; El-din, H.M.A.; Elberry, M.; Allam, N.G.; Hasanin, M.T.M.; Abdellah, A.M. Synthesis, Characterization and Cytotoxic Evaluation of Chitosan Nanoparticles: In Vitro Liver Cancer Model. Adv. Nat. Sci. Nanosci. Nanotechnol. 2016, 7, 035008. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.; Xi, X.; Shrestha, S.; Jiang, P.; Zhang, W.; Gao, C. Amino Acid-Modified Chitosan Nanoparticles for Cu2+chelation to Suppress CuO Nanoparticle Cytotoxicity. J. Mater. Chem. B 2017, 5, 3521–3530. [Google Scholar] [CrossRef]
- Grenha, A.; Grainger, C.I.; Dailey, L.A.; Seijo, B.; Martin, G.P.; Remuñán-López, C.; Forbes, B. Chitosan Nanoparticles Are Compatible with Respiratory Epithelial Cells in Vitro. Eur. J. Pharm. Sci. 2007, 31, 73–84. [Google Scholar] [CrossRef]
- Díaz, B.; Sánchez-Espinel, C.; Arruebo, M.; Faro, J.; De Miguel, E.; Magadán, S.; Yagüe, C.; Fernández-Pacheco, R.; Ibarra, M.R.; Santamaría, J.; et al. Assessing Methods for Blood Cell Cytotoxic Responses to Inorganic Nanoparticles and Nanoparticle Aggregates. Small 2008, 4, 2025–2034. [Google Scholar] [CrossRef] [PubMed]
- Powers, K.W.; Palazuelos, M.; Moudgil, B.M.; Roberts, S.M. Characterization of the Size, Shape, and State of Dispersion of Nanoparticles for Toxicological Studies. Nanotoxicology 2007, 1, 42–51. [Google Scholar] [CrossRef]
- Chithrani, B.D.; Chan, W.C. Elucidating the Mechanism of Cellular Uptake and Removal of Protein-Coated Gold Nanoparticles of Different Sizes and Shapes. TL—7. Nano Lett. 2007, 7, 1542–1550. [Google Scholar] [CrossRef]
- Chanphai, P.; Tajmir-Riahi, H.A. Encapsulation of Testosterone by Chitosan Nanoparticles. Int. J. Biol. Macromol. 2017, 98, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Madureira, A.R.; Pereira, A.; Pintado, M. Chitosan Nanoparticles Loaded with 2,5-Dihydroxybenzoic Acid and Protocatechuic Acid: Properties and Digestion. J. Food Eng. 2016, 174, 8–14. [Google Scholar] [CrossRef]
- Pham, T.; Nguyen, T.; Thi, T.; Nguyen, T.-T.; Le, T.; Vo, D.; Nguyen, D.; Nguyen, C.; Nguyen, D.; Nguyen, T.; et al. Investigation of Chitosan Nanoparticles Loaded with Protocatechuic Acid (PCA) for the Resistance of Pyricularia Oryzae Fungus against Rice Blast. Polymers (Basel) 2019, 11, 177. [Google Scholar] [CrossRef] [Green Version]
- Hassan, U.A.; Hussein, M.Z.; Alitheen, N.B.; Ariff, S.A.Y.; Masarudin, M.J. In Vitro Cellular Localization and Efficient Accumulation of Fluorescently Tagged Biomaterials from Monodispersed Chitosan Nanoparticles for Elucidation of Controlled Release Pathways for Drug Delivery Systems. Int. J. Nanomed. 2018, 13, 5075–5095. [Google Scholar] [CrossRef] [Green Version]











| Sample | Free PCA | CNP-PCA | pCNP-PCA | ||
|---|---|---|---|---|---|
| A296nm | A296nm | % EE | A296nm | % EE | |
| Replicate 1 | 0.79 | 0.53 | 32.91 | 0.42 | 46.84 |
| Replicate 2 | 0.83 | 0.51 | 38.55 | 0.33 | 60.24 |
| Replicate 3 | 0.82 | 0.54 | 34.15 | 0.36 | 56.10 |
| Average | 35.20 ± 1.71 | 54.39 ± 3.96 | |||
| Functional Group | Wavenumber (nm−1) | Percentage Transmittance (% T) | Sample |
|---|---|---|---|
| Hydrogen bond a (O—H) | 3228 | 49.26 | CS |
| 3360 | 71.24 | CNP | |
| 3383 | 55.82 | PCNP | |
| 3278 | 62.69 | PCA | |
| 3376 | 75.50 | CNP-PCA | |
| 3227 | 71.13 | PCNP-PCA | |
| Amine II group b (NH2) | 1600 | 10.00 | CS |
| 1629 | 45.68 | CNP | |
| 1635 | 35.77 | PCNP | |
| Inorganic Phosphate c (P=O) | 1201 | 35.74 | TPP |
| 1155 | 40.22 | CNP | |
| 1281 | 74.94 | pCNP | |
| Ether group d (C-O-C) | 1082 | 32.20 | CS |
| 1059 | 10 | CNP | |
| 1068 | 10 | pCNP | |
| Carbon double bond e (C=C) | 1658 | 15.56 | PCA |
| 1625 | 47.81 | CNP-PCA | |
| 1625 | 39.61 | pCNP-PCA | |
| Carbonyl group f (C=O) | 1300 | 69.14 | PCA |
| 1389 | 66.75 | CNP-PCA | |
| 1280 | 73.83 | pCNP-PCA |
| Time Point | 24 h | 48 h | 72 h |
|---|---|---|---|
| * IC50 value (µM) | |||
| PCA | N/A | N/A | N/A |
| CNP-PCA | 191.50 | 75.78 | 63.27 |
| pCNP-PCA | 53.71 | 110.70 | 48.34 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yee Kuen, C.; Galen, T.; Fakurazi, S.; Othman, S.S.; Masarudin, M.J. Increased Cytotoxic Efficacy of Protocatechuic Acid in A549 Human Lung Cancer Delivered via Hydrophobically Modified-Chitosan Nanoparticles As an Anticancer Modality. Polymers 2020, 12, 1951. https://doi.org/10.3390/polym12091951
Yee Kuen C, Galen T, Fakurazi S, Othman SS, Masarudin MJ. Increased Cytotoxic Efficacy of Protocatechuic Acid in A549 Human Lung Cancer Delivered via Hydrophobically Modified-Chitosan Nanoparticles As an Anticancer Modality. Polymers. 2020; 12(9):1951. https://doi.org/10.3390/polym12091951
Chicago/Turabian StyleYee Kuen, Cha, Tieo Galen, Sharida Fakurazi, Siti Sarah Othman, and Mas Jaffri Masarudin. 2020. "Increased Cytotoxic Efficacy of Protocatechuic Acid in A549 Human Lung Cancer Delivered via Hydrophobically Modified-Chitosan Nanoparticles As an Anticancer Modality" Polymers 12, no. 9: 1951. https://doi.org/10.3390/polym12091951