Individual Impact of Distinct Polysialic Acid Chain Lengths on the Cytotoxicity of Histone H1, H2A, H2B, H3 and H4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
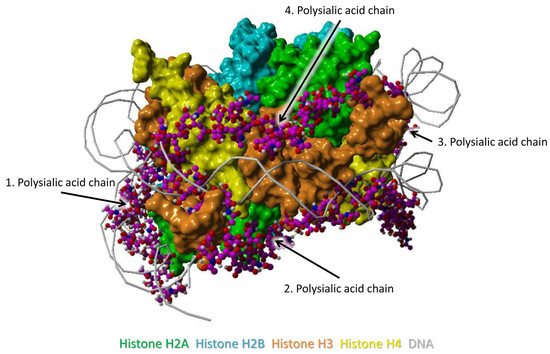
2.2. Binding Model
2.3. Fractionation and Determination of Sialic Acid Polymers by HPLC
2.4. Quantification of Sialic Acid Polymers by HPLC
2.5. Cell Culture Experiments
2.6. Agarose Gel-Electrophoresis
2.7. Statistical Analysis
3. Results & Discussion
3.1. Binding Model between PolySia and Histones
3.2. PolySia Interacts with Individual Histones
3.3. Cytotoxicity Effects Are Histone-Dependent
3.4. The Cytotoxicity of Histone H1, H2A, H2B, H3, and H4 Is Individually Influenced by PolySia
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zawrotniak, M.; Rapala-Kozik, M. Neutrophil extracellular traps (NETs)—Formation and implications. Acta Biochim. Pol. 2013, 60, 277–284. [Google Scholar] [PubMed]
- Miller, B.F.; Abrams, R.; Dorfman, A.; Klein, M. Antibacterial properties of protamine and histone. Science 1942, 96, 428–430. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Iwamuro, S. Potential roles of histones in host defense as antimicrobial agents. Infect. Disord. Drug Targets 2008, 8, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Hariton-Gazal, E.; Rosenbluh, J.; Graessmann, A.; Gilon, C.; Loyter, A. Direct translocation of histone molecules across cell membranes. J. Cell Sci. 2003, 116, 4577–4586. [Google Scholar] [CrossRef] [PubMed]
- Rosenbluh, J.; Singh, S.K.; Gafni, Y.; Graessmann, A.; Loyter, A. Non-endocytic penetration of core histones into petunia protoplasts and cultured cells: A novel mechanism for the introduction of macromolecules into plant cells. Biochim. Biophys. Acta 2004, 1664, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Patrzykat, A. Clinical development of cationic antimicrobial peptides: From natural to novel antibiotics. Curr. Drug Targets Infect. Disord. 2002, 2, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Urban, C.F.; Ermert, D.; Schmid, M.; Abu-Abed, U.; Goosmann, C.; Nacken, W.; Brinkmann, V.; Jungblut, P.R.; Zychlinsky, A. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against candida albicans. PLoS Pathog 2009, 5, e1000639. [Google Scholar] [CrossRef] [PubMed]
- Parseghian, M.H.; Luhrs, K.A. Beyond the walls of the nucleus: The role of histones in cellular signaling and innate immunity. Biochem. Cell Biol. 2006, 84, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, X.; Monestier, M.; Esmon, N.L.; Esmon, C.T. Extracellular histones are mediators of death through TLR2 and TLR4 in mouse fatal liver injury. J. Immunol. 2011, 187, 2626–2631. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Medzhitov, R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004, 5, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Krieg, A.M. Toll-like receptor 9 (TLR9) agonists in the treatment of cancer. Oncogene 2008, 27, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Boe, D.M.; Curtis, B.J.; Chen, M.M.; Ippolito, J.A.; Kovacs, E.J. Extracellular traps and macrophages: New roles for the versatile phagocyte. J. Leukoc. Biol. 2015, 97, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Leffler, J.; Martin, M.; Gullstrand, B.; Tyden, H.; Lood, C.; Truedsson, L.; Bengtsson, A.A.; Blom, A.M. Neutrophil extracellular traps that are not degraded in systemic lupus erythematosus activate complement exacerbating the disease. J. Immunol. 2012, 188, 3522–3531. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Li, C.; Wang, X.; Liu, X.; Han, C.; Jin, T.; Liu, S.; Zhang, X.; Zhang, H.; He, X.; et al. Increased toll-like receptors activity and TLR ligands in patients with autoimmune thyroid diseases. Front. Immunol. 2016, 7, 578. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, E.; Yalavarthi, S.; Berthier, C.C.; Hodgin, J.B.; Khandpur, R.; Lin, A.M.; Rubin, C.J.; Zhao, W.; Olsen, S.H.; Klinker, M.; et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J. Immunol. 2011, 187, 538–552. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, K.; Krumbholz, M.; Schonermarck, U.; Back, W.; Gross, W.L.; Werb, Z.; Grone, H.J.; Brinkmann, V.; Jenne, D.E. Netting neutrophils in autoimmune small-vessel vasculitis. Nat. Med. 2009, 15, 623–625. [Google Scholar] [CrossRef] [PubMed]
- Von Bruhl, M.L.; Stark, K.; Steinhart, A.; Chandraratne, S.; Konrad, I.; Lorenz, M.; Khandoga, A.; Tirniceriu, A.; Coletti, R.; Kollnberger, M.; et al. Monocytes, neutrophils, and platelets cooperate to initiate and propagate venous thrombosis in mice in vivo. J. Exp. Med. 2012, 209, 819–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, S.R.; Ma, A.C.; Tavener, S.A.; McDonald, B.; Goodarzi, Z.; Kelly, M.M.; Patel, K.D.; Chakrabarti, S.; McAvoy, E.; Sinclair, G.D.; et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat. Med. 2007, 13, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Marcos, V.; Zhou, Z.; Yildirim, A.O.; Bohla, A.; Hector, A.; Vitkov, L.; Wiedenbauer, E.M.; Krautgartner, W.D.; Stoiber, W.; Belohradsky, B.H.; et al. CXCR2 mediates nadph oxidase-independent neutrophil extracellular trap formation in cystic fibrosis airway inflammation. Nat. Med. 2010, 16, 1018–1023. [Google Scholar] [CrossRef] [PubMed]
- Saffarzadeh, M.; Juenemann, C.; Queisser, M.A.; Lochnit, G.; Barreto, G.; Galuska, S.P.; Lohmeyer, J.; Preissner, K.T. Neutrophil extracellular traps directly induce epithelial and endothelial cell death: A predominant role of histones. PLoS ONE 2012, 7, e32366. [Google Scholar] [CrossRef] [PubMed]
- Ulm, C.; Saffarzadeh, M.; Mahavadi, P.; Müller, S.; Prem, G.; Saboor, F.; Simon, P.; Middendorff, R.; Geyer, H.; Henneke, I.; et al. Soluble polysialylated NCAM: A novel player of the innate immune system in the lung. Cell. Mol. Life Sci. 2013, 70, 3695–3708. [Google Scholar] [CrossRef] [PubMed]
- Galuska, C.E.; Lutteke, T.; Galuska, S.P. Is polysialylated ncam not only a regulator during brain development but also during the formation of other organs? Biology 2017, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Colley, K.J.; Kitajima, K.; Sato, C. Polysialic acid: Biosynthesis, novel functions and applications. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 498–532. [Google Scholar] [CrossRef] [PubMed]
- Schnaar, R.L.; Gerardy-Schahn, R.; Hildebrandt, H. Sialic acids in the brain: Gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 2014, 94, 461–518. [Google Scholar] [CrossRef] [PubMed]
- Werneburg, S.; Buettner, F.F.; Erben, L.; Mathews, M.; Neumann, H.; Muhlenhoff, M.; Hildebrandt, H. Polysialylation and lipopolysaccharide-induced shedding of e-selectin ligand-1 and neuropilin-2 by microglia and THP-1 macrophages. Glia 2016, 64, 1314–1330. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Neumann, H. Alleviation of neurotoxicity by microglial human Siglec-11. J. Neurosci. 2010, 30, 3482–3488. [Google Scholar] [CrossRef] [PubMed]
- Shahraz, A.; Kopatz, J.; Mathy, R.; Kappler, J.; Winter, D.; Kapoor, S.; Schutza, V.; Scheper, T.; Gieselmann, V.; Neumann, H. Anti-inflammatory activity of low molecular weight polysialic acid on human macrophages. Sci. Rep. 2015, 5, 16800. [Google Scholar] [CrossRef] [PubMed]
- Curreli, S.; Arany, Z.; Gerardy-Schahn, R.; Mann, D.; Stamatos, N.M. Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J. Biol. Chem. 2007, 282, 30346–30356. [Google Scholar] [CrossRef] [PubMed]
- Stamatos, N.M.; Zhang, L.; Jokilammi, A.; Finne, J.; Chen, W.H.; El-Maarouf, A.; Cross, A.S.; Hankey, K.G. Changes in polysialic acid expression on myeloid cells during differentiation and recruitment to sites of inflammation: Role in phagocytosis. Glycobiology 2014, 24, 864–879. [Google Scholar] [CrossRef] [PubMed]
- Kiermaier, E.; Moussion, C.; Veldkamp, C.T.; Gerardy-Schahn, R.; de Vries, I.; Williams, L.G.; Chaffee, G.R.; Phillips, A.J.; Freiberger, F.; Imre, R.; et al. Polysialylation controls dendritic cell trafficking by regulating chemokine recognition. Science 2016, 351, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Simon, P.; Bäumner, S.; Busch, O.; Röhrich, R.; Kaese, M.; Richterich, P.; Wehrend, A.; Müller, K.; Gerardy-Schahn, R.; Mühlenhoff, M.; et al. Polysialic acid is present in mammalian semen as a post-translational modification of the neural cell adhesion molecule ncam and the polysialyltransferase ST8Siaii. J. Biol. Chem. 2013, 288, 18825–18833. [Google Scholar] [CrossRef] [PubMed]
- Galuska, S.P.; Galuska, C.E.; Tharmalingam, T.; Zlatina, K.; Prem, G.; Husejnov, F.C.O.; Rudd, P.M.; Vann, W.F.; Reid, C.; Vionnet, J.; et al. In vitro generation of polysialylated cervical mucins by bacterial polysialyltransferases to counteract cytotoxicity of extracellular histones. FEBS J. 2017, 284, 1688–1699. [Google Scholar] [CrossRef] [PubMed]
- Galuska, C.E.; Dambon, J.A.; Kuhnle, A.; Bornhofft, K.F.; Prem, G.; Zlatina, K.; Lutteke, T.; Galuska, S.P. Artificial polysialic acid chains as sialidase-resistant molecular-anchors to accumulate particles on neutrophil extracellular traps. Front. Immunol. 2017, 8, 1229. [Google Scholar] [CrossRef] [PubMed]
- Osley, M.A. Epigenetics: How to lose a tail. Nature 2008, 456, 885–886. [Google Scholar] [CrossRef] [PubMed]
- Hara, S.; Yamaguchi, M.; Takemori, Y.; Furuhata, K.; Ogura, H.; Nakamura, M. Determination of mono-O-acetylated N-acetylneuraminic acids in human and rat sera by fluorometric high-performance liquid chromatography. Anal. Biochem. 1989, 179, 162–166. [Google Scholar] [CrossRef]
- Hara, S.; Takemori, Y.; Yamaguchi, M.; Nakamura, M.; Ohkura, Y. Fluorometric high-performance liquid chromatography of N-acetyl- and N-glycolylneuraminic acids and its application to their microdetermination in human and animal sera, glycoproteins, and glycolipids. Anal. Biochem. 1987, 164, 138–145. [Google Scholar] [CrossRef]
- Galuska, S.P.; Geyer, H.; Weinhold, B.; Kontou, M.; Röhrich, R.C.; Bernard, U.; Gerardy-Schahn, R.; Reutter, W.; Münster-Kuhnel, A.; Geyer, R. Quantification of nucleotide-activated sialic acids by a combination of reduction and fluorescent labeling. Anal. Chem. 2010, 82, 4591–4598. [Google Scholar] [CrossRef] [PubMed]
- Bartel, J.; Feuerstacke, C.; Galuska, C.E.; Weinhold, B.; Gerardy-Schahn, R.; Geyer, R.; Munster-Kuhnel, A.; Middendorff, R.; Galuska, S.P. Laser microdissection of paraffin embedded tissue as a tool to estimate the sialylation status of selected cell populations. Anal. Chem. 2014, 86, 2326–2331. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Kusukawa, N. Sds agarose gels for analysis of proteins. Biotechniques 1998, 24, 676–678. [Google Scholar] [PubMed]
- Kim, R.; Yokota, H.; Kim, S.H. Electrophoresis of proteins and protein-protein complexes in a native agarose gel. Anal. Biochem. 2000, 282, 147–149. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; von der Ohe, M.; Schulze, C.; Bian, S.; Makhina, T.; Loers, G.; Kleene, R.; Schachner, M. Functional role of the interaction between polysialic acid and extracellular histone H1. J. Neurosci. 2010, 30, 12400–12413. [Google Scholar] [CrossRef] [PubMed]
- Papayannopoulos, V.; Metzler, K.D.; Hakkim, A.; Zychlinsky, A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell Biol. 2010, 191, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.H.; Sung, B.H.; Kim, S.C. Buforins: Histone H2A-derived antimicrobial peptides from toad stomach. Biochim. Biophys. Acta 2009, 1788, 1564–1569. [Google Scholar] [CrossRef] [PubMed]
- Koo, Y.S.; Kim, J.M.; Park, I.Y.; Yu, B.J.; Jang, S.A.; Kim, K.S.; Park, C.B.; Cho, J.H.; Kim, S.C. Structure-activity relations of parasin i, a histone H2A-derived antimicrobial peptide. Peptides 2008, 29, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Patat, S.A.; Carnegie, R.B.; Kingsbury, C.; Gross, P.S.; Chapman, R.; Schey, K.L. Antimicrobial activity of histones from hemocytes of the pacific white shrimp. Eur. J. Biochem. 2004, 271, 4825–4833. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.M.; Molle, G.; Kemp, G.D.; Smith, V.J. Isolation and characterisation of oncorhyncin II, a histone H1-derived antimicrobial peptide from skin secretions of rainbow trout, oncorhynchus mykiss. Dev. Comp. Immunol. 2004, 28, 127–138. [Google Scholar] [CrossRef]








| H1 | H2A | H2B | H3 | H4 | |
|---|---|---|---|---|---|
| Amino acids | 231 | 154 | 149 | 161 | 122 |
| pI | 10.11 | 10.95 | 9.76 | 11.02 | 11.3 |
| Mass [kDa] | 20.73 | 13.99 | 13.79 | 15.27 | 11.24 |
| Basic amino acids | 66 | 30 | 31 | 33 | 27 |
| Acidic amino acids | 9 | 9 | 10 | 11 | 10 |
| Relative amount of histone [%] 1 | - | 38.02 | 34.63 | 20.97 | 6.38 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zlatina, K.; Lütteke, T.; Galuska, S.P. Individual Impact of Distinct Polysialic Acid Chain Lengths on the Cytotoxicity of Histone H1, H2A, H2B, H3 and H4. Polymers 2017, 9, 720. https://doi.org/10.3390/polym9120720
Zlatina K, Lütteke T, Galuska SP. Individual Impact of Distinct Polysialic Acid Chain Lengths on the Cytotoxicity of Histone H1, H2A, H2B, H3 and H4. Polymers. 2017; 9(12):720. https://doi.org/10.3390/polym9120720
Chicago/Turabian StyleZlatina, Kristina, Thomas Lütteke, and Sebastian P. Galuska. 2017. "Individual Impact of Distinct Polysialic Acid Chain Lengths on the Cytotoxicity of Histone H1, H2A, H2B, H3 and H4" Polymers 9, no. 12: 720. https://doi.org/10.3390/polym9120720