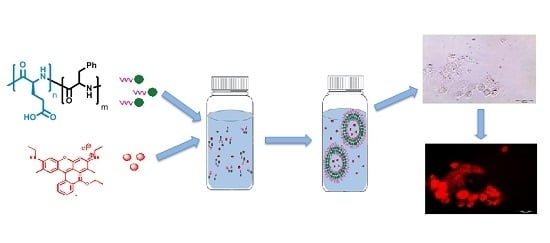
Preparation, Characterization, and Biological Evaluation of Poly(Glutamic Acid)-b-Polyphenylalanine Polymersomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instrumentation
2.3. Synthesis and Particles Preparation
2.4. Biodegradation study
2.5. Surface Modification
2.6. Encapsulation of Dyes
2.7. Cell Experiments
3. Results and Discussion
3.1. Synthesis of Poly(Amino Acid) Block-Copolymers
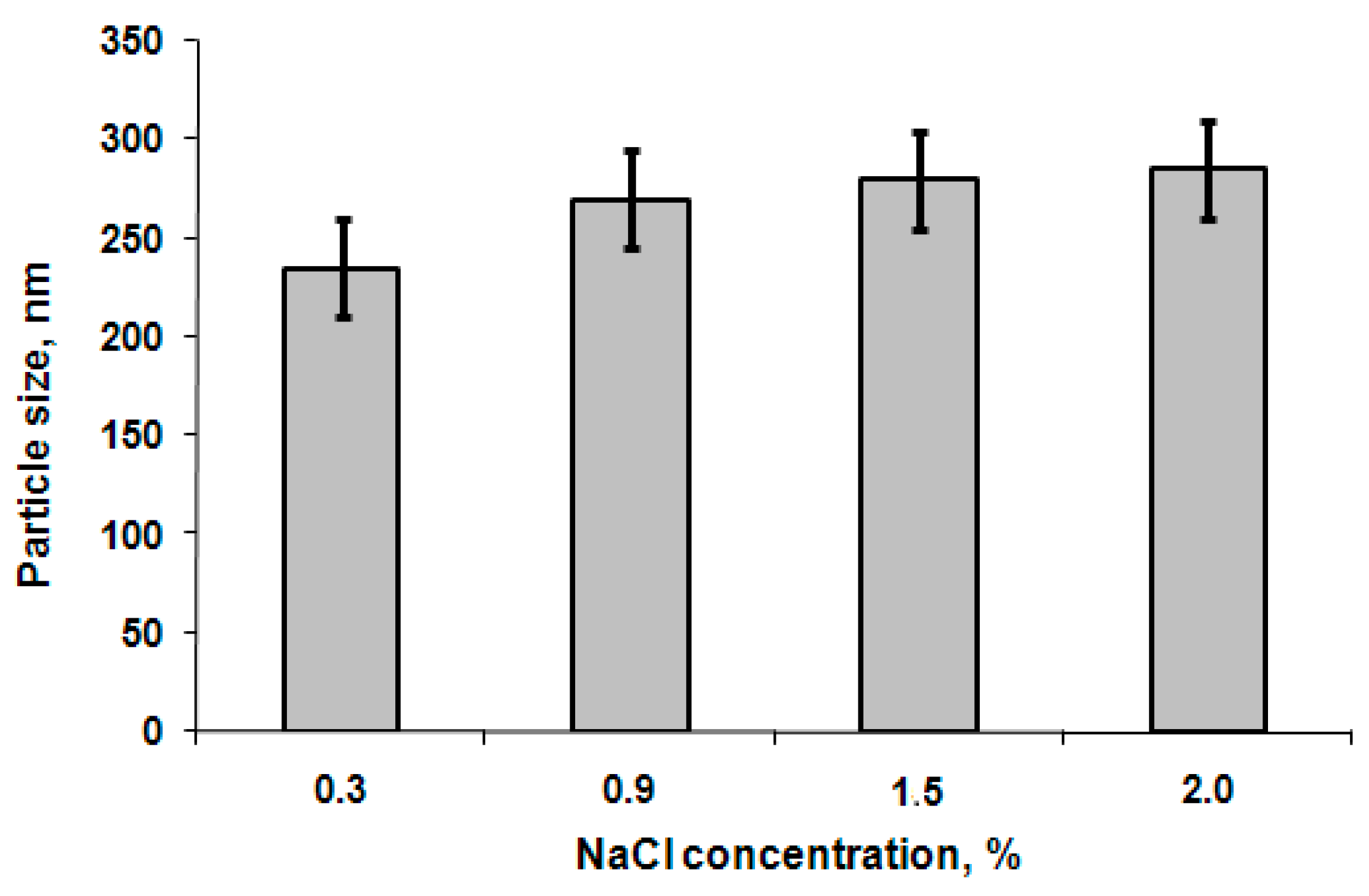
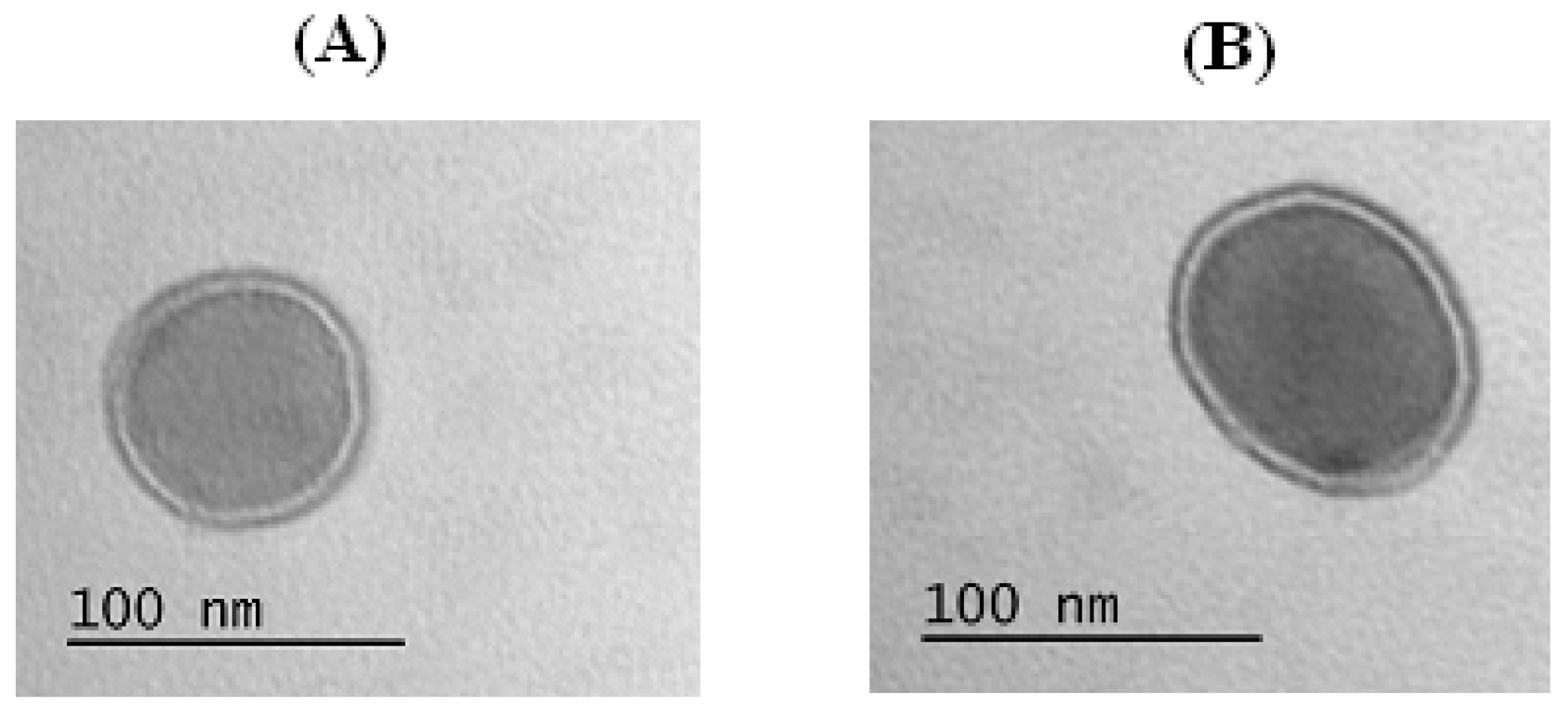
3.2. Characterization of Particles
3.3. Biodegradation
3.4. Surface Modification
3.5. Encapsulation of Model Compounds and Cell Experiments
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GPC | Gel permeation chromatography |
| TEM | Transmission electron microscopy |
| NCA | N-carboxyanhydrides |
| ROP | Ring-opening polymerization |
| HEXA | Hexylamine |
| DMF | Dimethylformamide |
| DMSO | Dimethyl sulfoxide |
| TFMSA | Trifluoromethane sulfonic acid |
| TFA | Trifluoroacetic acid |
| NHS | N-hydroxysuccinimide |
| CDI | 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| NMR | Nuclear magnetic resonance |
| DLS | Dynamic light scattering |
| HPLC | High performance liquid chromatography |
| HEK cells | Human embryonic kidney cells |
| Caco2 cells | Human epithelial colorectal adenocarcinoma cells |
References
- Christian, D.; Cai, S.; Bowen, D.M.; Kim, Y.; Pajerowski, J.D.; Discher, D.E. Polymersome carriers: From self-assembly to siRNA and protein therapeutics. Eur. J. Pharm. Biopharm. 2009, 71, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.J.; Kitchen, C.; Adams, S.; Furzeland, S.; Atkins, D.; Schuetz, P.; Fernyhough, C.M.; Tzokova, N.; Ryan, A.J.; Butler, M.F. On the mechanism of formation of vesicles from poly(ethylene oxide)-block-poly(caprolactone) copolymers. Soft Matter. 2009, 5, 3086–3096. [Google Scholar] [CrossRef]
- Carlsen, A.; Lecommandoux, S. Self-assembly of polypeptide-based block copolymer amphiphiles. Curr. Opin. Colloid Interface Sci. 2009, 14, 329–339. [Google Scholar] [CrossRef]
- Ding, J.; Xu, W.; Zhang, Y.; Sun, D.; Xiao, C.; Liu, D.; Zhu, X.; Chen, X. Self-reinforced endocytoses of smart polypeptide nanogels for “on-demand” drug delivery. J. Control. Release 2013, 172, 444–455. [Google Scholar] [CrossRef] [PubMed]
- Discher, B.M.; Won, Y.Y.; Ege, D.S.; Lee, J.C.; Bates, F.S.; Disher, D.E.; Hammer, D.A. Polymersomes: Tough vesicles made from diblock copolymers. Science 1999, 284, 1143–1146. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Feijen, J. Polymersomes for drug delivery: Design, formation and characterization. J. Control. Release 2012, 161, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Engbers, G.H.M.; Feijen, J. Biodegradable polymersomes as a basis for artificial cells: Encapsulation, release and targeting. J. Control. Release 2005, 101, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Hubina, A.V.; Pogodaev, A.A.; Sharoyko, V.V.; Vlakh, E.G.; Tennikova, T.B. Self-assembled spin-labeled nanoparticles based on poly(amino acids). React. Funct. Polym. 2016, 100, 173–180. [Google Scholar] [CrossRef]
- LoPresti, C.; Lomas, H.; Massignani, M.; Smart, T.; Battaglia, G. Polymersomes: Nature inspired nanometer sized compartments. J. Mater. Chem. 2009, 19, 3576–3590. [Google Scholar] [CrossRef]
- Nardin, C.; Hirt, T.; Leukel, J.; Meier, W. Polymerized ABA triblock copolymer vesicles. Langmuir 2000, 16, 1035–1041. [Google Scholar] [CrossRef]
- Bermudez, H.; Brannan, A.K.; Hammer, D.; Bates, F.S.; Discher, D.E. Molecular weight dependence of polymersome membrane structure, elasticity, and stability. Macromolecules 2002, 35, 8203–8208. [Google Scholar] [CrossRef]
- Meng, F.; Zhong, Z.; Feijen, J. Stimuli-responsive polymersomes for programmed drug delivery. Biomacromolecules 2009, 10, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Klok, H.A.; Lecommandoux, S. Supramolecular materials via block copolymer self-assembly. Adv. Mater. 2001, 13, 1217–1229. [Google Scholar] [CrossRef]
- Deng, J.; Gao, N.; Wang, Y.; Yi, H.; Fang, S.; Ma, Y.; Cai, L. Self-assembled cationic micelles based on PEG-PLL-PLLeu hybrid polypeptides as highly effective gene vectors. Biomacromolecules 2012, 13, 3795–3804. [Google Scholar] [CrossRef] [PubMed]
- Ghoroghchian, P.P.; Li, G.; Levine, D.H.; Davis, K.P.; Bates, F.S.; Hammer, D.A.; Therien, M.J. Bioresorbable vesicles formed through spontaneous self-assembly of amphiphilic poly(ethylene oxide)-block-polycaprolactone. Macromolecules 2006, 39, 1673–1675. [Google Scholar] [CrossRef] [PubMed]
- Checot, F.; Lecommandoux, S.; Klok, H.A.; Gnanou, Y. From supramolecular polymersomes to stimuli-responsive nano-capsules based on poly(diene-b-peptide) diblock copolymers. Eur. Phys. J. E 2003, 10, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Liu, M. Self-assembly and morphology control of new l-glutamic acid-based amphiphilic random copolymers: Giant vesicles, vesicles, spheres, and honeycomb Film. Langmuir 2011, 27, 12844–12850. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, N.; Wang, K.; Shi, C.; Zhang, L.; Luan, Y. A review of polypeptide-based polymersomes. Biomaterials 2014, 35, 1284–1301. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Huang, Y.; Shi, Q.; Chen, X.; Jing, X. Oxygen carrier based on hemoglobin/poly(l-lysine)-block-poly(l-phenylalanine) vesicles. Langmuir 2009, 25, 13726–13729. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhu, J.; Chen, T.; Lin, S.; Cai, C.; Zhang, L.; Zhuang, Y.; Wang, X.S. Drug releasing behavior of hybrid micelles containing polypeptide triblock copolymer. Biomaterials 2009, 30, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Amblard, M.; Fehrentz, J.; Martinez, J.; Subra, G. Methods and protocols of modern solid phase peptide synthesis. Molec. Biotech. 2006, 33, 239–254. [Google Scholar] [CrossRef]
- Kricheldorf, H.R. Polypeptides and 100 years of chemistry of a-amino acid N-carboxyanhydrides. Angew. Chem. Int. Ed. 2006, 45, 5752–5784. [Google Scholar] [CrossRef] [PubMed]
- Deming, T.J. Synthetic polypeptides for biomedical applications. Prog. Polym. Sci. 2007, 32, 858–875. [Google Scholar] [CrossRef]
- Kim, M.S.; Dayananda, K.; Choi, E.K.; Park, H.J.; Kim, J.S.; Lee, D.S. Synthesis and characterization of poly(l-glutamic acid)-block-poly(l-phenylalanine). Polymer 2009, 50, 2252–2257. [Google Scholar] [CrossRef]
- Holowka, E.P.; Sun, V.Z.; Kamei, D.T.; Deming, T.J. Polyarginine segments in block copolypeptides drive both vesicular assembly and intracellular delivery. Nat. Mater. 2007, 6, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Yang, Y.S.; Lai, T.Y.; Jan, J.S. Lysine-block-tyrosine block copolypeptides: Self-assembly, cross-linking, and conjugation of targeted ligand for drug encapsulation. Polymer 2012, 53, 913–922. [Google Scholar] [CrossRef]
- Lee, B.S.; Yip, A.T.; Thach, A.V.; Rodriguez, A.R.; Deming, T.J.; Kamei, D.T. The targeted delivery of doxorubicin with transferrin-conjugated block copolypeptide vesicles. Int. J. Pharm. 2015, 496, 903–911. [Google Scholar]
- Yang, C.-Y.; Song, B.; Ao, Y.; Nowak, A.P.; Abelowitz, R.B.; Korsak, R.A.; Havton, L.A.; Deming, T.J.; Sofroniew, M.V. Biocompatibility of amphiphilic diblock copolypeptide hydrogels in the central nervous system. Biomaterials 2009, 30, 2881–2898. [Google Scholar] [CrossRef] [PubMed]
- Birke, A.; Huesmann, D.; Kelsch, A.; Weilbächer, M.; Xie, J.; Bros, M.; Bopp, T.; Becker, C.; Landfester, K.; Barz, M. Polypeptoid-block-polypeptide copolymers: Synthesis, characterization, and application of amphiphilic block copolypept(o)ides in drug formulations and miniemulsion techniques. Biomacromolecules 2014, 15, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Wilder, R.; Mobashery, S. The use of triphosgene in the preparation of N-carboxy-α-amino acid anhydrides. J. Org. Chem. 1992, 57, 2755–2756. [Google Scholar] [CrossRef]
- Randall, R.J.; Lewis, A. The folin by oliver. Readings 1951, 193, 265–275. [Google Scholar]
- Benčina, M.; Babič, J.; Podgornik, A. Preparation and characterisation of ribonuclease monolithic bioreactor. J. Chromatogr. A 2007, 1144, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Turrado, C.; Puig, T.; García-Carceles, J.; Artola, V.; Benhamu, B.; Ortega-Gutierrez, S.; Relat, J.; Oliveras, G.; Blancafort, A.; Haro, D.; et al. New synthetic inhibitors of fatty acid synthase with anticancer Activity. J. Med. Chem. 2012, 55, 5013–5023. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.P.; Chen, Y.Y.; Hsieh, M.F. Biofabrication and in vitro study of hydroxyapatite/mPEG-PCL-mPEG scaffolds for bone tissue engineering using air pressure-aided deposition technology. Mater. Sci. Eng. C 2013, 33, 680–690. [Google Scholar] [CrossRef] [PubMed]
- Spaeth, J.R.; Kevrekidis, I.G.; Panagiotopoulos, A.Z. Dissipative particle dynamics simulations of polymer-protected nanoparticle self-assembly dissipative particle dynamics simulations of polymer-protected nanoparticle self-assembly. J. Chem. Phys. 2011, 135, 184903. [Google Scholar] [CrossRef] [PubMed]
- Chécot, F.; Brûlet, A.; Oberdisse, J.; Gnanou, Y.; Mondain-Monval, O.; Lecommandoux, S. Structure of polypeptide-based diblock copolymers in solution: Stimuli-responsive vesicles and micelles. Langmuir 2005, 21, 4308–4315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prokop, A.; Iwasaki, Y.; Harada, A. (Eds.) Intracellular delivery II: Fundamentals and Applications; Springer: Heidelberg, Germany, 2014; p. 479.
- Rypaeek, F.; Pytela, J.; Kotva, R.; Skarda, V. Biodegradation of poly(amino acid)s: Evaluation methods and structure to funaction relationships. Macromol. Symp. 1997, 123, 9–24. [Google Scholar] [CrossRef]
- Farkhani, S.M.; Valizadeh, A.; Karami, H.; Mohammadi, S.; Sohrabi, N.; Badrzadeh, F. Cell penetrating peptides: Efficient vectors for delivery of nanoparticles, nanocarriers, therapeutic and diagnostic molecules. Peptides 2014, 57, 78–94. [Google Scholar] [CrossRef] [PubMed]
- Ponomareva, E.A.; Kartuzova, V.E.; Vlakh, E.G.; Tennikova, T.B. Monolithic bioreactors: effect of chymotrypsin immobilization on its biocatalytic properties. J. Chromatogr. B 2010, 878, 567–574. [Google Scholar] [CrossRef] [PubMed]









| Sample | Mn | Mw | Mw/Mn | n |
|---|---|---|---|---|
| BG1 | 13,800 | 16,600 | 1.2 | 62 |
| BG2 | 25,800 | 36,100 | 1.4 | 117 |
| Sample | Mn (PGlu) | n | Mn (PPhe) | m | Hydrophilic block content, % |
|---|---|---|---|---|---|
| GP1 | 8,000 | 62 | 4,600 | 31 | 64 |
| GP2 | 8,000 | 62 | 12,100 | 82 | 40 |
| GP3 | 15,100 | 117 | 11,900 | 81 | 56 |
| GP4 | 15,100 | 117 | 24,200 | 165 | 38 |
| Biocatalyst form | Activity, µmol·min−1·mg−1 | KM, mM |
|---|---|---|
| Free ribonuclease A | 2.4 | 27 |
| Immobilized ribonuclease A | 2.2 | 18 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vlakh, E.; Ananyan, A.; Zashikhina, N.; Hubina, A.; Pogodaev, A.; Volokitina, M.; Sharoyko, V.; Tennikova, T. Preparation, Characterization, and Biological Evaluation of Poly(Glutamic Acid)-b-Polyphenylalanine Polymersomes. Polymers 2016, 8, 212. https://doi.org/10.3390/polym8060212
Vlakh E, Ananyan A, Zashikhina N, Hubina A, Pogodaev A, Volokitina M, Sharoyko V, Tennikova T. Preparation, Characterization, and Biological Evaluation of Poly(Glutamic Acid)-b-Polyphenylalanine Polymersomes. Polymers. 2016; 8(6):212. https://doi.org/10.3390/polym8060212
Chicago/Turabian StyleVlakh, Evgenia, Anastasiia Ananyan, Natalia Zashikhina, Anastasiia Hubina, Aleksander Pogodaev, Mariia Volokitina, Vladimir Sharoyko, and Tatiana Tennikova. 2016. "Preparation, Characterization, and Biological Evaluation of Poly(Glutamic Acid)-b-Polyphenylalanine Polymersomes" Polymers 8, no. 6: 212. https://doi.org/10.3390/polym8060212