The Anti-Inflammatory Effect of Lactose-Modified Hyaluronic Acid Molecules on Primary Bronchial Fibroblasts of Smokers
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Silico Methods
2.1.1. Protein Retrieval and Protein and Ligand Preparation
2.1.2. Molecular Docking
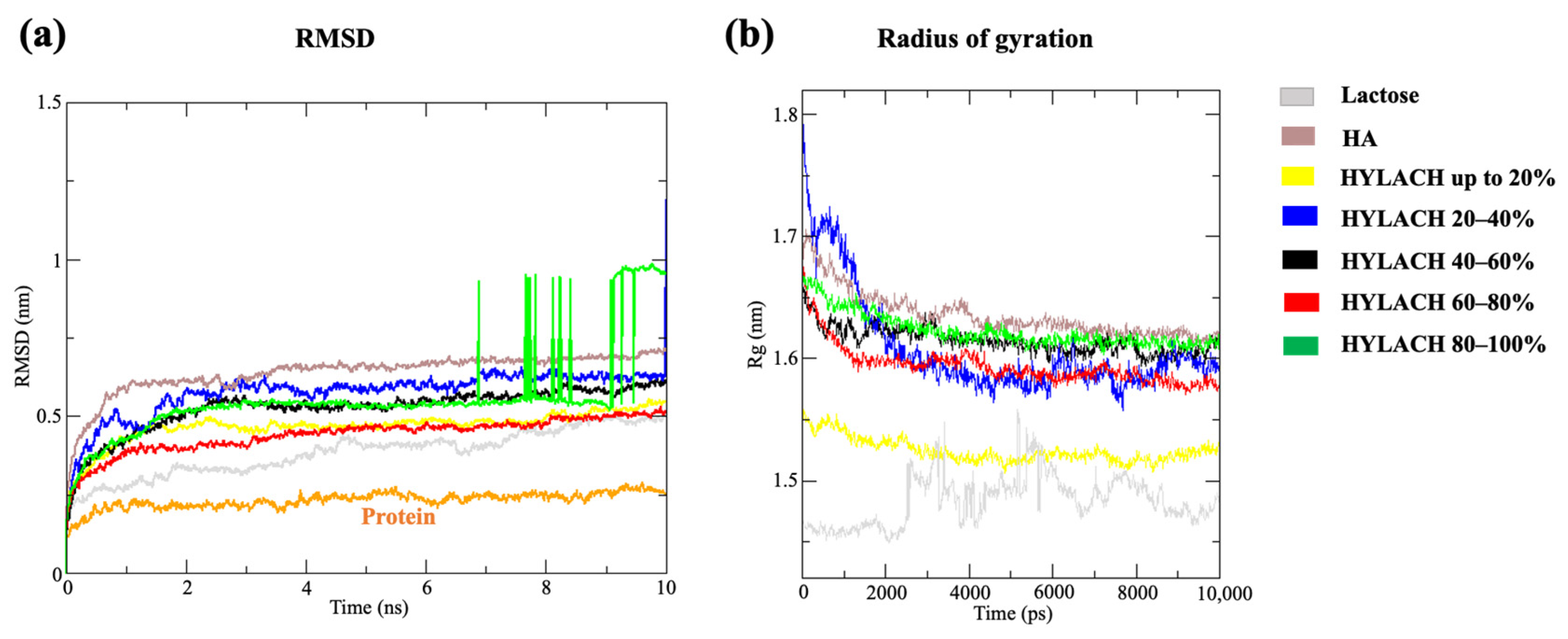
2.1.3. Molecular Dynamics Simulations
2.1.4. Methods of Analysis
2.2. In Vitro Methods
2.2.1. Drugs and Chemicals
2.2.2. Primary Human Bronchial Fibroblasts and U937 Monocytes
2.2.3. Activated U937 Monocyte Conditioned Medium
2.2.4. Evaluation of the Effects of HYLACH on Primary Human Bronchial Fibroblast Viability
2.2.5. Analysis of Anti-Inflammatory and Antioxidative Effects Induced by HYLACH
- -
- Detection of intracellular ROS generation. Human bronchial fibroblasts were seeded in 96-well plates with a density of 10,000 cells/well, allowed to adhere overnight, and then, exposed to activated U937 CM to induce intracellular ROS generation. Cells were then cultivated in the presence or absence of HA or HYLACH for 4 h. Subsequently, the medium was removed, the cells were rinsed twice with PBS and then loaded with 5 µM 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA; Molecular Probes, ThermoFisher, Waltham, MA, USA) diluted in PBS for 1 h at 37 °C. H2DCFDA is a nonfluorescent probe that, in the presence of intracellular ROS, rapidly oxidizes to the fluorescent 2′,7′-dichlorofluorescein. The cells were washed three times with PBS before measuring fluorescence intensity with a microtiter plate reader (Infinite 2000, Tecan, Milan, Italy) at 485 and 535 nm for excitation and emission, respectively.
- -
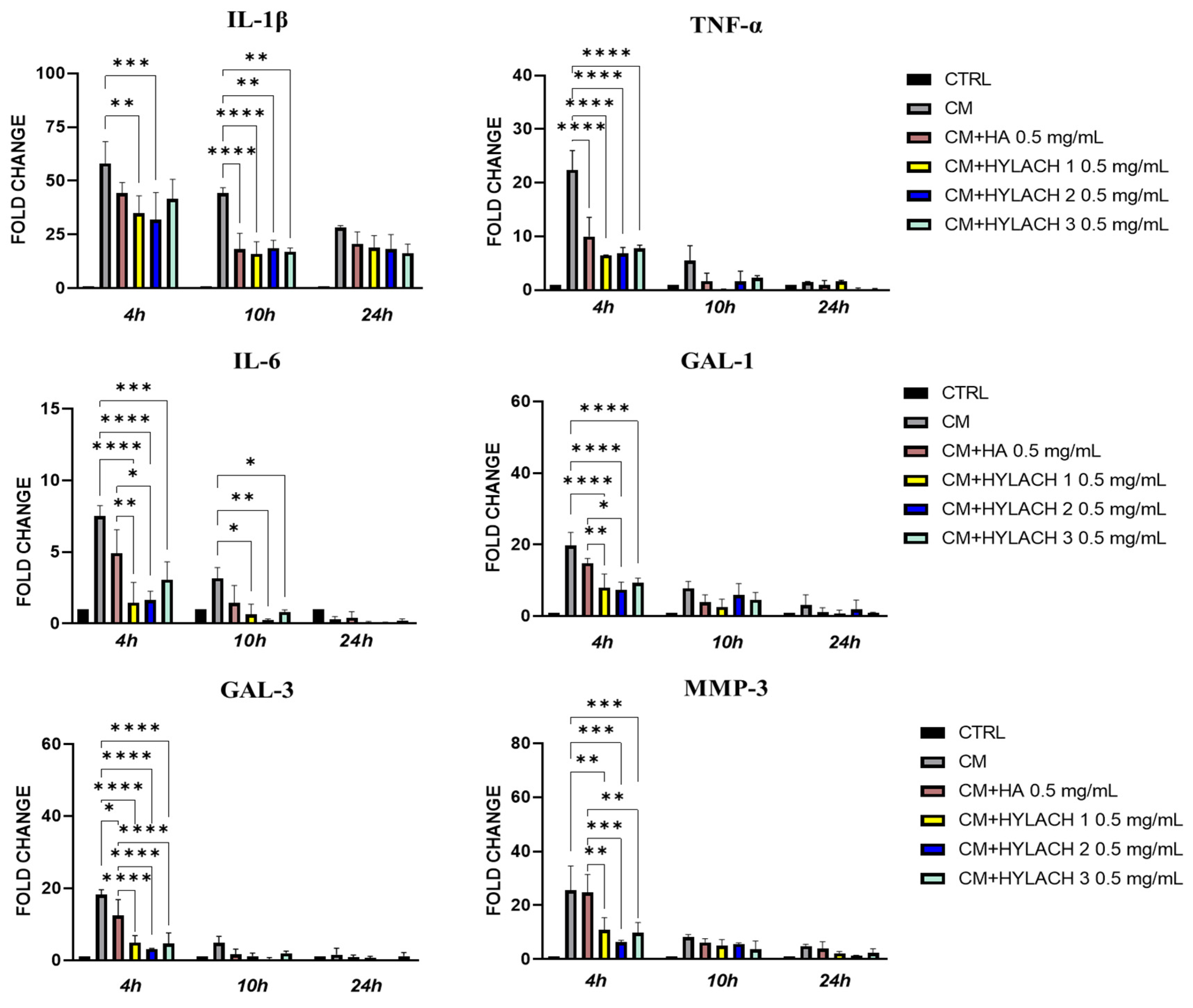
- Gene expression of inflammatory molecules. In primary human bronchial fibroblasts exposed or not to activated U937 CM and treated with HA and HYLACH for 4, 10, and 24 h, differences in the gene expression of IL-1β, IL-6, TNF-α, Gal-1, Gal-3, and MMP-3 were examined by qPCR. Total RNA was purified using TRIzol (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s instructions and the Nanodrop 2000c spectrophotometer (Thermo Scientific) was used to evaluate RNA quality by measuring absorbance at 260/280 nm. To remove DNA from the samples, total RNA was treated for 15 min with DNAse I (Thermo Fisher). Five hundred ng of total RNA were reverse transcribed with oligo-dT and Superscript II (Life Technologies, Carlsbad, CA, USA) to generate cDNA. Gene expression levels of cytokines and MMP-3 were evaluated with a Rotor-Gene RG-3000A (QIAGEN, Hilden, Germany) using the Xpert fast SYBR (GRISP, Porto, Portugal). The expression of target genes was normalized to the endogenous levels of peptidylprolyl isomerase A (PPIA). The 2ΔCt method, where dCt = Ct PPIA − Ct target gene, was used to assess gene expression. Table 1 contains a list of the primers used in the qPCR analysis.
- -
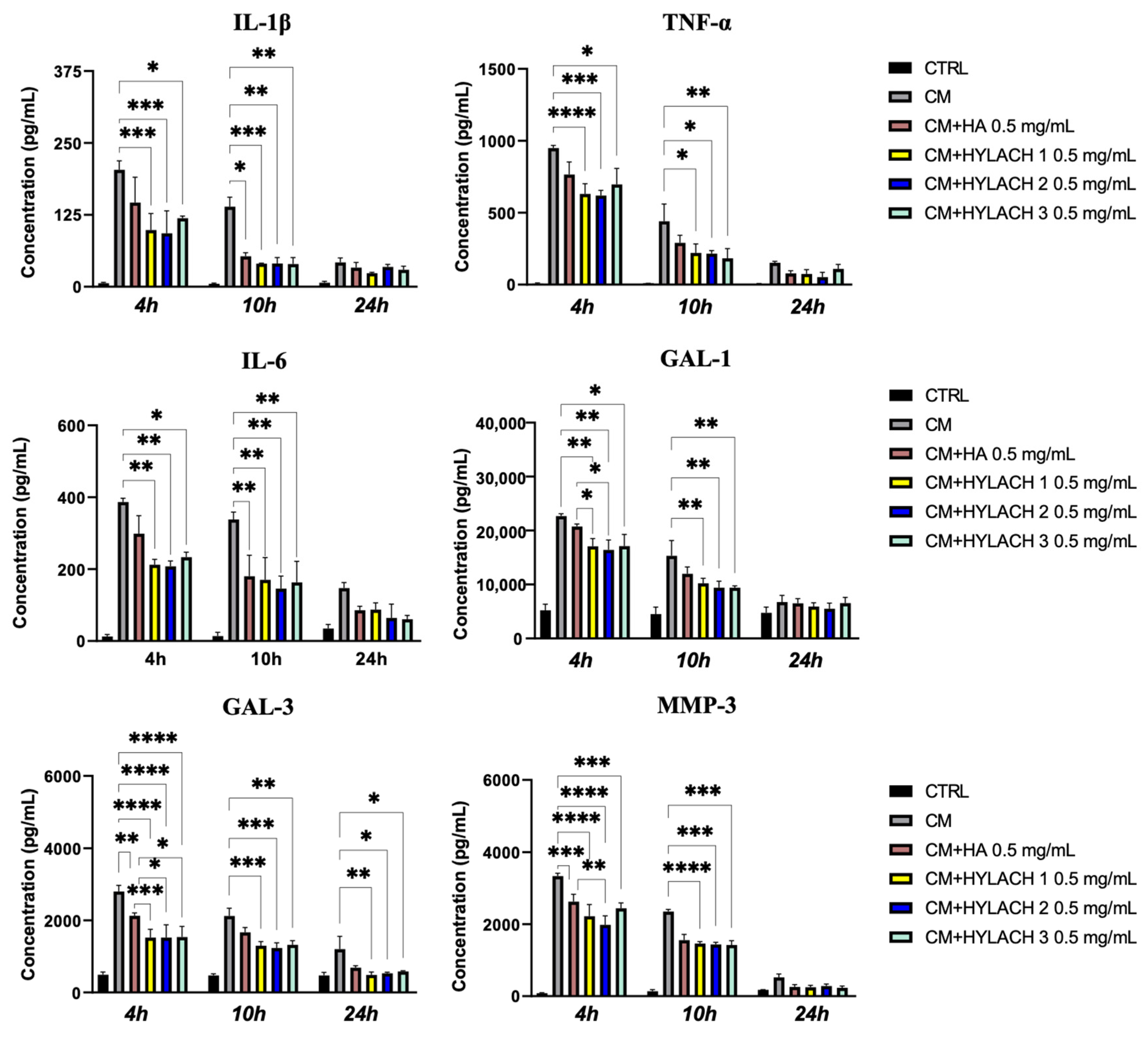
- Enzyme-linked immunosorbent assay (ELISA). Supernatants were collected from primary human bronchial fibroblast cultures stimulated or not with activated U937 CM for 24 h and subsequently treated with HA and HYLACH. IL-1β, TNF-α, and MMP-3 were quantified using ELISA kits from R&D Systems (Minneapolis, MN, USA), Gal-1 with the ELISA Kit from Ray Biotech (Ray Biotech, Inc., Peachtree Corners, GA, USA), IL-6 with the human ELISA kit from Boster Bio (Pleasanton, TX, USA) and Gal-3 with Sino Biological (Sino Biological, Wayne, PA, USA) according to the suppliers’ protocols. Each experiment was carried out in triplicate.
2.2.6. Statistical Analysis
3. Results
3.1. In Silico Results
3.1.1. HYLACH Compounds Show a Better Docking Score Than Nonfunctionalized HA
3.1.2. HYLACH with a Percentage of Lactose-Derived Residues of up to 30% and Lactose Represent the Best Ligands for Gal-3
3.2. In Vitro Results
3.2.1. HA and HYLACH Do Not Induce Changes in the Viability of Primary Human Bronchial Fibroblasts from Smokers
3.2.2. HYLACH Exerts a Higher Anti-Inflammatory Effect Than HA on Primary Human Bronchial Fibroblasts
3.2.3. HYLACH 1 and HYLACH 2 Exert an Antioxidant Effect on Primary Human Bronchial Fibroblasts Cultures Exposed to CM
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Rabe, K.F.; Watz, H. Chronic obstructive pulmonary disease. Lancet 2017, 389, 1931–1940. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The Top 10 Causes of Death 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 9 January 2023).
- Belchamber, K.B.R.; Walker, E.M.; Stockley, R.A.; Sapey, E. Monocytes and Macrophages in Alpha-1 Antitrypsin Deficiency. Int. J. Chron. Obstruct. Pulmon. Dis. 2020, 15, 3183–3192. [Google Scholar] [CrossRef] [PubMed]
- D’Anna, S.E.; Maniscalco, M.; Carriero, V.; Gnemmi, I.; Caramori, G.; Nucera, F.; Righi, L.; Brun, P.; Balbi, B.; Adcock, I.M.; et al. Evaluation of innate immune mediators related to respiratory viruses in the lung of stable COPD patients. J. Clin. Med. 2020, 9, 1807. [Google Scholar] [CrossRef]
- Zhou, H.; Wu, Y.; Jin, Y.; Zhou, J.; Zhang, C.; Che, L.; Jing, J.; Chen, Z.; Li, W.; Shen, H. Genetic polymorphism of matrix metalloproteinase family and chronic obstructive pulmonary disease susceptibility: A meta-analysis. Sci. Rep. 2013, 3, 2818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraen, M.; Frantz, S.; Nihlén, U.; Engström, G.; Löfdahl, C.G.; Wollmer, P.; Dencker, M. Matrix Metalloproteinases in COPD and atherosclerosis with emphasis on the effects of smoking. PLoS ONE 2019, 14, e0211987. [Google Scholar] [CrossRef] [Green Version]
- Johannes, L.; Jacob, R.; Leffler, H. Galectins at a glance. J. Cell Sci. 2018, 131, jcs208884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.T.; Yang, R.Y.; Hsu, D.K. Galectins in acute and chronic inflammation. Ann. N. Y. Acad. Sci. 2012, 1253, 80–91. [Google Scholar] [CrossRef]
- Sundblad, V.; Morosi, L.G.; Geffner, J.R.; Rabinovich, G.A. Galectin-1: A Jack-of-All-Trades in the Resolution of Acute and Chronic Inflammation. J. Immunol. 2017, 199, 3721–3730. [Google Scholar] [CrossRef] [Green Version]
- Erriah, M.; Pabreja, K.; Fricker, M.; Baines, K.J.; Donnelly, L.E.; Bylund, J.; Karlsson, A.; Simpson, J.L. Galectin-3 enhances monocyte-derived macrophage efferocytosis of apoptotic granulocytes in asthma. Respir. Res. 2019, 20, 1. [Google Scholar] [CrossRef]
- Humphries, D.C.; Mills, R.; Dobie, R.; Henderson, N.C.; Sethi, T.; Mackinnon, A.C. Selective Myeloid Depletion of Galectin-3 Offers Protection Against Acute and Chronic Lung Injury. Front. Pharmacol. 2021, 12, 715986. [Google Scholar] [CrossRef]
- Barnes, P.J. COPD 2020: New directions needed. Am. J. Physiol. Lung Cell Mol. Physiol. 2020, 319, L884–L886. [Google Scholar] [CrossRef] [PubMed]
- Cosio, M.G.; Saetta, M.; Agusti, A. Immunologic aspects of chronic obstructive pulmonary disease. N. Engl. J. Med. 2009, 360, 2445–2454. [Google Scholar] [CrossRef] [PubMed]
- D’Anna, S.E.; Balbi, B.; Cappello, F.; Carone, M.; Di Stefano, A. Bacterial-viral load and the immune response in stable and exacerbated COPD: Significance and therapeutic prospects. Int. J. Chron. Obstruct. Pulmon. Dis. 2016, 11, 445–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brun, P.; Zavan, B.; Vindigni, V.; Schiavinato, A.; Pozzuoli, A.; Iacobellis, C.; Abatangelo, G. In vitro response of osteoarthritic chondrocytes and fibroblast-like synoviocytes to a 500-730 kDa hyaluronan amide derivative. J. Biomed. Mater. Res. B Appl. Biomater. 2012, 100, 2073–2081. [Google Scholar] [CrossRef]
- Marinho, A.; Nunes, C.; Reis, S. Hyaluronic Acid: A Key Ingredient in the Therapy of Inflammation. Biomolecules 2021, 11, 1518. [Google Scholar] [CrossRef]
- Tarricone, E.; Mattiuzzo, E.; Belluzzi, E.; Elia, R.; Benetti, A.; Venerando, R.; Vindigni, V.; Ruggieri, P.; Brun, P. Anti-Inflammatory Performance of Lactose-Modified Chitosan and Hyaluronic Acid Mixtures in an In Vitro Macrophage-Mediated Inflammation Osteoarthritis Model. Cells 2020, 9, 1328. [Google Scholar] [CrossRef]
- Marcon, P.; Marsich, E.; Vetere, A.; Mozetic, P.; Campa, C.; Donati, I.; Vittur, F.; Gamini, A.; Paoletti, S. The role of Galectin-1 in the interaction between chondrocytes and a lactose-modified chitosan. Biomaterials 2005, 26, 4975–4984. [Google Scholar] [CrossRef]
- Wang, D.D.; Zhu, M.; Yan, H. Computationally predicting binding affinity in protein-ligand complexes: Free energy-based simulations and machine learning-based scoring functions. Brief Bioinform. 2021, 22, bbaa107. [Google Scholar] [CrossRef]
- Schrödinger. Schrödinger Release 2022-3: Maestro; LLC: New York, NY, USA, 2021.
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Schuttelkopf, A.W.; van Aalten, D.M.F. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 1355–1363. [Google Scholar] [CrossRef] [Green Version]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R. Amber 2021; University of California: San Francisco, CA, USA, 2021. [Google Scholar]
- Martínez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. PACKMOL: A package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; Delano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Di Stefano, A.; Caramori, G.; Barczyk, A.; Vicari, C.; Brun, P.; Zanini, A.; Cappello, F.; Garofano, E.; Padovani, A.; Contoli, M.; et al. Innate immunity but not NLRP3 inflammasome activation correlates with severity of stable COPD. Thorax 2014, 69, 516–524. [Google Scholar] [CrossRef] [Green Version]
- Denizot, F.; Lang, R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 1986, 89, 271–277. [Google Scholar] [CrossRef]
- Saraboji, K.; Håkansson, M.; Genheden, S.; Diehl, C.; Qvist, J.; Weininger, U.; Nilsson, U.J.; Leffler, H.; Ryde, U.; Akke, M.; et al. The carbohydrate-binding site in galectin-3 is preorganized to recognize a sugarlike framework of oxygens: Ultra-high-resolution structures and water dynamics. Biochemistry 2012, 51, 296–306. [Google Scholar] [CrossRef]
- Durham, A.L.; Adcock, I.M. The relationship between COPD and lung cancer. Lung Cancer 2015, 90, 121–127. [Google Scholar] [CrossRef] [Green Version]
- Abatangelo, G.; Vindigni, V.; Avruscio, G.; Pandis, L.; Brun, P. Hyaluronic Acid: Redefining Its Role. Cells 2020, 9, 1743. [Google Scholar] [CrossRef] [PubMed]
- Salamanna, F.; Giavaresi, G.; Parrilli, A.; Martini, L.; Aldini, N.N.; Abatangelo, G.; Frizziero, A.; Fini, M. Effects of intra-articular hyaluronic acid associated to Chitlac (arty-duo®) in a rat knee osteoarthritis model. J. Orthop. Res. 2019, 37, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Wang, L.; Zheng, R.; Guo, L.; Chen, Y.; Qian, X. Research and progress of chemical depolymerization of waste PET and high-value application of its depolymerization products. RSC Adv. 2022, 12, 31564–31576. [Google Scholar] [CrossRef]
- Máiz Carro, L.; Martínez-García, M.A. Use of Hyaluronic Acid (HA) in Chronic Airway Diseases. Cells 2020, 9, 2210. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallorini, M.; Antonetti Lamorgese Passeri, C.; Cataldi, A.; Berardi, A.C.; Osti, L. Hyaluronic Acid Alleviates Oxidative Stress and Apoptosis in Human Tenocytes via Caspase 3 and 7. Int. J. Mol. Sci. 2022, 23, 8817. [Google Scholar] [CrossRef]
- Yao, H.; Irfan, R. Current concepts on the role of inflammation in COPD and lung cancer. Curr. Opin. Pharmacol. 2009, 9, 375–383. [Google Scholar] [CrossRef] [Green Version]
- Donato, A.; Belluzzi, E.; Mattiuzzo, E.; Venerando, R.; Cadamuro, M.; Ruggieri, P.; Vindigni, V.; Brun, P. Anti-Inflammatory and Pro-Regenerative Effects of Hyaluronan-Chitlac Mixture in Human Dermal Fibroblasts: A Skin Ageing Perspective. Polymers 2022, 14, 1817. [Google Scholar] [CrossRef]
- Cantor, J.; Ma, S.; Turino, G. A pilot clinical trial to determine the safety and efficacy of aerosolized hyaluronan as a treatment for COPD. Int. J. Chron. Obstruct. Pulmon. Dis. 2017, 12, 2747–2752. [Google Scholar] [CrossRef] [Green Version]
- Brivio, A.; Conese, M.; Gambazza, S.; Biffi, A.; Tirelli, A.S.; Russo, M.; Foà, M.; Colombo, C. Pilot randomized controlled trial evaluating the effect of hypertonic saline with and without hyaluronic acid in reducing inflammation in cystic fibrosis. J. Aerosol. Med. Pulm. Drug Deliv. 2016, 29, 482–489. [Google Scholar] [CrossRef]
- Bassiouni, W.; Ali, M.A.M.; Schulz, R. Multifunctional intracellular matrix metalloproteinases: Implications in disease. FEBS J. 2021, 288, 7162–7182. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhong, X.; Li, J.; Liu, Z.; Cheng, L. Inorganic nanomaterials with rapid clearance for biomedical applications. Chem. Soc. Rev. 2021, 50, 8669–8742. [Google Scholar] [CrossRef] [PubMed]





| Gene (Accession Number) | Name | Primer Sequences |
|---|---|---|
| IL-1β (NM_000576.3) | Interleukin 1 beta | Fw 5′-GAATCTCCGACCACCACTACAG-3′ Rv 5′-TGATCGTACAGGTGCATCGTG-3′ |
| LSGALS1 (NM_002305.4) | Galectin 1 | Fw 5′-TCTCGGGTGGAGTCTTCTGA-3′ Rv 5′-GTTCAGCACGAAGCTCTTAGC-3′ |
| LSGALS3 (NM_002306.4) | Galectin 3 | Fw 5′-CTGCTGGGGCACTGATTGT-3′ Rv 5′-TGTTTGCATTGGGCTTCACC-3′ |
| MMP-3 (NM_002422.5) | Matrix metalloproteinase 3 | Fw 5′-TCACTCACAGACCTGACTCG -3′ Rv 5′-AAAGCAGGATCACAGTTGGC-3′ |
| IL-6 (NM_000501.4) | Interleukin 6 | Fw 5′-ATGAACTCCTTCTCCACAAGCG-3′ Rv 5′-CTCCTTTCTCAGGGCTGAG-3′ |
| PPIA (NM_021130.5) | Peptidylprolyl Isomerase A | Fw 5′-GGGCTTTAGGCTGTAGGTCAA-3′ Rv 5′-AACCAAAGCTAGGGAGAGGC-3′ |
| TNF-α (NM_000594.3) | TNF-alpha | Fw 5′-AAGCCTGTAGCCCATGTTGT-3′ Rv 5′-GGACCTGGGAGTAGATGAGGT-3′ |
| Compound | % of Lactosylation | Average Binding Free Energy (kJ/mol) |
|---|---|---|
| HA | - | 4138 |
| LACTOSE | - | −39 |
| HYLACH | 10% | −302 |
| 20% | −236 | |
| 30% | −284 | |
| 40% | −87 | |
| 50% | −166 | |
| 60% | −197 | |
| 70% | −218 | |
| 80% | −222 | |
| 90% | −212 | |
| 100% | −282 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donato, A.; Fontana, F.; Venerando, R.; Di Stefano, A.; Brun, P. The Anti-Inflammatory Effect of Lactose-Modified Hyaluronic Acid Molecules on Primary Bronchial Fibroblasts of Smokers. Polymers 2023, 15, 1616. https://doi.org/10.3390/polym15071616
Donato A, Fontana F, Venerando R, Di Stefano A, Brun P. The Anti-Inflammatory Effect of Lactose-Modified Hyaluronic Acid Molecules on Primary Bronchial Fibroblasts of Smokers. Polymers. 2023; 15(7):1616. https://doi.org/10.3390/polym15071616
Chicago/Turabian StyleDonato, Alice, Federico Fontana, Rina Venerando, Antonino Di Stefano, and Paola Brun. 2023. "The Anti-Inflammatory Effect of Lactose-Modified Hyaluronic Acid Molecules on Primary Bronchial Fibroblasts of Smokers" Polymers 15, no. 7: 1616. https://doi.org/10.3390/polym15071616





