Deep Learning for Cell Migration in Nonwoven Materials and Evaluating Gene Transfer Effects following AAV6-ND4 Transduction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Electrospun Technique
2.2. Scanning Electron Microscopy
2.3. Dynamic Mechanical Analysis
2.4. Laser Scanning Confocal Microscopy
2.5. AAV Production
2.6. Fibroblast Transduction by AAV
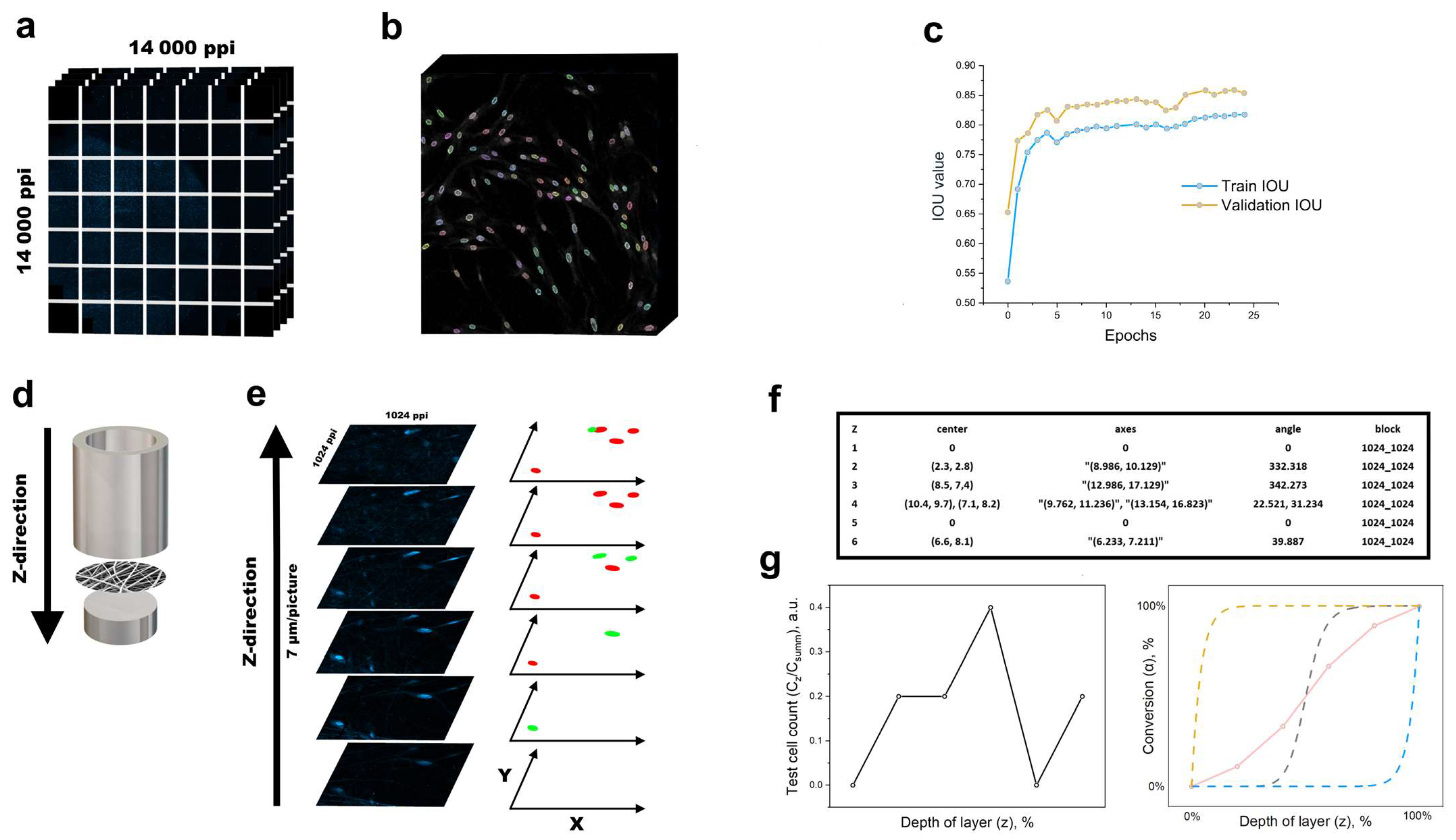
2.7. Convolutional Neural Network (CNN)
3. Results and Discussion
3.1. Mechanical Properties of Microenvironment
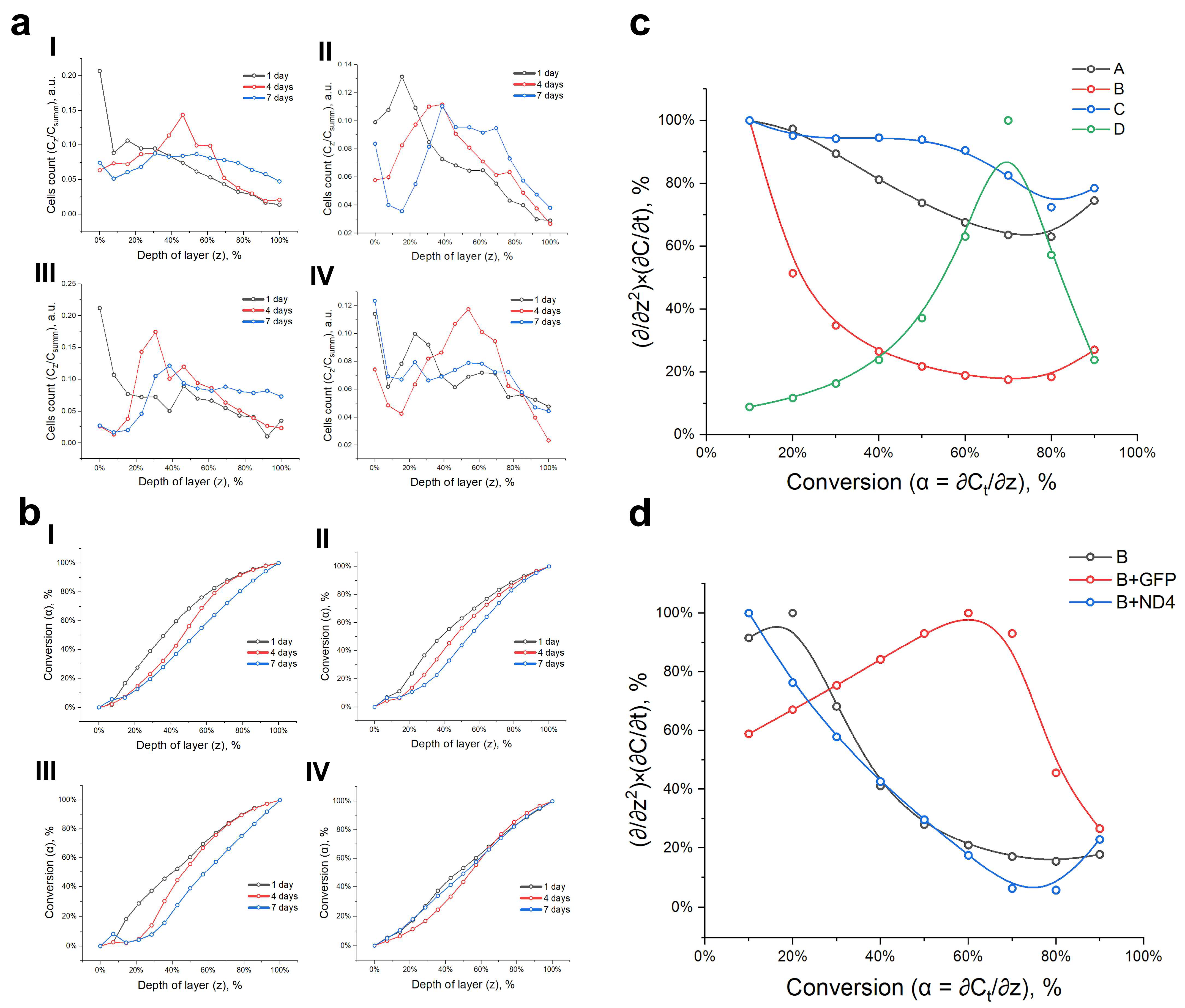
3.2. Proliferation and Changes in Nuclear Morphology during Cell Migration and Homing
3.3. Model of Cell Migration on Three-Dimensional Systems
4. Conclusions
4.1. Fibroblast Mechanobiology
4.2. Conclusion and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stegemann, J.P.; Wagner, W.R.; Göbel, U.; Perets, E.; Leong, K.W.; Leach, J.K.; Gilbert, J. The Year 2022 in biomaterials research: A perspective from the editors of six leading journals. J. Biomed. Mater. Res. Part A 2023, 111, 1298–1308. [Google Scholar] [CrossRef] [PubMed]
- Stupin, V.A.; Gabitov, R.B.; Sinelnikova, T.G.; Silina, E.V. Biological mechanisms of chronic wound and diabetic foot healing: The role of collagen. Serbian J. Exp. Clin. Res. 2018, 19, 373–382. [Google Scholar] [CrossRef]
- Pina, S.; Oliveira, J.M.; Reis, R.L. Natural-based nanocomposites for bone tissue engineering and regenerative medicine: A review. Adv. Mater. 2015, 27, 1143–1169. [Google Scholar] [CrossRef] [PubMed]
- Diller, R.B.; Tabor, A.J. The role of the extracellular matrix (ECM) in wound healing: A review. Biomimetics 2022, 7, 87. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, S.; Stojadinovic, O.; Golinko, M.S.; Brem, H.; Tomic-Canic, M. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008, 16, 585–601. [Google Scholar] [CrossRef]
- Przekora, A. Current trends in fabrication of biomaterials for bone and cartilage regeneration: Materials modifications and biophysical stimulations. Int. J. Mol. Sci. 2019, 20, 435. [Google Scholar] [CrossRef]
- Manea, L.R.; Hristian, L.; Leon, A.L.; Popa, A. Recent advances of basic materials to obtain electrospun polymeric nanofibers for medical applications. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2016; Volume 145, p. 032006. [Google Scholar] [CrossRef]
- Bogdanova, A.S.; Sokolova, A.I.; Pavlova, E.R.; Klinov, D.V.; Bagrov, D.V. Investigation of cellular morphology and proliferation on patterned electrospun PLA-gelatin mats. J. Biol. Phys. 2021, 47, 205–214. [Google Scholar] [CrossRef]
- Liu, S.; Yu, J.-M.; Gan, Y.-C.; Qiu, X.-Z.; Gao, Z.-C.; Wang, H.; Chen, S.-X.; Xiong, Y.; Liu, G.-H.; Lin, S.-E.; et al. Biomimetic natural biomaterials for tissue engineering and regenerative medicine: New biosynthesis methods, recent advances, and emerging applications. Mil. Med. Res. 2023, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Z.; Huang, C.H.; Zhang, X.Y.; Shen, G.L. The clinical application effects of artificial dermis scaffold and autologous split-thickness skin composite grafts combined with vacuum-assisted closure in refractory wounds. Int. Wound J. 2023, 20, 2113–2120. [Google Scholar] [CrossRef]
- Žunar, B.; Ito, T.; Mosrin, C.; Sugahara, Y.; Bénédetti, H.; Guégan, R.; Vallée, B. Confocal imaging of biomarkers at a single-cell resolution: Quantifying ‘living’ in 3D-printable engineered living material based on Pluronic F-127 and yeast Saccharomyces cerevisiae. Biomater. Res. 2022, 26, 85. [Google Scholar] [CrossRef]
- Man, P.Y.W.; Turnbull, D.M.; Chinnery, P.F. Leber hereditary optic neuropathy. J. Med. Genet. 2002, 39, 162–169. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, Y.-R.; Wang, A.-G.; Chen, Y.-T.; Yarmishyn, A.A.; Buddhakosai, W.; Yang, T.-C.; Hwang, D.-K.; Yang, Y.-P.; Shen, C.-N.; Lee, H.-C.; et al. Bioactivity and gene expression profiles of hiPSC-generated retinal ganglion cells in MT-ND4 mutated Leber’s hereditary optic neuropathy. Exp. Cell Res. 2018, 363, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Omura, T. Mitochondria-targeting sequence, a multi-role sorting sequence recognized at all steps of protein import into mitochondria. J. Biochem. 1998, 123, 1010–1016. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, C.; Augustin, S.; Ellouze, S.; Bénit, P.; Bouaita, A.; Rustin, P.; Sahel, J.-A.; Corral-Debrinski, M. The optimized allotopic expression of ND1 or ND4 genes restores respiratory chain complex I activity in fibroblasts harboring mutations in these genes. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2008, 1783, 1707–1717. [Google Scholar] [CrossRef] [PubMed]
- Howard, D.B.; Powers, K.; Wang, Y.; Harvey, B.K. Tropism and toxicity of adeno-associated viral vector serotypes 1, 2, 5, 6, 7, 8, and 9 in rat neurons and glia in vitro. Virology 2008, 372, 24–34. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Cui, W.; Li, X.; Jin, Y. Electrospun fibrous mats with high porosity as potential scaffolds for skin tissue engineering. Biomacromolecules 2008, 9, 1795–1801. [Google Scholar] [CrossRef] [PubMed]
- ASTM D638; Standard Test Method for Tensile Properties of Plastics. ASTM International: West Conshohocken, PA, USA, 2022.
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Part III 18; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Data Science Bowl. 2018. Available online: https://www.kaggle.com/c/data-science-bowl-2018 (accessed on 21 December 2021).
- Mumuni, A.; Mumuni, F. Data augmentation: A comprehensive survey of modern approaches. Array 2022, 16, 100258. [Google Scholar] [CrossRef]
- Joodaki, H.; Panzer, M.B. Skin mechanical properties and modeling: A review. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2018, 232, 323–343. [Google Scholar] [CrossRef]
- Carter, M.; Shieh, J. Cell culture techniques. In Guide to Research Techniques in Neuroscience; Elsevier: Amsterdam, The Netherlands, 2015; pp. 295–310. [Google Scholar]
- Bambole, V.; Yakhmi, J.V. Tissue engineering: Use of electrospinning technique for recreating physiological functions. In Nanobiomaterials in Soft Tissue Engineering; William Andrew Publishing: Norwich, NY, USA, 2016; pp. 387–455. [Google Scholar]
- Ellis, B.L.; Hirsch, M.L.; Barker, J.C.; Connelly, J.P.; Steininger, R.J.; Porteus, M.H. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virol. J. 2013, 10, 74. [Google Scholar] [CrossRef]
- Ansari, A.M.; Ahmed, A.K.; Matsangos, A.E.; Lay, F.; Born, L.J.; Marti, G.; Harmon, J.W.; Sun, Z. Cellular GFP toxicity and immunogenicity: Potential confounders in in vivo cell tracking experiments. Stem Cell Rev. Rep. 2016, 12, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Mamaghani, P.; Donald, A.M. The impact of fibronection stripe patterns on the cellular and nuclear morphology of fibroblasts. bioRxiv 2018. [Google Scholar] [CrossRef]
- Hanson, L.; Zhao, W.; Lou, H.-Y.; Lin, Z.C.; Lee, S.W.; Chowdary, P.; Cui, Y.; Cui, B. Vertical nanopillars for in situ probing of nuclear mechanics in adherent cells. Nat. Nanotechnol. 2015, 10, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Kulangara, K.; Adler, A.F.; Wang, H.; Chellappan, M.; Hammett, E.; Yasuda, R.; Leong, K.W. The effect of substrate topography on direct reprogramming of fibroblasts to induced neurons. Biomaterials 2014, 35, 5327–5336. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Ricotta, V.; Simon, M.; Clark, R.A.F.; Rafailovich, M.H. Continual cell deformation induced via attachment to oriented fibers enhances fibroblast cell migration. PLoS ONE 2015, 10, e0119094. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Sun, S.X. Cellular pressure and volume regulation and implications for cell mechanics. Biophys. J. 2013, 105, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.T.; Horwitz, A.R.; Schwartz, M.A. Cell adhesion: Integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 2010, 11, 633–643. [Google Scholar] [CrossRef]
- Kim, D.-H.; Li, B.; Si, F.; Philips, J.; Wirtz, D.; Sun, S.X. Volume regulation and shape bifurcation in the cell nucleus. J. Cell Sci. 2015, 128, 3375–3385. [Google Scholar] [CrossRef] [PubMed]
- Luthria, G.; Li, R.; Wang, S.; Prytyskach, M.; Kohler, R.H.; Lauffenburger, D.A.; Mitchison, T.J.; Weissleder, R.; Miller, M.A. In vivo microscopy reveals macrophage polarization locally promotes coherent microtubule dynamics in migrating cancer cells. Nat. Commun. 2020, 11, 3521. [Google Scholar] [CrossRef]
- Stringer, C.; Wang, T.; Michaelos, M.; Pachitariu, M. Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods 2021, 18, 100–106. [Google Scholar] [CrossRef]
- Borchardt, H.J.; Daniels, F. Differential thermal analysis of inorganic hydrates. J. Phys. Chem. 1957, 61, 917–921. [Google Scholar] [CrossRef]
- Ozawa, T.J. Calorimetric Measurements of Grain growth in Ultrafine-Grained nichel. Therm. Anal. 1970, 2, 301. [Google Scholar] [CrossRef]
- Wang, T.; Chen, Y.; Dong, W.; Liu, Y.; Shi, L.; Chen, R.; Pan, T. Fractal Characteristics of Porosity of Electrospun Nanofiber Membranes. Math. Probl. Eng. 2020, 2020, 2503154. [Google Scholar] [CrossRef]
- Chen, Y.; Qi, L.; Liu, Y.; Zhang, L.; Zhu, R.; Chen, R. Study of fractal dimension of electrospun nanofibres non-woven fabrics and the relationship between it and air resistance. J. Innov. Technol. Educ. 2015, 2, 15–28. [Google Scholar] [CrossRef]
- Wang, T.; Dong, W.; Chen, Y.; Pan, T.; Chen, R. Study on porosity of electrospun nanofiber membrane by neural network. Appl. Math. Sci. 2018, 12, 1059–1074. [Google Scholar] [CrossRef]
- Hong, Z.-X.; Zhu, S.-T.; Li, H.; Luo, J.-Z.; Yang, Y.; An, Y.; Wang, X.; Wang, K. Bioengineered skin organoids: From development to applications. Mil. Med. Res. 2023, 10, 40. [Google Scholar] [CrossRef]
- Veron, P.; Allo, V.; Rivière, C.; Bernard, J.; Douar, A.-M.; Masurier, C. Major subsets of human dendritic cells are efficiently transduced by self-complementary adeno-associated virus vectors 1 and 2. J. Virol. 2007, 81, 5385–5394. [Google Scholar] [CrossRef]
- Zhang, Y.; Chirmule, N.; Gao, G.-P.; Wilson, J. CD40 ligand-dependent activation of cytotoxic T lymphocytes by adeno-associated virus vectors in vivo: Role of immature dendritic cells. J. Virol. 2000, 74, 8003–8010. [Google Scholar] [CrossRef]



| σbreak | σσ | εbreak | σε | |
|---|---|---|---|---|
| 1 h | 1.75 | 0.23 | 2.81 | 0.68 |
| 2 h | 1.39 | 0.25 | 2.86 | 0.22 |
| 4 h | 1.14 | 0.24 | 2.03 | 0.41 |
| 6 h | 0.83 | 0.12 | 1.67 | 0.40 |
| 24 h | 1.04 | 0.07 | 2.27 | 0.20 |
| Control + UV | 1.03 | 0.43 | 2.54 | 1.13 |
| Control | 1.18 | 0.40 | 10.17 | 1.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larin, I.I.; Shatalova, R.O.; Laktyushkin, V.S.; Rybtsov, S.A.; Lapshin, E.V.; Shevyrev, D.V.; Karabelsky, A.V.; Moskalets, A.P.; Klinov, D.V.; Ivanov, D.A. Deep Learning for Cell Migration in Nonwoven Materials and Evaluating Gene Transfer Effects following AAV6-ND4 Transduction. Polymers 2024, 16, 1187. https://doi.org/10.3390/polym16091187
Larin II, Shatalova RO, Laktyushkin VS, Rybtsov SA, Lapshin EV, Shevyrev DV, Karabelsky AV, Moskalets AP, Klinov DV, Ivanov DA. Deep Learning for Cell Migration in Nonwoven Materials and Evaluating Gene Transfer Effects following AAV6-ND4 Transduction. Polymers. 2024; 16(9):1187. https://doi.org/10.3390/polym16091187
Chicago/Turabian StyleLarin, Ilya I., Rimma O. Shatalova, Victor S. Laktyushkin, Stanislav A. Rybtsov, Evgeniy V. Lapshin, Daniil V. Shevyrev, Alexander V. Karabelsky, Alexander P. Moskalets, Dmitry V. Klinov, and Dimitry A. Ivanov. 2024. "Deep Learning for Cell Migration in Nonwoven Materials and Evaluating Gene Transfer Effects following AAV6-ND4 Transduction" Polymers 16, no. 9: 1187. https://doi.org/10.3390/polym16091187







