Translational Pigeonpea Genomics Consortium for Accelerating Genetic Gains in Pigeonpea (Cajanus cajan L.)
Abstract
:1. Introduction
2. Advances in Genetics and Genomics
2.1. Novel Breeding and Genetic Materials
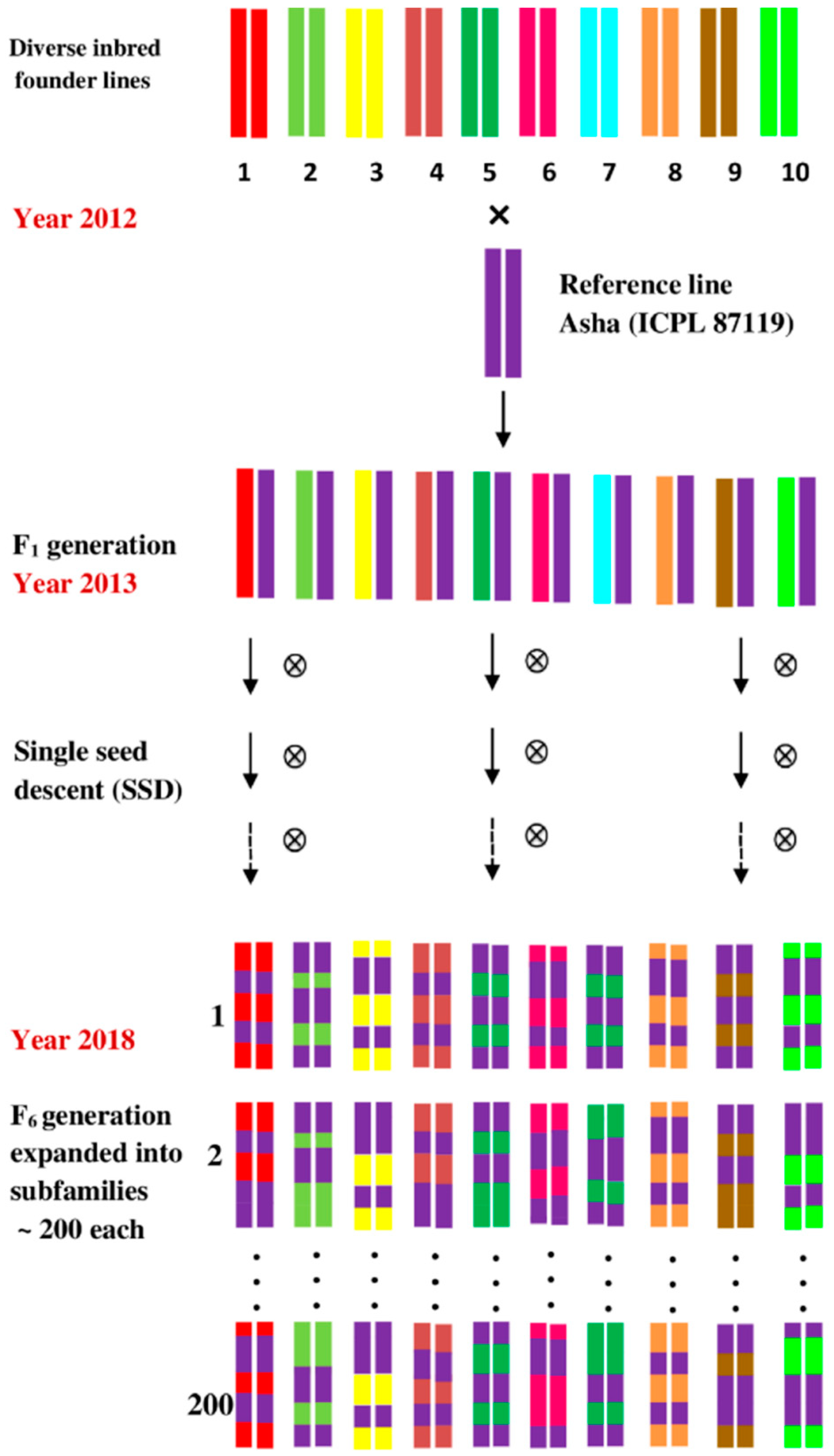
2.2. Nested Association Mapping Population (NAM)
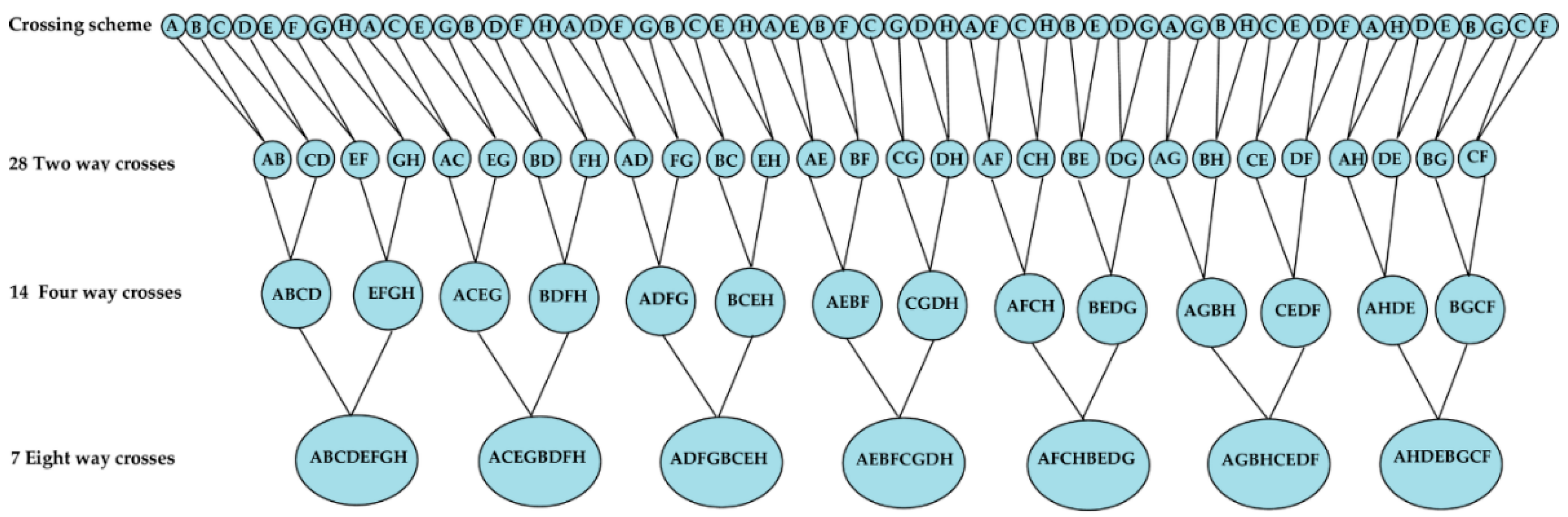
2.3. Multi-Parent Advanced Generation Inter-Cross (MAGIC)
3. Genomics Advances and Genomics-Assisted Breeding
3.1. Marker Assisted Back-Crossing for Fusarium Wilt (FW) and Sterility Mosaic Diseases (SMD) Resistance
3.2. Development of Trait-Associated Markers
4. Promising Pigeonpea Lines Identified through Multi-Location Trials
4.1. Performance and Stability of Genotype and Environment for Grain Yield
4.2. Promising Lines in Super-Early Duration Trial
4.3. Promising Lines in Early Duration Trial
4.4. Promising Lines in Medium Duration Trial
5. Enhancing Varietal Adoption through Farmer Participatory Varietal Selection (FPVS) Trials
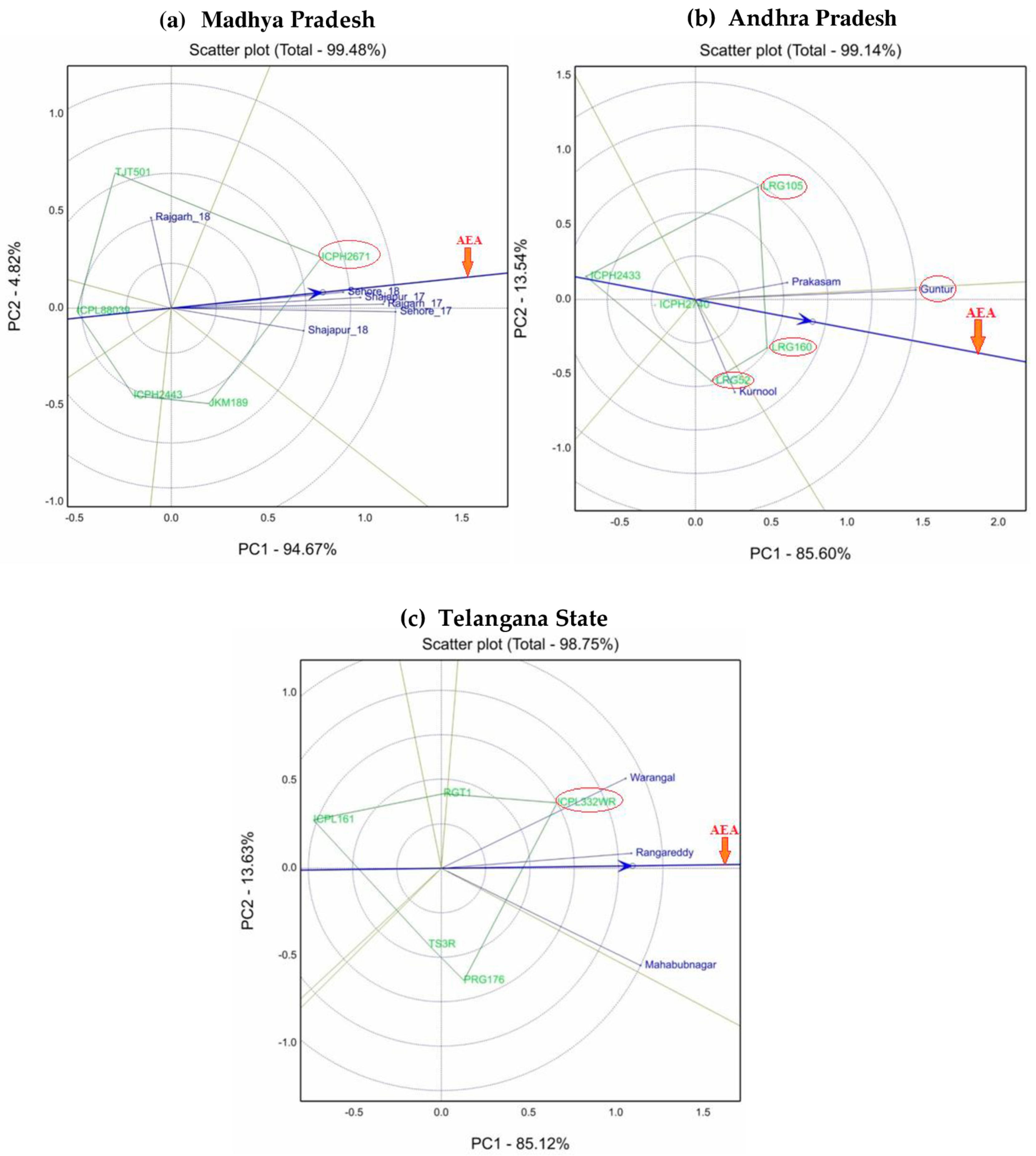
5.1. Performance and Stability of Genotype and Environment for Grain Yield
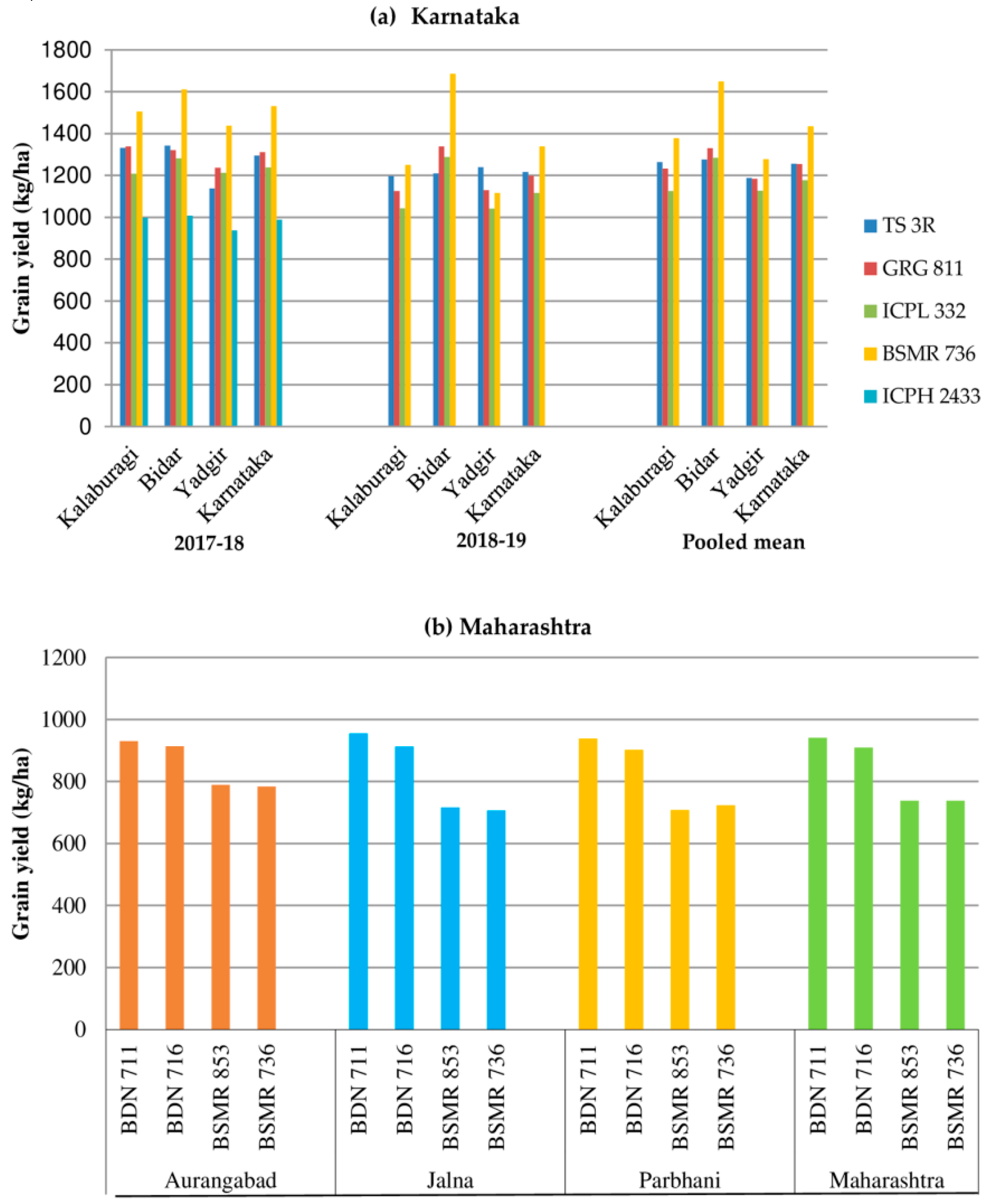
5.2. Selection of High-Performing Varieties in Different States
6. Summary and Outlook
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations Database (FAOSTAT). 2020. Available online: http://faostat.fao.org/database (accessed on 16 January 2020).
- Bhatia, V.S.; Singh, P.; Wani, S.P.; Rao, A.K.; Srinivas, K. Yield Gap Analysis of Soybean, Groundnut, Pigeonpea and Chickpea in India Using Simulation Modeling: Global Theme on Agroecosystems; Report no. 31. Patancheru 502 324; International Crops Research Institute for the Semi-Arid Tropics (ICRISAT): Andhra Pradesh, India, 2006; p. 156. [Google Scholar]
- Yang, D.; Tang, J.; Yang, D.; Chen, Y.; Ali, J.; Mou, T. Improving rice blast resistance of Feng39S through molecular marker-assisted backcrossing. Rice 2019, 12, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yugander, A.; Sundaram, R.M.; Singh, K.; Ladhalakshmi, D.; Rao, L.V.S.; Madhav, M.S.; Badri, J.; Prasad, M.S.; Laha, G.S. Incorporation of the novel bacterial blight resistance gene Xa38 into the genetic background of elite rice variety Improved Samba Mahsuri. PLoS ONE 2018, 13, e0198260. [Google Scholar] [CrossRef] [PubMed]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Latif, M.A. Marker-assisted introgression of broad-spectrum blast resistance genes into the cultivated MR219 rice variety. J. Sci. Food Agric. 2017, 97, 2810–2818. [Google Scholar] [CrossRef] [PubMed]
- Linh, L.H.; Linh, T.H.; Xuan, T.D.; Ham, L.H.; Ismail, A.M.; Khanh, T.D. Molecular breeding to improve salt tolerance of rice (Oryza sativa L.) in the Red River Delta of Vietnam. Int. J. Plant Genom. 2012, 2012. [Google Scholar] [CrossRef] [Green Version]
- Randhawa, M.S.; Bains, N.S.; Sohu, V.S.; Chhuneja, P.; Trethowan, R.M.; Bariana, H.S.; Bansal, U. Marker assisted transfer of stripe rust and stem rust resistance genes into four wheat cultivars. Agronomy 2019, 9, 497. [Google Scholar] [CrossRef] [Green Version]
- Tyagi, S.; Mir, R.R.; Kaur, H.; Chhuneja, P.; Ramesh, B.; Balyan, H.S.; Gupta, P.K. Marker-assisted pyramiding of eight QTLs/genes for seven different traits in common wheat (Triticum aestivum L.). Mol. Breed. 2014, 34, 167–175. [Google Scholar] [CrossRef]
- Charpe, A.; Koul, S.; Gupta, S.K.; Singh, A.; Pallavi, J.K.; Prabhu, K.V. Marker assisted gene pyramiding of leaf rust resistance genes Lr 9, Lr 24 and Lr 28 in a bread wheat cultivar HD 2329. J. Wheat Res. 2012, 4, 20–28. [Google Scholar]
- Kumar, J.; Jaiswal, V.; Kumar, A.; Kumar, N.; Mir, R.R.; Kumar, S.; Dhariwal, R.; Tyagi, S.; Khandelwal, M.; Prabhu, K.V.; et al. Introgression of a major gene for high grain protein content in some Indian bread wheat cultivars. Field Crops Res. 2011, 123, 226–233. [Google Scholar] [CrossRef]
- Barloy, D.; Lemoine, J.; Abelard, P.; Tanguy, A.M.; Rivoal, R.; Jahier, J. Marker-assisted pyramiding of two cereal cyst nematode resistance genes from Aegilops variabilis in wheat. Mol. Breed. 2007, 20, 31–40. [Google Scholar] [CrossRef]
- Afolayan, G.; Aladele, S.E.; Deshpande, S.P.; Oduoye, O.T.; Nwosu, D.J.; Michael, C.; Blay, E.T.; Danquah, E.Y. Marker assisted foreground selection for identification of striga resistant backcross lines in Sorghum Bicolor. Covenant J. Phys. Life Sci. 2019, 7, 29–36. [Google Scholar]
- Kadam, S.R.; Fakrudin, B. Marker assisted pyramiding of root volume QTLs to improve drought tolerance in rabi sorghum. Res. Crops 2017, 18, 683–692. [Google Scholar] [CrossRef]
- Ngugi, K.; Kimani, W.; Kiambi, D. Introgression of stay-green trait into a Kenyan farmer prefered sorghum variety. Afr. Crop Sci. J. 2010, 18, 141–146. [Google Scholar] [CrossRef]
- Krishna, M.S.R.; Reddy, S.S.; Satyanarayana, S.D. Marker-assisted breeding for introgression of opaque-2 allele into elite maize inbred line BML-7. 3 Biotech 2017, 7, 165. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, V.; Hossain, F.; Thirunavukkarasu, N.; Choudhary, M.; Saha, S.; Bhat, J.S.; Prasanna, B.M.; Gupta, H.S. Development of β-carotene rich maize hybrids through marker-assisted introgression of β-carotene hydroxylase allele. PLoS ONE 2014, 9, e113583. [Google Scholar] [CrossRef] [Green Version]
- Babu, R.; Nair, S.K.; Kumar, A.; Venkatesh, S.; Sekhar, J.C.; Singh, N.N.; Srinivasan, G.; Gupta, H.S. Two-generation marker-aided backcrossing for rapid conversion of normal maize lines to quality protein maize (QPM). Theor. Appl. Genet. 2005, 111, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Taunk, J.; Rani, A.; Yadav, N.R.; Yadav, D.V.; Yadav, R.C.; Raj, K.; Kumar, R.; Yadav, H.P. Molecular breeding of ameliorating commercial pearl millet hybrid for downy mildew resistance. J. Genet. 2018, 97, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Hash, C.T.; Sharma, A.; Kolesnikova-Allen, M.A.; Singh, S.D.; Thakur, R.P.; Raj, A.G.B.; Rao, M.N.V.R.; Nijhawan, D.C.; Beniwal, C.R.; Sagar, P.; et al. Teamwork delivers biotechnology products to Indian small-holder crop-livestock producers: Pearl millet hybrid “HHB 67 Improved” enters seed delivery pipeline. SAT Agric. Res. 2006, 2, 1–3. [Google Scholar]
- Mannur, D.M.; Babbar, A.; Thudi, M.; Sabbavarapu, M.M.; Roorkiwal, M.; Sharanabasappa, B.Y.; Bansal, V.P.; Jayalakshmi, S.K.; Yadav, S.S.; Rathore, A.; et al. Super Annigeri 1 and improved JG 74: Two Fusarium wilt-resistant introgression lines developed using marker-assisted backcrossing approach in chickpea (Cicer arietinum L.). Mol. Breed. 2019, 39, 2. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Mohan, S.M.; Gaur, P.M.; Chamarthi, S.K.; Singh, V.K.; Srinivasan, S.; Swapna, N.; Sharma, M.; Pande, S.; Singh, S.; et al. Marker-assisted backcrossing to introgress resistance to Fusarium wilt race 1 and Ascochyta blight in C 214, an elite cultivar of chickpea. Plant Genome 2014, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Gaur, P.M.; Chamarthi, S.K.; Krishnamurthy, L.; Tripathi, S.; Kashiwagi, J.; Samineni, S.; Singh, V.K.; Thudi, M.; Jaganathan, D. Fast-track introgression of “QTL-hotspot” for root traits and other drought tolerance traits in JG 11, an elite and leading variety of chickpea. Plant Genome 2013, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Taran, B.; Warkentin, T.D.; Vandenberg, A. Fast track genetic improvement of ascochyta blight resistance and double podding in chickpea by marker-assisted backcrossing. Theor. Appl. Genet. 2013, 126, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K. Exciting journey of 10 years from genomes to fields and markets: Some success stories of genomics-assisted breeding in chickpea, pigeonpea and groundnut. Plant Sci. 2016, 242, 98–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, T.; Khang, D.T.; Tuyen, P.T.; Toan, T.B.; Huong, N.N.; Lang, N.T.; Buu, B.C.; Xuan, T.D. Development of new drought tolerant breeding lines for Vietnam using marker-assisted backcrossing. Int. Lett. Nat. Sci. 2016, 59, 1–13. [Google Scholar]
- Kumar, V.; Rani, A.; Rawal, R.; Mourya, V. Marker assisted accelerated introgression of null allele of kunitz trypsin inhibitor in soybean. Breed Sci. 2015, 65, 447–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landau-Ellis, D.; Pantalone, V.R. Marker-assisted backcrossing to incorporate two low phytate alleles into the Tennessee soybean cultivar 5601T. In Induced Plant Mutations in the Genomics Era, Proceedings of an International Joint FAO/IAEA Symposium; Food and Agriculture Organization of the United Nations: Rome, Italy, 2009; pp. 316–318. [Google Scholar]
- Varshney, R.K.; Chen, W.; Li, Y.; Bharti, A.K.; Saxena, R.K.; Schlueter, J.A.; Donoghue, M.T.; Azam, S.; Fan, G.; Whaley, A.M.; et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 2012, 30, 83. [Google Scholar] [CrossRef] [Green Version]
- Saxena, R.K.; Prathima, C.; Saxena, K.B.; Hoisington, D.A.; Singh, N.K.; Varshney, R.K. Novel SSR markers for polymorphism detection in pigeonpea (Cajanus spp.). Plant Breed. 2010, 129, 142–148. [Google Scholar] [CrossRef] [Green Version]
- Bohra, A.; Saxena, K.B.; Varshney, R.K.; Saxena, R.K. Genomics-assisted breeding for pigeonpea improvement. Theor. Appl. Genet. 2020, 133, 1–17. [Google Scholar] [CrossRef]
- Saxena, R.K.; Kale, S.M.; Kumar, V.; Parupali, S.; Joshi, S.; Singh, V.; Garg, V.; Das, R.R.; Sharma, M.; Yamini, K.N.; et al. Genotyping-by-sequencing of three mapping populations for identification of candidate genomic regions for resistance to sterility mosaic disease in pigeonpea. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Saxena, R.K.; Rathore, A.; Bohra, A.; Yadav, P.; Das, R.R.; Khan, A.W.; Singh, V.K.; Chitikineni, A.; Singh, I.P.; Kumar, C.V.; et al. Development and application of high-density axiom Cajanus SNP array with 56k SNPs to understand the genome architecture of released cultivars and founder genotypes. Plant Genome 2018, 11, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Parupalli, S.; Saxena, R.K.; Kumar, C.V.; Sharma, M.; Singh, V.K.; Vechalapu, S.; Kavikishor, P.B.; Saxena, K.B.; Varshney, R.K. Genetics of fusarium wilt resistance in pigeonpea as revealed by phenotyping of RILs. J. Food Legumes 2017, 30, 241–244. [Google Scholar]
- Daspute, A.; Fakrudin, B.; Bhairappanavar, S.B.; Kavil, S.P.; Narayana, Y.D.; Muniswamy Kaumar, A.; Krishnaraj, P.U.; Yerimani, A.; Khadi, B.M. Inheritance of pigeonpea sterility mosaic disease resistance in pigeonpea. Plant Pathol. J. 2014, 30, 188–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshney, R.K.; Penmetsa, R.V.; Dutta, S.; Kulwal, P.L.; Saxena, R.K.; Datta, S.; Sharma, T.R.; Rosen, B.; Carrasquilla-Garcia, N.; Farmer, A.D.; et al. Pigeonpea genomics initiative (PGI): An international effort to improve crop productivity of pigeonpea (Cajanus cajan L.). Mol. Breed. 2010, 26, 393–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upadhyaya, H.D.; Reddy, K.N.; Sharma, S.; Varshney, R.K.; Bhattacharjee, R.; Singh, S.; Gowda, C.L.L. Pigeonpea composite collection and identification of germplasm for use in crop improvement programmes. Plant Genet. Resour. 2011, 9, 97–108. [Google Scholar] [CrossRef] [Green Version]
- Reddy, L.J.; Upadhyaya, H.D.; Gowda, C.L.L.; Singh, S. Development of core collection in pigeonpea [Cajanus cajan (L.) Millspaugh] using geographic and qualitative morphological descriptors. Genet. Resour. Crop Evol. 2005, 52, 1049–1056. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Reddy, L.J.; Gowda, C.L.L.; Reddy, K.N.; Singh, S. Development of a mini core subset for enhanced and diversified utilization of pigeonpea germplasm resources. Crop Sci. 2006, 46, 2127–2132. [Google Scholar] [CrossRef] [Green Version]
- Saxena, R.K.; Kale, S.; Mir, R.R.; Mallikarjuna, N.; Yadav, P.; Das, R.R.; Molla, J.; Sonnappa, M.; Ghanta, A.; Narasimhan, Y.; et al. Genotyping-by-sequencing and multilocation evaluation of two interspecific backcross populations identify QTLs for yield-related traits in pigeonpea. Theor. Appl. Genet. 2020, 133, 737–749. [Google Scholar] [CrossRef]
- IBPGR and ICRISAT. Descriptors for Pigeonpea [Cajanus cajan (L.) Millsp.]; IBPGR: Rome, Italy; ICRISAT: Patancheru, India, 1993.
- Cavanagh, C.; Morell, M.; Mackay, I.; Powell, W. From mutations to MAGIC: Resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 2008, 11, 215–221. [Google Scholar] [CrossRef]
- Prasad, P.S.; Muhammad, S.; Mahesh, M.; Kumar, G.N.V. Management of pigeonpea wilt caused by Fusarium udum Butler through integrated approaches. J. Biol. Control. 2012, 26, 361–367. [Google Scholar]
- Reddy, M.V.; Raju, T.N.; Lenne, J.M. Diseases of pigeonpea. In The Pathology of Food and Pasture Legumes; Allen, D.J., Lenne, J.M., Eds.; CAB International: Wallingford, UK, 1998; pp. 517–558. [Google Scholar]
- Reddy, M.V.; Sharma, S.B.; Nene, Y.L. Pigeonpea: Disease management. In The Pigeonpea; Nene, Y.L., Hall, S.D., Sheila, V.K., Eds.; CAB International: Wallingford, UK, 1990; pp. 303–348. [Google Scholar]
- Okiror, M.A. Genetics of resistance to Fusarium udum in pigeonpea [Cajanus cajan (L.) Millsp.]. Ind. J. Genet. Plant Breed. 2002, 62, 218–220. [Google Scholar]
- Gnanesh, B.N.; Ganapathy, K.N.; Ajay, B.C.; Byre Gowda, M. Inheritance of sterility mosaic disease resistance to Bangalore and Patancheru isolates in pigeonpea (Cajanus cajan (L.) Millsp.). Electron. J. Plant Breed. 2011, 2, 218–223. [Google Scholar]
- Kulkarni, N.K.; Reddy, A.S.; Kumar, P.L.; Vijayanarasimha, J.; Rangaswamy, K.T.; Muniyappa, V.; Reddy, L.J.; Saxena, K.B.; Jones, A.T.; Reddy, D.V. Broad based resistance to pigeonpea sterility mosaic disease in accessions of Cajanus scarbaeoides (L.) Benth. Ind. J. Plant Pro. 2003, 31, 6–11. [Google Scholar]
- Reddy, M.V.; Raju, T.N.; Nene, Y.L.; Ghanekar, A.M.; Amin, K.S.; Arjunan, G.; Astaputre, J.V.; Sinha, B.K.; Reddy, S.V.; Gupta, R.P.; et al. Variability in sterility mosaic pathogen in pigeonpea in India. Indian Phytopathol. 1993, 46, 206–212. [Google Scholar]
- Yadav, P.; Saxena, K.B.; Hingane, A.; Kumar, C.S.; Kandalkar, V.S.; Varshney, R.K.; Saxena, R.K. An “Axiom Cajanus SNP Array” based high density genetic map and QTL mapping for high-selfing flower and seed quality traits in pigeonpea. BMC Genom. 2019, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Obala, J.; Saxena, R.K.; Singh, V.K.; Kumar, C.S.; Saxena, K.B.; Tongoona, P.; Sibiya, J.; Varshney, R.K. Development of sequence-based markers for seed protein content in pigeonpea. Mol. Genet. Genom. 2019, 294, 57–68. [Google Scholar] [CrossRef]
- Obala, J.; Saxena, R.K.; Singh, V.K.; Kale, S.M.; Garg, V.; Kumar, C.S.; Saxena, K.B.; Tongoona, P.; Sibiya, J.; Varshney, R.K. Seed protein content and its relationships with agronomic traits in pigeonpea is controlled by both main and epistatic effects QTLs. Sci. Rep. 2020, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Panse, V.G.; Sukhatme, P.V. Statistical Methods for Agricultural Workers, 3rd ed.; ICAR: New Delhi, India, 1954; p. 58. [Google Scholar]
- SAS Institute Inc. SAS/STAT® 9.2 User’s Guide; SAS Institute Inc.: Cary, NC, USA, 2008. [Google Scholar]
- Yan, W.; Hunt, L.A.; Sheng, Q.; Szlavnics, Z. Cultivar evaluation and mega-environment investigation based on the GGE biplot. Crop Sci. 2000, 40, 597–605. [Google Scholar] [CrossRef]
- Ladejobi, O.; Elderfield, J.; Gardner, K.A.; Gaynor, R.C.; Hickey, J.; Hibberd, J.M.; Mackay, I.J.; Bentley, A.R. Maximizing the potential of multi-parental crop populations. Appl. Transl. Genom. 2016, 11, 9–17. [Google Scholar] [CrossRef] [Green Version]
- Janila, P.; Pandey, M.K.; Shasidhar, Y.; Variath, M.T.; Sriswathi, M.; Khera, P.; Manohar, S.S.; Nagesh, P.; Vishwakarma, M.K.; Mishra, G.P.; et al. Molecular breeding for introgression of fatty acid desaturase mutant alleles (ahFAD2A and ahFAD2B) enhances oil quality in high and low oil containing peanut genotypes. Plant Sci. 2016, 242, 203–213. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Pandey, M.K.; Janila, P.; Nigam, S.N.; Sudini, H.; Gowda, M.V.C.; Sriswathi, M.; Radhakrishnan, T.; Manohar, S.S.; Nagesh, P. Marker-assisted introgression of a QTL region to improve rust resistance in three elite and popular varieties of peanut (Arachis hypogaea L.). Theor. Appl. Genet. 2014, 127, 1771–1781. [Google Scholar] [CrossRef] [Green Version]
- Yan, W.; Tinker, N.A. Biplot analysis of multi-environment trial data: Principles and applications. Can. J. Plant Sci. 2006, 86, 623–645. [Google Scholar] [CrossRef] [Green Version]
- Muniswamy, S.; Lokesha, R.; Diwan, J.R. Stability for disease, genotype × environment interaction for yield and its components in pigeonpea [Cajanus cajan (L.) Millsp.]. Legume Res. 2017, 40, 624–629. [Google Scholar]
- Arunkumar, B.; Muniswamy, S.; Dharmaraj, P.S. Interpretation of genotype × environment interaction and stability analysis for grain yield of pigeon pea (Cajanus cajan L.). J. Appl. Nat. Sci. 2014, 6, 744–747. [Google Scholar] [CrossRef]




| Genotypes | Features |
|---|---|
| Nested Parent | |
| Asha (ICPL 87119) | Genome sequence available, leading variety, resistant to FW and SMD |
| Founder Parents | |
| HPL 24 | High protein content, medium duration, compact, susceptible to FW and resistant to SMD, inter-specific derivative |
| ICP 7035 | Medium duration, SMD resistant, large purple seed, high sugar |
| ICP 8863 | Mid-late, highly resistant to FW and susceptible to SMD, an extensively grown variety in Northern Karnataka and Maharashtra region of India |
| ICPL 87 | Early duration, determinate, short, high combiner |
| ICPL 88039 | Extra early maturity, indeterminate, good yield |
| ICPL 85063 | Medium duration, indeterminate, good yield, more branching |
| MN 1 | Super early, small seeded, determinate |
| ICP 28 | Early maturity, local variety |
| ICPL 85010 | Early maturity, local variety |
| ICP 7263 | Determinate, long podded, white seeded |
| Genotypes | Features |
|---|---|
| ICP 7426 | High pod numbers, medium duration |
| HPL 24 | High protein content, medium duration, compact, susceptible to FW and resistant to SMD, inter-specific derivative |
| ICP 11605 | Early flowering, germplasm line |
| ICP 14209 | High number of pods, germplasm line |
| ICP 14486 | Early flowering, germplasm line |
| ICP 5529 | Medium duration, obcordate leaves, compact plant, modified flower |
| ICP 7035 | Medium duration, SMD resistant, large purple seed, high sugar content |
| ICP 8863 | Mid-late, highly resistant to FW and susceptible to SMD, an extensively grown variety in Northern Karnataka and Maharashtra region of India |
| Varieties | # Centre | Introgression of the Trait |
|---|---|---|
| LRG 41 | RARS-Lam | FW and SMD |
| LRG 52 | RARS-Lam | FW and SMD |
| ICPL 88039 | ICRISAT | FW |
| UPAS 120 | ICRISAT | FW |
| ICP 8863 | ARS-Kalaburagi | SMD |
| TS 3R | ARS-Kalaburagi | SMD |
| TJT 501 | RAKCA-Sehore | FW |
| JKM 189 | RAKCA-Sehore | FW |
| BDN 711 | ARS-Badnapur | FW |
| PRG 176 | PJTSAU | FW and SMD |
| Bahar | ICAR-IIPR | FW |
| S. No. | Recurrent Parent | Donor Parent | F1 Plants | F1s Confirmed | No. of BC1F1s Grown | No. of BC1F1s Confirmed |
|---|---|---|---|---|---|---|
| 1 | BDN 711 | ICPL 20096 | 17 | 17 | 92 | 85 |
| 2 | ICP 8863 | ICPL 20096 | 30 | 21 | 92 | 78 |
| 3 | TS 3R | ICPL 20096 | 26 | 22 | 92 | 80 |
| 4 | TJT 501 | ICPL 20096 | 28 | 27 | 19 | 18 |
| 5 | JKM 189 | ICPL 20096 | 25 | 7 | 60 | 52 |
| 6 | Bahar | ICPL 20096 | 30 | 28 | 92 | 89 |
| 7 | PRG 176 | ICPL 20096 | 11 | 9 | 5 | 4 |
| 8 | LRG 41 | ICPL 20096 | 11 | 11 | 5 | 5 |
| 9 | LRG 52 | ICPL 20096 | 4 | 2 | 7 | 6 |
| 10 | ICPL 88039 | ICPL 20096 | 11 | 11 | 132 | 122 |
| 11 | UPAS 120 | ICPL 20096 | 28 | 15 | 88 | 65 |
| Trait | Number of QTLs/Genomic Segments Identified | # PVE Range (%) | Reference |
|---|---|---|---|
| Cleistogamy | 5 | 9.10–50.60 | Yadav et al. 2019 |
| Seed shape | 3 | 11.80–37.20 | Yadav et al. 2019 |
| Seed size | 2 | 29.50–33.90 | Yadav et al. 2019 |
| Seed protein content | 19 | 2.20–23.50 | Obala et al. 2019; 2020 |
| 100 seed weight | 18 | 10.10–46.60 | Obala et al. 2020 |
| Seed yield | 18 | 10.20–53.00 | Obala et al. 2020 |
| Growth habit | 21 | 10.90–91.30 | Obala et al. 2020 |
| Days to first flowering | 28 | 10.90–47.60 | Obala et al. 2020 |
| Trial | Effects | ||||
|---|---|---|---|---|---|
| Environment | Rep (Env) | Genotype | Genotype × Environment | ||
| Super-early (30 genotypes) | df | 7.0 | 16.0 | 28.0 | 196.0 |
| F | 2245.8 ** | 10.7 ** | 14.2 ** | 17.9 ** | |
| Early (30 genotypes) | df | 7.0 | 14.0 | 29.0 | 203.0 |
| F | 1305.0 * | 7.9 ** | 24.8 ** | 15.6 ** | |
| Medium (40 genotypes) | df | 8.0 | 15.0 | 34.0 | 272.0 |
| F | 1093.2 ** | 11.7 ** | 22.0 ** | 14.1 ** | |
| SN | State | Districts | Variety/Hybrids # | No. of FPVS | Total FPVS | |
|---|---|---|---|---|---|---|
| 2017–2018 | 2018–2019 | |||||
| 1 | Andhra Pradesh | Guntur, Prakasam, and Kurnool | LRG 52, LRG 105, LRG 160, ICPH 2433, and ICPH 2740 | 30 | 30 | 60 |
| 2 | Karnataka | Kalaburagi, Bida, r and Yadgir | TS 3R, GRG 811, ICPL 332, BSMR 736 and ICPH 2433 | 25 | 30 | 55 |
| 3 | Madhya Pradesh | Sehore, Shajapur, and Rajgarh | ICPH 2671, ICPH 2443, JKM 189, TJT 501, and ICPL 88039 | 25 | 30 | 55 |
| 4 | Maharashtra | Aurangabad, Jalna, and Parbhani | BDN 711, BDN 716, BSMR 853, and BSMR 736 | - | 28 | 28 |
| 5 | Telangana State | Mahabubnagar, Rangareddy, and Warangal | ICPL 161, ICPL 99050, ICPL 332 WR, PRG 176, and RGT 1 | 30 | 30 | 60 |
| 6 | Uttar Pradesh | Kanpur, Banda, and Chitrakoot | ICPH 2740, IPA 203, JKM 189, and ICPL 88039 | 25 | 20 | 45 |
| FPVS Trial | Effects | |||
|---|---|---|---|---|
| Environment | Genotype | Genotype × Environment | ||
| Andhra Pradesh | df | 2.0 | 4.0 | 8.0 |
| F | 145.8 ** | 23.78 ** | 6.6 ** | |
| Maharashtra | df | 2.0 | 3.0 | 6.0 |
| F | 0.09 NS | 63.9 ** | 1.36 NS | |
| Madhya Pradesh | df | 2.0 | 4.0 | 8.0 |
| F | 13.5 ** | 109.9 ** | 5.1 ** | |
| Karnataka | df | 2.0 | 3.0 | 6.0 |
| F | 1.3 NS | 9.8 ** | 1.4 NS | |
| Telangana | df | 2.0 | 4.0 | 8.0 |
| F | 4.11 * | 9.56 ** | 14.58 ** | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saxena, R.K.; Hake, A.; Hingane, A.J.; Kumar, C.V.S.; Bohra, A.; Sonnappa, M.; Rathore, A.; Kumar, A.V.; Mishra, A.; Tikle, A.N.; et al. Translational Pigeonpea Genomics Consortium for Accelerating Genetic Gains in Pigeonpea (Cajanus cajan L.). Agronomy 2020, 10, 1289. https://doi.org/10.3390/agronomy10091289
Saxena RK, Hake A, Hingane AJ, Kumar CVS, Bohra A, Sonnappa M, Rathore A, Kumar AV, Mishra A, Tikle AN, et al. Translational Pigeonpea Genomics Consortium for Accelerating Genetic Gains in Pigeonpea (Cajanus cajan L.). Agronomy. 2020; 10(9):1289. https://doi.org/10.3390/agronomy10091289
Chicago/Turabian StyleSaxena, Rachit K., Anil Hake, Anupama J. Hingane, C. V. Sameer Kumar, Abhishek Bohra, Muniswamy Sonnappa, Abhishek Rathore, Anil V. Kumar, Anil Mishra, A. N. Tikle, and et al. 2020. "Translational Pigeonpea Genomics Consortium for Accelerating Genetic Gains in Pigeonpea (Cajanus cajan L.)" Agronomy 10, no. 9: 1289. https://doi.org/10.3390/agronomy10091289







