Potentiating Gilteritinib Efficacy Using Nanocomplexation with a Hyaluronic Acid–Epigallocatechin Gallate Conjugate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
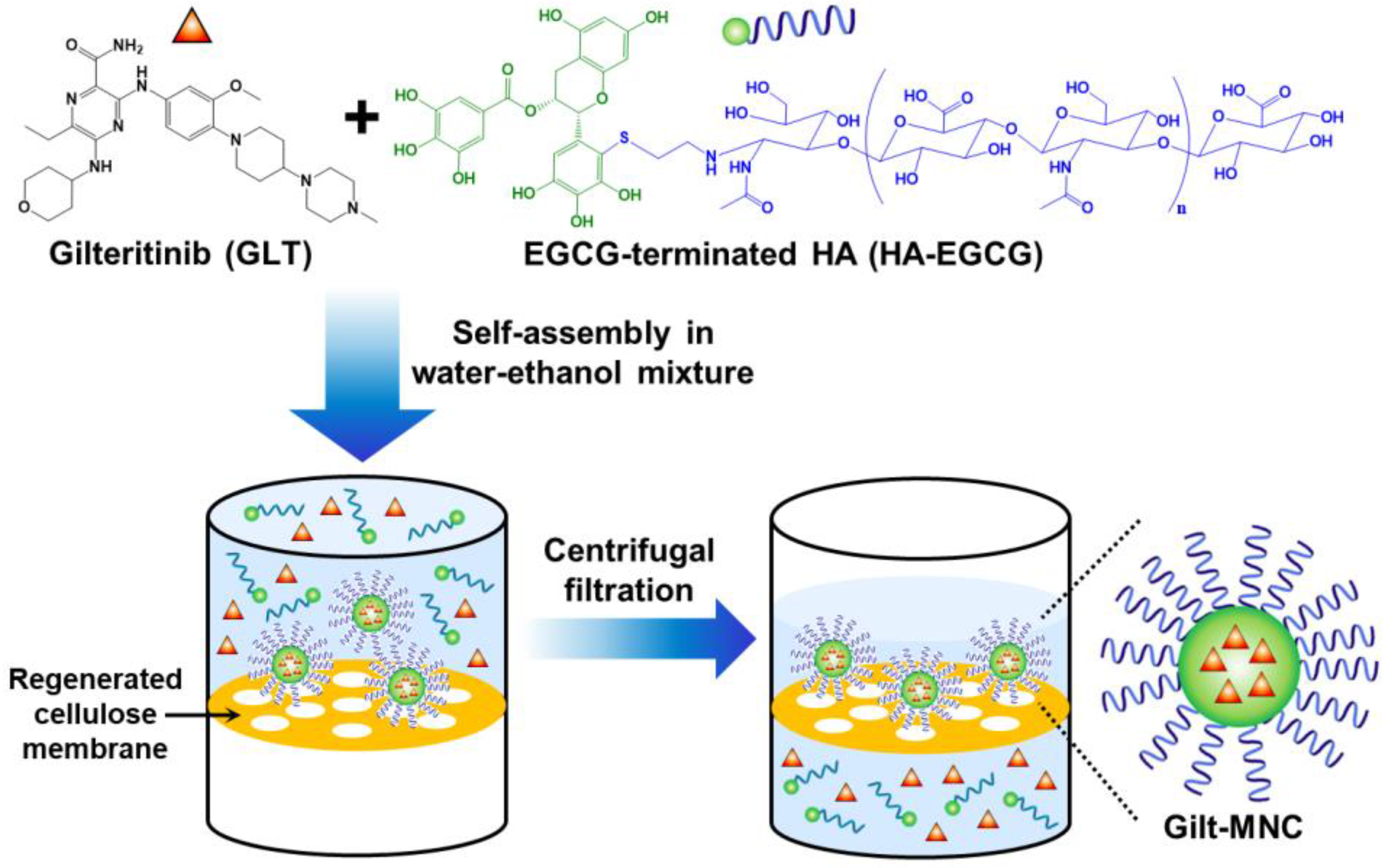
2.2. Preparation of Gilt-MNC
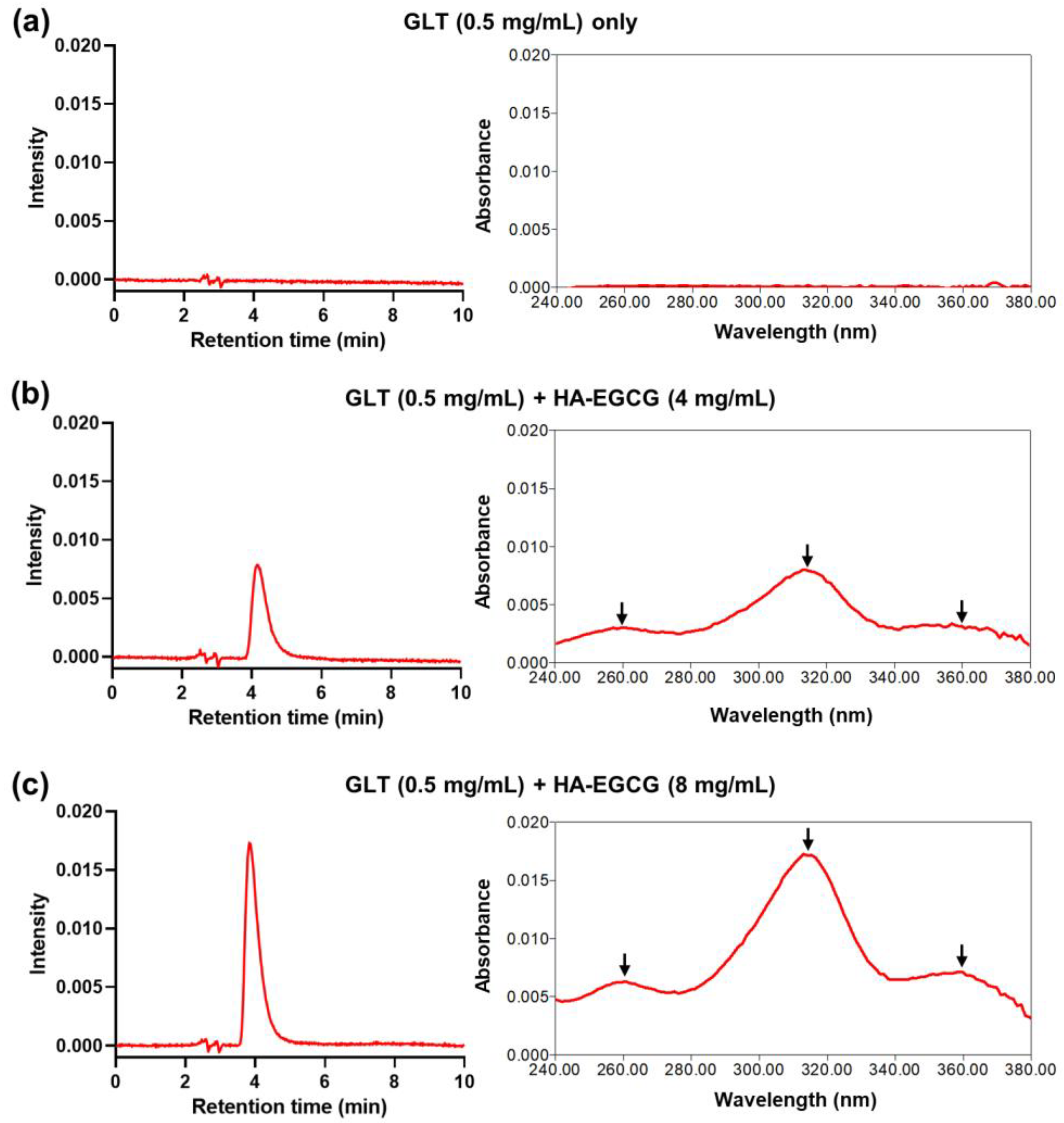
2.3. Reversed-Phase High-Performance Liquid Chromatography (RP-HPLC)
2.4. Physicochemical Characterization
2.5. DyLight 488 Tagging of Gilt-MNC
2.6. Flow Cytometry Analysis
2.7. Evaluation of Anti-Leukemic Activity
2.8. Assessment of Combination Effects
2.9. Measurement of Cellular ROS Production
2.10. Visualization of Cellular Apoptosis
2.11. Quantification of Caspase-3/7 Activity
2.12. Statistical Analysis Methods
3. Results and Discussion
3.1. Self-Assembly and Purification of Gilt-MNC
3.2. Physicochemical Characterization of Gilt-MNC
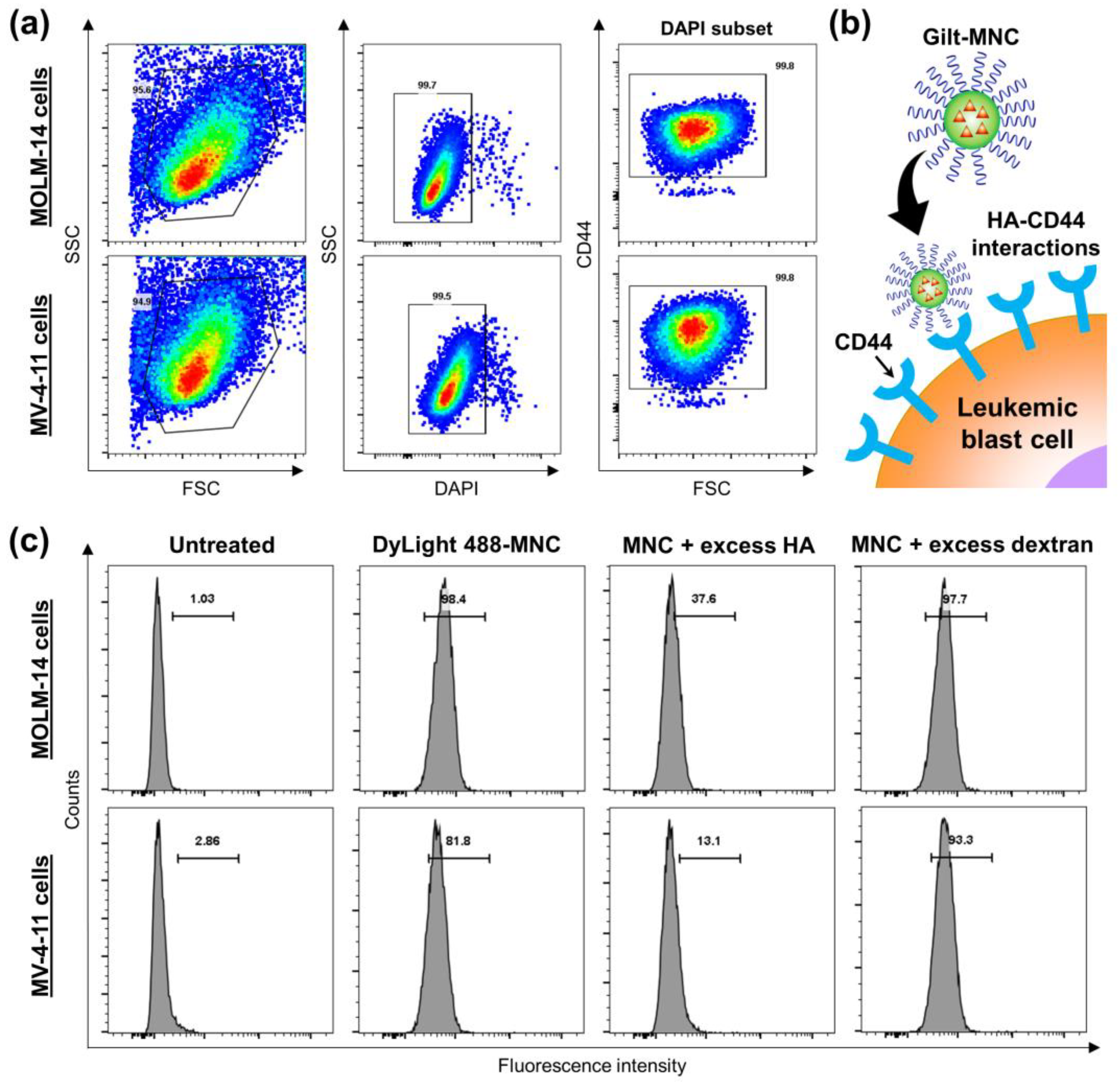
3.3. CD44-Targeting and Anti-Leukemic Effects of Gilt-MNC
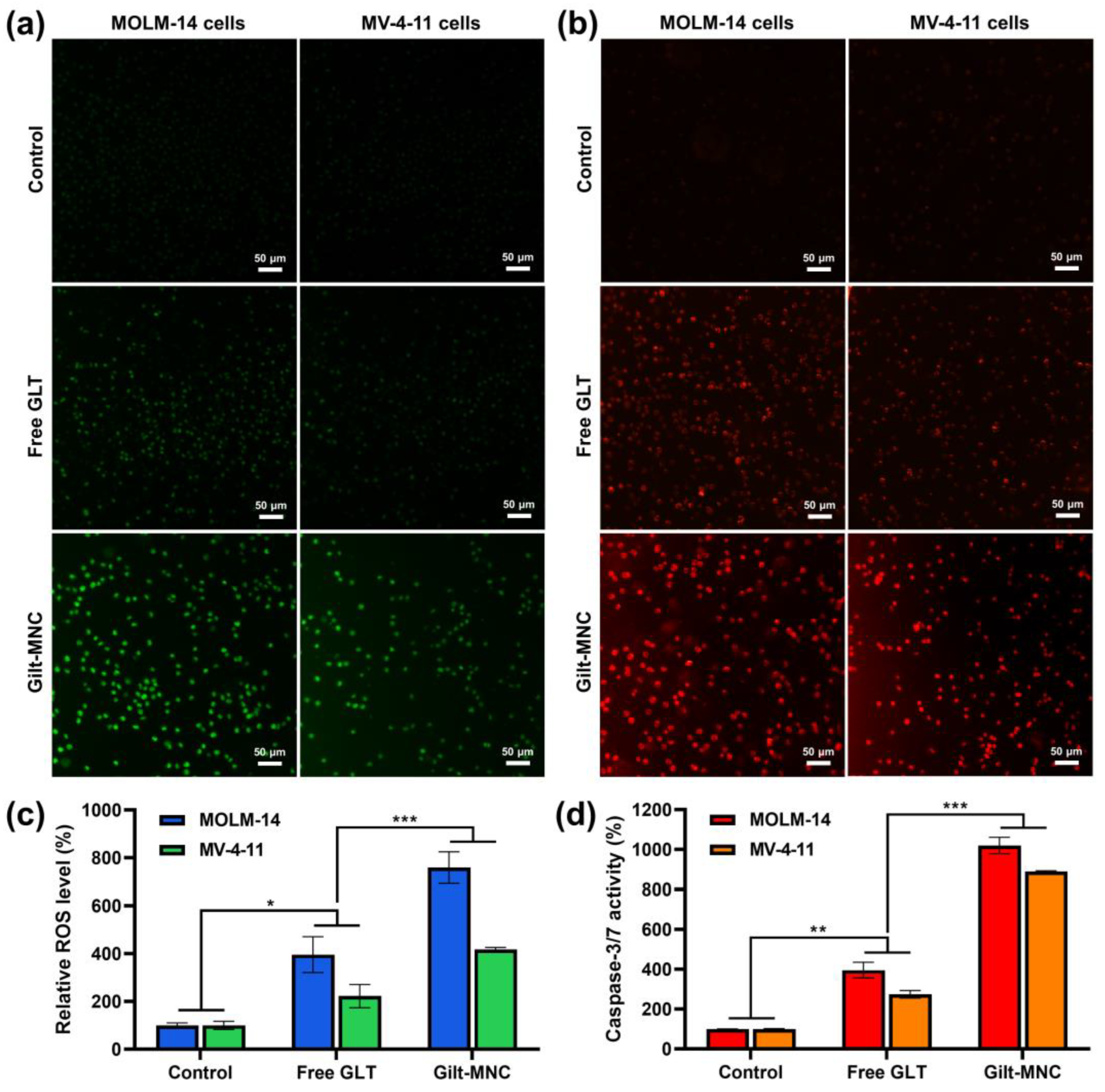
3.4. Evaluation of Cellular ROS Levels and Caspase-3/7 Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lai, C.; Doucette, K.; Norsworthy, K. Recent Drug Approvals for Acute Myeloid Leukemia. J. Hematol. Oncol. 2019, 12, 100. [Google Scholar] [CrossRef] [PubMed]
- Hassanein, M.; Almahayni, M.H.; Ahmed, S.O.; Gaballa, S.; El Fakih, R. FLT3 Inhibitors for Treating Acute Myeloid Leukemia. Clin. Lymphoma Myeloma Leuk. 2016, 16, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, P.; Wang, Y.; Shen, Y. Sitravatinib as a Potent FLT3 Inhibitor Can Overcome Gilteritinib Resistance in Acute Myeloid Leukemia. Biomark. Res. 2023, 11, 8. [Google Scholar] [CrossRef]
- Perl, A.E.; Martinelli, G.; Cortes, J.E.; Neubauer, A.; Berman, E.; Paolini, S.; Montesinos, P.; Baer, M.R.; Larson, R.A.; Ustun, C.; et al. Gilteritinib or Chemotherapy for Relapsed or Refractory FLT3 -Mutated AML. N. Engl. J. Med. 2019, 381, 1728–1740. [Google Scholar] [CrossRef] [PubMed]
- Molica, M.; Perrone, S.; Rossi, M. Gilteritinib: The Story of a Proceeding Success into Hard-to-Treat FLT3-Mutated AML Patients. J. Clin. Med. 2023, 12, 3647. [Google Scholar] [CrossRef]
- Zhang, S.; Dong, J.; Pan, R.; Xu, Z.; Li, M.; Zang, R. Structures, Properties, and Bioengineering Applications of Alginates and Hyaluronic Acid. Polymers 2023, 15, 2149. [Google Scholar] [CrossRef]
- Fakhari, A.; Berkland, C. Applications and Emerging Trends of Hyaluronic Acid in Tissue Engineering, as a Dermal Filler and in Osteoarthritis Treatment. Acta Biomater. 2013, 9, 7081–7092. [Google Scholar] [CrossRef]
- Lee, E.J.; Kang, E.; Kang, S.-W.; Huh, K.M. Thermo-Irreversible Glycol Chitosan/Hyaluronic Acid Blend Hydrogel for Injectable Tissue Engineering. Carbohydr. Polym. 2020, 244, 116432. [Google Scholar] [CrossRef]
- Tsanaktsidou, E.; Kammona, O.; Kiparissides, C. Recent Developments in Hyaluronic Acid-Based Hydrogels for Cartilage Tissue Engineering Applications. Polymers 2022, 14, 839. [Google Scholar] [CrossRef]
- Jin, L.; Hope, K.J.; Zhai, Q.; Smadja-Joffe, F.; Dick, J.E. Targeting of CD44 Eradicates Human Acute Myeloid Leukemic Stem Cells. Nat. Med. 2006, 12, 1167–1174. [Google Scholar] [CrossRef]
- Qiu, J.; Cheng, R.; Zhang, J.; Sun, H.; Deng, C.; Meng, F.; Zhong, Z. Glutathione-Sensitive Hyaluronic Acid-Mercaptopurine Prodrug Linked via Carbonyl Vinyl Sulfide: A Robust and CD44-Targeted Nanomedicine for Leukemia. Biomacromolecules 2017, 18, 3207–3214. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhou, J.-K.; Zhao, L.; Zheng, Z.-Y.; Li, J.; Pu, W.; Liu, S.; Liu, X.-S.; Liu, S.-J.; Zheng, Y.; et al. Novel Curcumin Liposome Modified with Hyaluronan Targeting CD44 Plays an Anti-Leukemic Role in Acute Myeloid Leukemia in Vitro and in Vivo. ACS Appl. Mater. Interfaces 2017, 9, 16857–16868. [Google Scholar] [CrossRef] [PubMed]
- Della Via, F.I.; Alvarez, M.C.; Basting, R.T.; Saad, S.T.O. The Effects of Green Tea Catechins in Hematological Malignancies. Pharmaceuticals 2023, 16, 1021. [Google Scholar] [CrossRef] [PubMed]
- Cornwall, S.; Cull, G.; Joske, D.; Ghassemifar, R. Green Tea Polyphenol “Epigallocatechin-3-Gallate”, Differentially Induces Apoptosis in CLL B-and T-Cells but Not in Healthy B-and T-Cells in a Dose Dependant Manner. Leuk. Res. 2016, 51, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.; Bae, K.H.; Nambu, A.; Dutta, B.; Chung, J.E.; Osato, M.; Kurisawa, M. A Two-Pronged Anti-Leukemic Agent Based on a Hyaluronic Acid-Green Tea Catechin Conjugate for Inducing Targeted Cell Death and Terminal Differentiation. Biomater. Sci. 2020, 8, 497–505. [Google Scholar] [CrossRef]
- Bae, K.H.; Lai, F.; Mong, J.; Niibori-Nambu, A.; Chan, K.H.; Her, Z.; Osato, M.; Tan, M.-H.; Chen, Q.; Kurisawa, M. Bone Marrow-Targetable Green Tea Catechin-Based Micellar Nanocomplex for Synergistic Therapy of Acute Myeloid Leukemia. J. Nanobiotechnol. 2022, 20, 481. [Google Scholar] [CrossRef]
- Nakazato, T.; Sagawa, M.; Yamato, K.; Xian, M.; Yamamoto, T.; Suematsu, M.; Ikeda, Y.; Kizaki, M. Myeloperoxidase Is a Key Regulator of Oxidative Stress-Mediated Apoptosis in Myeloid Leukemic Cells. Clin. Cancer Res. 2007, 13, 5436–5445. [Google Scholar] [CrossRef]
- Tyagi, T.; Treas, J.N.; Mahalingaiah, P.K.S.; Singh, K.P. Potentiation of Growth Inhibition and Epigenetic Modulation by Combination of Green Tea Polyphenol and 5-Aza-2′-Deoxycytidine in Human Breast Cancer Cells. Breast Cancer Res. Treat. 2015, 149, 655–668. [Google Scholar] [CrossRef]
- Yao, S.; Zhong, L.; Chen, M.; Zhao, Y.; Li, L.; Liu, L.; Xu, T.; Xiao, C.; Gan, L.; Shan, Z.; et al. Epigallocatechin-3-Gallate Promotes All-Trans Retinoic Acid-Induced Maturation of Acute Promyelocytic Leukemia Cells via PTEN. Int. J. Oncol. 2017, 51, 899–906. [Google Scholar] [CrossRef]
- Bae, K.H.; Tan, S.; Yamashita, A.; Ang, W.X.; Gao, S.J.; Wang, S.; Chung, J.E.; Kurisawa, M. Hyaluronic Acid-Green Tea Catechin Micellar Nanocomplexes: Fail-Safe Cisplatin Nanomedicine for the Treatment of Ovarian Cancer without off-Target Toxicity. Biomaterials 2017, 148, 41–53. [Google Scholar] [CrossRef]
- Topel, Ö.; Çakır, B.A.; Budama, L.; Hoda, N. Determination of Critical Micelle Concentration of Polybutadiene-Block-Poly(Ethyleneoxide) Diblock Copolymer by Fluorescence Spectroscopy and Dynamic Light Scattering. J. Mol. Liq. 2013, 177, 40–43. [Google Scholar] [CrossRef]
- Ganesh, S.; Iyer, A.K.; Morrissey, D.V.; Amiji, M.M. Hyaluronic Acid Based Self-Assembling Nanosystems for CD44 Target Mediated SiRNA Delivery to Solid Tumors. Biomaterials 2013, 34, 3489–3502. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C. Preclinical versus Clinical Drug Combination Studies. Leuk. Lymphoma 2008, 49, 2059–2080. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.-C.; Talalay, P. Quantitative Analysis of Dose-Effect Relationships: The Combined Effects of Multiple Drugs or Enzyme Inhibitors. Adv. Enzyme Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Bae, K.H.; Lai, F.; Oruc, B.; Osato, M.; Chen, Q.; Kurisawa, M. Self-Assembled Daunorubicin/Epigallocatechin Gallate Nanocomplex for Synergistic Reversal of Chemoresistance in Leukemia. Int. J. Mol. Sci. 2022, 24, 381. [Google Scholar] [CrossRef] [PubMed]
- Botten, D.; Fugallo, G.; Fraternali, F.; Molteni, C. Structural Properties of Green Tea Catechins. J. Phys. Chem. B 2015, 119, 12860–12867. [Google Scholar] [CrossRef]
- Liang, K.; Chung, J.E.; Gao, S.J.; Yongvongsoontorn, N.; Kurisawa, M. Highly Augmented Drug Loading and Stability of Micellar Nanocomplexes Composed of Doxorubicin and Poly(Ethylene Glycol)–Green Tea Catechin Conjugate for Cancer Therapy. Adv. Mater. 2018, 30, 1–8. [Google Scholar] [CrossRef]
- Shang, L.; Nienhaus, K.; Nienhaus, G.U. Engineered Nanoparticles Interacting with Cells: Size Matters. J. Nanobiotechnol. 2014, 12, 5. [Google Scholar] [CrossRef]
- Hoshyar, N.; Gray, S.; Han, H.; Bao, G. The Effect of Nanoparticle Size on in Vivo Pharmacokinetics and Cellular Interaction. Nanomedicine 2016, 11, 673–692. [Google Scholar] [CrossRef]
- Gu, L.; Faig, A.; Abdelhamid, D.; Uhrich, K. Sugar-Based Amphiphilic Polymers for Biomedical Applications: From Nanocarriers to Therapeutics. Acc. Chem. Res. 2014, 47, 2867–2877. [Google Scholar] [CrossRef]
- Alves, A.C.; Ribeiro, D.; Nunes, C.; Reis, S. Biophysics in Cancer: The Relevance of Drug-Membrane Interaction Studies. Biochim. Biophys. Acta-Biomembr. 2016, 1858, 2231–2244. [Google Scholar] [CrossRef] [PubMed]
- Mukerjee, S.; Saeedan, A.S.; Ansari, M.N.; Singh, M. Polyunsaturated Fatty Acids Mediated Regulation of Membrane Biochemistry and Tumor Cell Membrane Integrity. Membranes 2021, 11, 479. [Google Scholar] [CrossRef] [PubMed]
- Zavorka Thomas, M.E.; Lu, X.; Talebi, Z.; Jeon, J.Y.; Buelow, D.R.; Gibson, A.A.; Uddin, M.E.; Brinton, L.T.; Nguyen, J.; Collins, M.; et al. Gilteritinib Inhibits Glutamine Uptake and Utilization in FLT3-ITD–Positive AML. Mol. Cancer Ther. 2021, 20, 2207–2217. [Google Scholar] [CrossRef] [PubMed]
- Earnshaw, W.C.; Martins, L.M.; Kaufmann, S.H. Mammalian Caspases: Structure, Activation, Substrates, and Functions During Apoptosis. Ann. Rev. Biochem. 1999, 68, 383–424. [Google Scholar] [CrossRef]
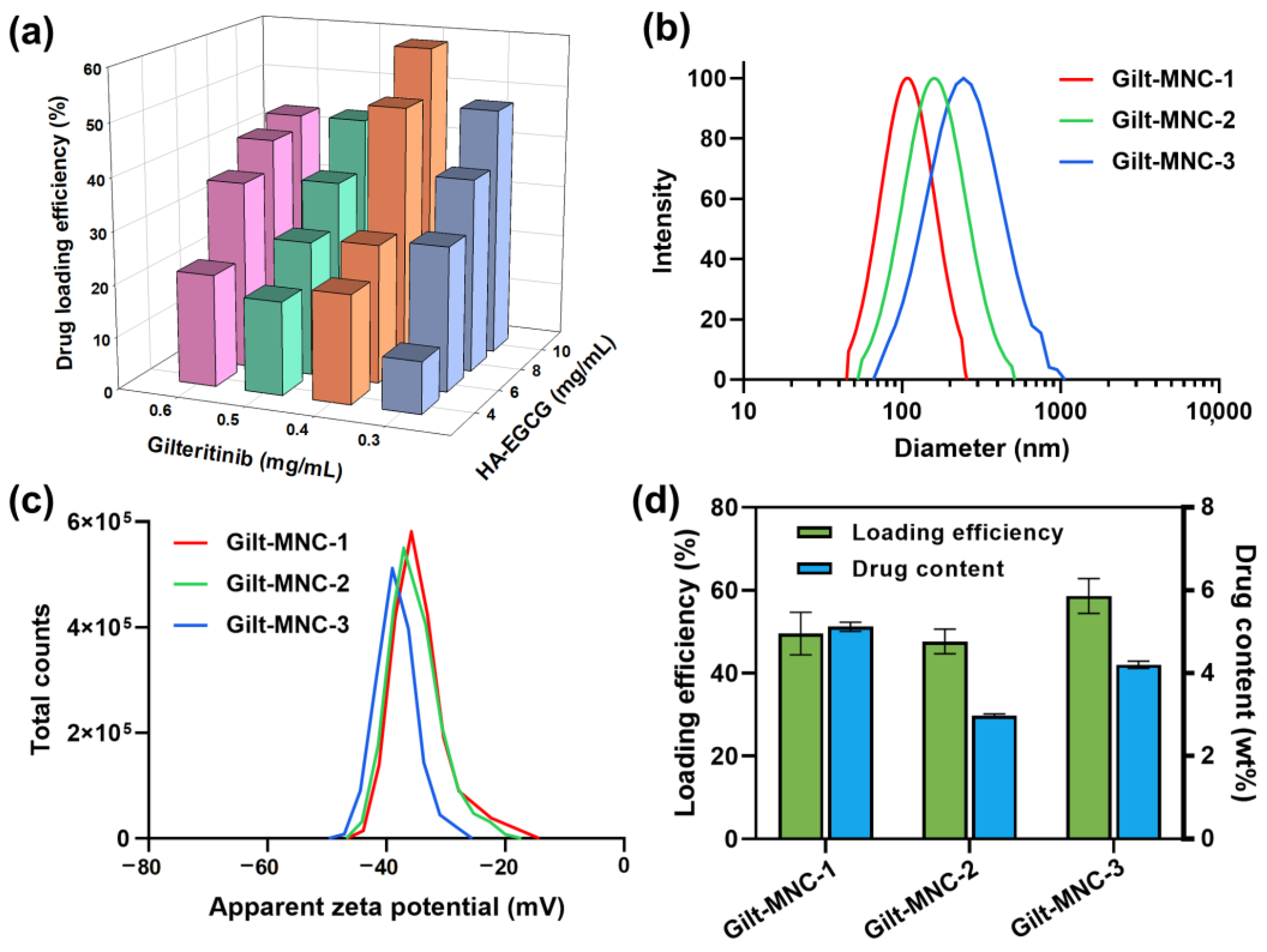


| Sample Code | Final Feeding Concentration of HA-EGCG (mg/mL) | Final Feeding Concentration of GLT (mg/mL) | Effective Diameter (nm) | Zeta Potential (mV) |
|---|---|---|---|---|
| Gilt-MNC-1 | 8 | 0.4 | 110.8 ± 4.0 | −34.1 ± 5.2 |
| Gilt-MNC-2 | 10 | 0.3 | 148.6 ± 7.9 | −35.2 ± 4.3 |
| Gilt-MNC-3 | 10 | 0.4 | 176.1 ± 14.4 | −38.2 ± 3.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bae, K.H.; Lai, F.; Chen, Q.; Kurisawa, M. Potentiating Gilteritinib Efficacy Using Nanocomplexation with a Hyaluronic Acid–Epigallocatechin Gallate Conjugate. Polymers 2024, 16, 225. https://doi.org/10.3390/polym16020225
Bae KH, Lai F, Chen Q, Kurisawa M. Potentiating Gilteritinib Efficacy Using Nanocomplexation with a Hyaluronic Acid–Epigallocatechin Gallate Conjugate. Polymers. 2024; 16(2):225. https://doi.org/10.3390/polym16020225
Chicago/Turabian StyleBae, Ki Hyun, Fritz Lai, Qingfeng Chen, and Motoichi Kurisawa. 2024. "Potentiating Gilteritinib Efficacy Using Nanocomplexation with a Hyaluronic Acid–Epigallocatechin Gallate Conjugate" Polymers 16, no. 2: 225. https://doi.org/10.3390/polym16020225





