Phenotypic and Nodule Microbial Diversity among Crimson Clover (Trifolium incarnatum L.) Accessions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Field Evaluation
2.3. Laboratory Evaluation
2.4. Data Analysis
3. Results
3.1. Fall Emergence
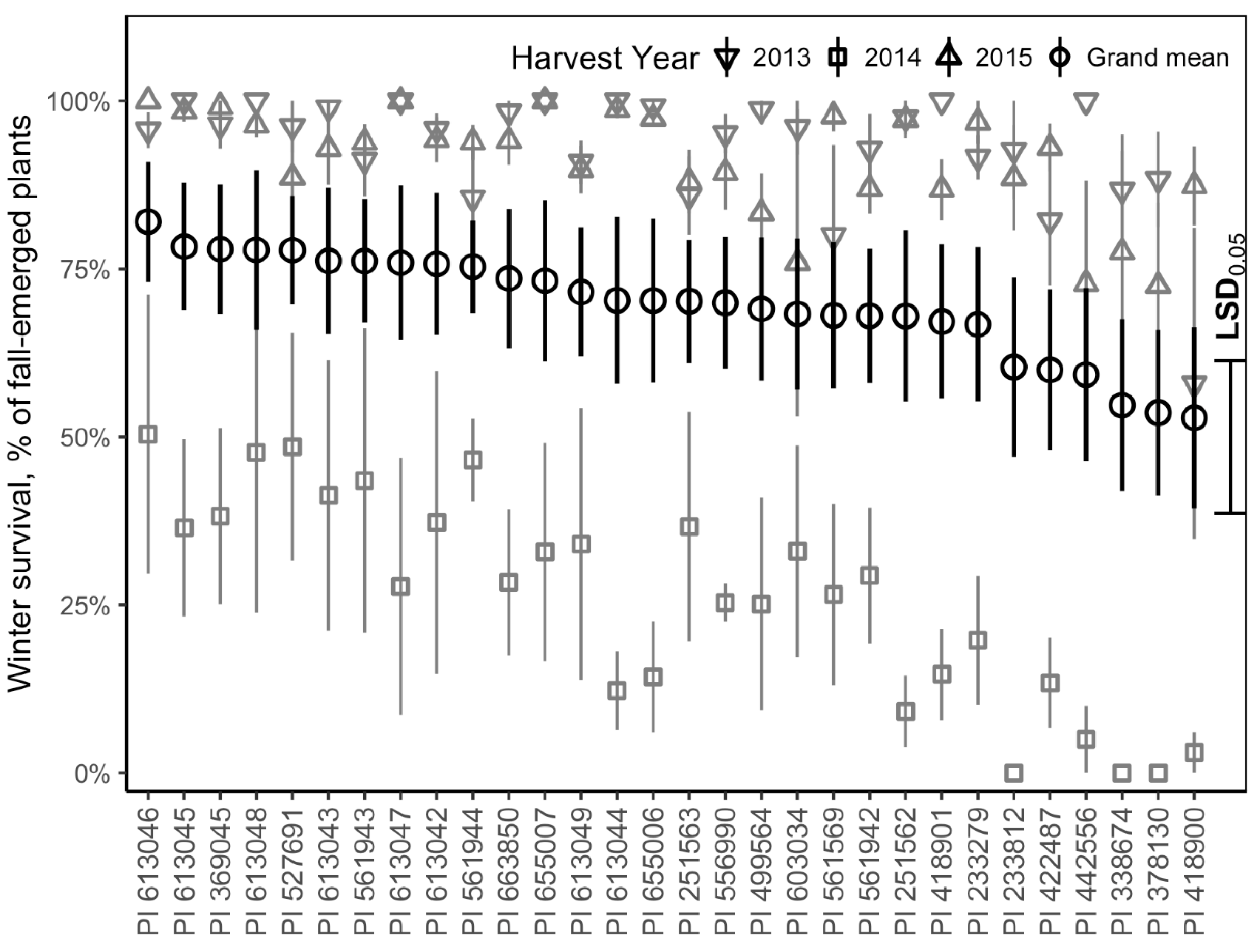
3.2. Winter Survival
3.3. Flowering Time
3.4. Biomass per Plant
3.5. Nitrogen Content
3.6. Proportion of Plant Nitrogen as BNF
3.7. Nodule Metagenome
3.8. Correlation among Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Drinkwater, L.E.; Wagoner, P.; Sarrantonio, M. Legume-based cropping systems have reduced carbon and nitrogen losses. Nature 1998, 396, 262–265. [Google Scholar] [CrossRef]
- Lu, Y.C.; Watkins, B.; Teasdale, J.; Abdul-Baki, A. Cover crops in sustainable food production. Food Rev. Int. 2000, 16, 121–157. [Google Scholar] [CrossRef]
- Altieri, M.; Nicholls, C. Biodiversity and Pest Management in Agroecosystems, 2nd ed.; CRC Press: New York, NY, USA, 2004; ISBN 978-1-56022-923-0. [Google Scholar]
- Gardner, J.B.; Drinkwater, L.E. The fate of nitrogen in grain cropping systems: A meta-analysis of 15N field experiments. Ecol. Appl. 2009, 19, 2167–2184. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, J.L.; Garrido, A.; Quemada, M. Cover crops effect on farm benefits and nitrate leaching: Linking economic and environmental analysis. Agric. Syst. 2013, 121, 23–32. [Google Scholar] [CrossRef]
- Schipanski, M.E.; Barbercheck, M.; Douglas, M.R.; Finney, D.M.; Haider, K.; Kaye, J.P.; Kemanian, A.R.; Mortensen, D.A.; Ryan, M.R.; Tooker, J.; et al. A framework for evaluating ecosystem services provided by cover crops in agroecosystems. Agric. Syst. 2014, 125, 12–22. [Google Scholar] [CrossRef]
- Malone, R.W.; Jaynes, D.B.; Kaspar, T.C.; Thorp, K.R.; Kladivko, E.; Ma, L.; James, D.E.; Singer, J.; Morin, X.K.; Searchinger, T. Cover crops in the upper midwestern United States: Simulated effect on nitrate leaching with artificial drainage. J. Soil Water Conserv. 2014, 69, 292–305. [Google Scholar] [CrossRef] [Green Version]
- Singer, J.W.; Nusser, S.M.; Alf, C.J. Are cover crops being used in the US corn belt? J. Soil Water Conserv. 2007, 62, 353–358. [Google Scholar]
- Wallander, S. While Crop Rotations Are Common, Cover Crops Remain Rare. In Amber Waves: The Economics of Food, Farming, Natural Resources, and Rural America; United States Department of Agriculture (USDA) Economic Research Service (ERS): Washington, DC, USA, 2013. [Google Scholar]
- CTIC. Report of the 2016–17 National Cover Crop Survey; Joint publication of the Conservation Technology Information Center (CTIC): West Lafayette, IN, USA; The North Central Region Sustainable Agriculture Research and Education (SARE) Program: St. Paul, MN, USA; The American Seed Trade Association: Alexandria, VA, USA, 2017. [Google Scholar]
- Wilke, B.J.; Snapp, S.S. Winter cover crops for local ecosystems: Linking plant traits and ecosystem function. J. Sci. Food Agric. 2008, 88, 551–557. [Google Scholar] [CrossRef]
- Wayman, S.; Kucek, L.K.; Mirsky, S.B.; Ackroyd, V.; Cordeau, S.; Ryan, M.R. Organic and conventional farmers differ in their perspectives on cover crop use and breeding. Renew. Agric. Food Syst. 2017, 32, 376–385. [Google Scholar] [CrossRef]
- Duvick, D.N. Genetic Contributions to Yield Gains of U.S. Hybrid Maize, 1930 to 1980. In Genetic Contributions to Yield Gains of Five Major Crop Plants; John Wiley and Sons, Ltd.: Hoboken, NJ, USA, 1984; pp. 15–47. ISBN 978-0-89118-586-4. [Google Scholar]
- Duvick, D.N. Plant breeding: Past achievements and expectations for the future. Econ. Bot. 1986, 40, 289–297. [Google Scholar] [CrossRef]
- Meredith, W.R.; Bridge, R.R. Genetic Contributions to Yield Changes in Upland Cotton. In Genetic Contributions to Yield Gains of Five Major Crop Plants; John Wiley and Sons, Ltd.: Hoboken, NJ, USA, 1984; pp. 75–87. ISBN 978-0-89118-586-4. [Google Scholar]
- Miller, F.R.; Kebede, Y. Genetic Contributions to Yield Gains in Sorghum, 1950 to 1980. In Genetic Contributions to Yield Gains of Five Major Crop Plants; John Wiley and Sons, Ltd.: Hoboken, NJ, USA, 1984; pp. 1–14. ISBN 978-0-89118-586-4. [Google Scholar]
- Schmidt, J.W. Genetic Contributions to Yield Gains in Wheat. In Genetic Contributions to Yield Gains of Five Major Crop Plants; John Wiley and Sons, Ltd.: Hoboken, NJ, USA, 1984; pp. 89–101. ISBN 978-0-89118-586-4. [Google Scholar]
- Specht, J.E.; Williams, J.H. Contribution of Genetic Technology to Soybean Productivity—Retrospect and Prospect. In Genetic Contributions to Yield Gains of Five Major Crop Plants; John Wiley and Sons, Ltd.: Hoboken, NJ, USA, 1984; pp. 49–74. ISBN 978-0-89118-586-4. [Google Scholar]
- Russell, G.E. (Ed.) Progress in Plant Breeding—1; Butterworth-Heinemann: Oxford, UK, 1985. [Google Scholar]
- Brummer, E.C.; Barber, W.T.; Collier, S.M.; Cox, T.S.; Johnson, R.; Murray, S.C.; Olsen, R.T.; Pratt, R.C.; Thro, A.M. Plant breeding for harmony between agriculture and the environment. Front. Ecol. Environ. 2011, 9, 561–568. [Google Scholar] [CrossRef] [Green Version]
- Yeater, K.M.; Bollero, G.A.; Bullock, D.; Rayburn, A.L.; Rodriguez-Zas, S.L. Assessment of Genetic Variation in Hairy Vetch Using Canonical Discriminant Analysis. Crop Sci. 2004, 44, 185–189. [Google Scholar] [CrossRef] [Green Version]
- Maul, J.; Mirsky, S.; Emche, S.; Devine, T. Evaluating a Germplasm Collection of the Cover Crop Hairy Vetch for Use in Sustainable Farming Systems. Crop Sci. Madison 2011, 51, 2615–2625. [Google Scholar] [CrossRef]
- Clark, A. (Ed.) Managing Cover Crops Profitably, 3rd ed.; SARE Outreach: College Park, MD, USA, 2007; ISBN 978-1-888626-12-4. [Google Scholar]
- Steiner, J.; Piccioni, E.; Falcinelli, M.; Liston, A. Germplasm Diversity among Cultivars and the NPGS Crimson Clover Collection. Crop Sci. 1998, 38, 263–271. [Google Scholar] [CrossRef]
- Mothapo, N.V.; Grossman, J.M.; Maul, J.E.; Shi, W.; Isleib, T. Genetic diversity of resident soil rhizobia isolated from nodules of distinct hairy vetch (Vicia villosa Roth) genotypes. Appl. Soil Ecol. 2013, 64, 201–213. [Google Scholar] [CrossRef] [Green Version]
- Mothapo, N.V.; Grossman, J.M.; Sooksa-nguan, T.; Maul, J.; Bräuer, S.L.; Shi, W. Cropping history affects nodulation and symbiotic efficiency of distinct hairy vetch (Vicia villosa Roth.) genotypes with resident soil rhizobia. Biol. Fertil. Soils 2013, 49, 871–879. [Google Scholar] [CrossRef]
- Baxter, L.L.; Grey, T.L.; Tucker, J.J.; Hancock, D.W. Optimizing Temperature Requirements for Clover Seed Germination. Agrosyst. Geosci. Environ. 2019, 2, 180059. [Google Scholar] [CrossRef] [Green Version]
- Shearer, G.; Kohl, D.H. N2-Fixation in Field Settings: Estimations Based on Natural 15N Abundance. Funct. Plant Biol. 1986, 13, 699–756. [Google Scholar] [CrossRef]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D. NLME: Linear and Nonlinear Mixed Effects Models. R Package Version 2019. Available online: https://www.researchgate.net/publication/303803175_Nlme_Linear_and_Nonlinear_Mixed_Effects_Models (accessed on 30 January 2019).
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [Green Version]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Overbeek, R.; Olson, R.; Pusch, G.; Olsen, G.; Davis, J.; Disz, T.; Edwards, R.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2013, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, W.E. Crimson Clover. In Clover Science and Technology; John Wiley and Sons, Ltd.: Hoboken, NJ, USA, 1985; pp. 491–502. ISBN 978-0-89118-218-4. [Google Scholar]
- Williams, W.A.; Elliott, J.R. Ecological Significance of Seed Coat Impermeability to Moisture in Crimson, Subterreanean and Rose Clovers in a Mediterranean-Type Climate. Ecology 1960, 41, 733–742. [Google Scholar] [CrossRef]
- Bennett, H.W. The Effectiveness of Selection for the Hard Seeded Character in Crimson Clover1. Agron. J. 1959, 51, 15–16. [Google Scholar] [CrossRef]
- Fowler, D. Selection for Winterhardiness in Wheat. II. Variation within Field Trials. Crop Sci. 1979, 19, 773–775. [Google Scholar] [CrossRef]
- Gusta, L.V.; Wisniewski, M. Understanding plant cold hardiness: An opinion. Physiol. Plant. 2013, 147, 4–14. [Google Scholar] [CrossRef]
- Nair, R.M.; Craig, A.D.; Rowe, T.D.; Biggins, S.R.; Hunt, C.H. Genetic variability and heritability estimates for hardseededness and flowering in balansa clover (Trifolium michelianum Savi) populations. Euphytica 2004, 138, 197–203. [Google Scholar] [CrossRef]
- Salisbury, P.A.; Aitken, Y.; Halloran, G.M. Genetic control of flowering time and its component processes in subterranean clover (Trifolium subterraneum L.). Euphytica 1987, 36, 887–902. [Google Scholar] [CrossRef]
- Abberton, M.T.; Marshall, A.H. Progress in breeding perennial clovers for temperate agriculture. J. Agric. Sci. 2005, 143, 117–135. [Google Scholar] [CrossRef]
- Gaby, J.C.; Buckley, D.H. A Comprehensive Evaluation of PCR Primers to Amplify the nifH Gene of Nitrogenase. PLoS ONE 2012, 7, e42149. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.J.; Nedwell, D.B.; Dong, L.F.; Osborn, A.M. Diversity and Abundance of Nitrate Reductase Genes (narG and napA), Nitrite Reductase Genes (nirS and nrfA), and Their Transcripts in Estuarine Sediments. Appl. Environ. Microbiol. 2007, 73, 3612–3622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamazaki, M.; Thorne, L.; Mikolajczak, M.; Armentrout, R.W.; Pollock, T.J. Linkage of genes essential for synthesis of a polysaccharide capsule in Sphingomonas strain S88. J. Bacteriol. 1996, 178, 2676–2687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozsoki, Z.; Cheng, J.; Feng, F.; Gysel, K.; Vinther, M.; Andersen, K.R.; Oldroyd, G.; Blaise, M.; Radutoiu, S.; Stougaard, J. Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proc. Natl. Acad. Sci. USA 2017, 114, E8118–E8127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Yang, J.; Dou, Y.; Chen, M.; Ping, S.; Peng, J.; Lu, W.; Zhang, W.; Yao, Z.; Li, H.; et al. Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc. Natl. Acad. Sci. USA 2008, 105, 7564–7569. [Google Scholar] [CrossRef] [Green Version]
- Di Cesare, A.; Cabello-Yeves, P.J.; Chrismas, N.A.M.; Sánchez-Baracaldo, P.; Salcher, M.M.; Callieri, C. Genome analysis of the freshwater planktonic Vulcanococcus limneticus sp. nov. reveals horizontal transfer of nitrogenase operon and alternative pathways of nitrogen utilization. BMC Genom. 2018, 19, 259. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.-C.; Lin, R.-F.; Chu, M.-K.; Chen, H.-M. Organization and expression of nitrogen-fixation genes in the aerobic nitrogen-fixing unicellular cyanobacterium Synechococcus sp. strain RF-1. Microbiol. Read. 1999, 145, 743–753. [Google Scholar] [CrossRef]
- Liu, D.; Liberton, M.; Yu, J.; Pakrasi, H.B.; Bhattacharyya-Pakrasi, M. Engineering Nitrogen Fixation Activity in an Oxygenic Phototroph. mBio 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Riday, H. Correlations between visual biomass scores and forage yield in space planted red clover (Trifolium pratense L.) breeding nurseries. Euphytica Dordr. 2009, 170, 339–345. [Google Scholar] [CrossRef]
- Riday, H.; Brummer, E.C. Forage Yield Heterosis in Alfalfa. Crop Sci. 2002, 42, 716–723. [Google Scholar] [CrossRef]
- Terpolilli, J.; Rui, T.; Yates, R.; Howieson, J.; Poole, P.; Munk, C.; Tapia, R.; Han, C.; Markowitz, V.; Tatiparthi, R.; et al. Genome sequence of Rhizobium leguminosarum bv trifolii strain WSM1689, the microsymbiont of the one flowered clover Trifolium uniflorum. Stand. Genom. Sci. 2014, 9, 527. [Google Scholar] [CrossRef] [Green Version]
- Mcewan, C.E.A.; Gatherer, D.; Mcewan, N.R. Nitrogen-Fixing Aerobic Bacteria have Higher Genomic GC Content than Non-Fixing Species within the Same Genus. Hereditas 1998, 128, 173–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Provorov, N.A.; Andronov, E.E.; Onishchuk, O.P. Forms of natural selection controlling the genomic evolution in nodule bacteria. Russ. J. Genet. 2017, 53, 411–419. [Google Scholar] [CrossRef]
- Muresu, R.; Porceddu, A.; Sulas, L.; Squartini, A. Nodule-associated microbiome diversity in wild populations of Sulla coronaria reveals clues on the relative importance of culturable rhizobial symbionts and co-infecting endophytes. Microbiol. Res. 2019, 221, 10–14. [Google Scholar] [CrossRef]
- Caetano-Anollés, G.; Gresshoff, P.M. Plant genetic control of nodulation. Annu. Rev. Microbiol. 1991, 45, 345–382. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, S.; Kim, J.; Muñoz, A.; Heckmann, A.B.; Downie, J.A.; Oldroyd, G.E.D. GRAS Proteins Form a DNA Binding Complex to Induce Gene Expression during Nodulation Signaling in Medicago truncatula. Plant Cell 2009, 21, 545–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, S.A.; Viprey, V.; Downie, J.A. Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by Nod factors and chitin oligomers. Proc. Natl. Acad. Sci. USA 2000, 97, 13413–13418. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Growing Season | |||
|---|---|---|---|
| Measurement | 2012–2013 | 2013–2014 | 2014–2015 |
| Growing degree days (GDD) | 2208.9 | 2102.8 | 2198.2 |
| Freezing degree days (FDD) | 84 | 115 | 102 |
| Days below freezing without snow cover | 80 | 86 | 72 |
| Total precipitation (mm) | 596.0 | 802.8 | 616.2 |
| Minimum low temperature (°C) | −11.0 | −16.5 | −17.0 |
| Term | Fall Emergence | Winter Survival | Flowering Time | Biomass per Plant | N Content | Plant N from BNF |
|---|---|---|---|---|---|---|
| F value | ||||||
| Accession | 49.30 ** | 30.63 *** | 2819.88 *** | 22.66 *** | 140.30 *** | 21.61 *** |
| Year | 4.30 * | 144.41 *** | 16.58 *** | 317.17 *** | 92.43 *** | 77.34 *** |
| Accession × Year | 1.72 ** | 1.04 | 1.81 ** | 1.18 | 2.16 *** | 2.32 *** |
| chi square | ||||||
| Replicate (Year) | 0.20 | 384.86 *** | 1.10 | 357.61 *** | 75.77 *** | 443.61 *** |
| Variance decomposition | ||||||
| Fixed effects | 0.75 | 0.33 | 0.83 | 0.38 | 0.43 | 0.45 |
| Random effects | 0.02 | 0.72 | 0.03 | 0.74 | 0.37 | 0.87 |
| Coefficient of Variation (CV) | 0.20 | 0.23 | 0.03 | 0.34 | 0.13 | 0.08 |
| Taxa Identified by 16S Operon | nifH | narG | hlyD | hesB |
|---|---|---|---|---|
| Acidovorax | X | |||
| Bacillus/Psychrobacillus/Paenibacillus | X | X | X | X |
| Microbacterium | X | |||
| Pelamonas puraquae | X | |||
| Propionibacterium | X | X | ||
| Pseudomonas (fluorescens, koreensis) | X | X | ||
| Rhizobium | X | |||
| Rhizobium leguminosarum | X | X | X | |
| Sphingomonas | X | |||
| Sporosarcina | X | X |
| Fall Emergence | Winter Survival | Biomass per Plant | Flowering Time | N Content | Plant N from BNF | Library Size | Contigs | GC Content | |
|---|---|---|---|---|---|---|---|---|---|
| Winter Survival | 0.59 *** | 1 | |||||||
| Biomass per Plant | −0.60 *** | −0.48 ** | 1 | ||||||
| Flowering Time | −0.15 | −0.23 | 0.15 | 1 | |||||
| N Content | −0.12 | −0.09 | 0.06 | −0.25 | 1 | ||||
| Plant N from BNF | −0.42 * | −0.65 *** | 0.24 | 0.24 | −0.27 | 1 | |||
| Library Size | 0.12 | 0.09 | −0.09 | 0.05 | 0.07 | 0.18 | 1 | ||
| Contigs | −0.12 | −0.05 | −0.04 | −0.35 | −0.26 | −0.06 | −0.16 | 1 | |
| GC Content | −0.10 | 0.12 | −0.04 | 0.05 | −0.26 | 0.29 | 0.46 * | −0.03 | 1 |
| Microbial Diversity | −0.14 | −0.20 | 0.17 | −0.05 | −0.10 | 0.25 | 0.60 ** | 0.37 | 0.58 ** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moore, V.; Davis, B.; Poskaitis, M.; Maul, J.E.; Kissing Kucek, L.; Mirsky, S. Phenotypic and Nodule Microbial Diversity among Crimson Clover (Trifolium incarnatum L.) Accessions. Agronomy 2020, 10, 1434. https://doi.org/10.3390/agronomy10091434
Moore V, Davis B, Poskaitis M, Maul JE, Kissing Kucek L, Mirsky S. Phenotypic and Nodule Microbial Diversity among Crimson Clover (Trifolium incarnatum L.) Accessions. Agronomy. 2020; 10(9):1434. https://doi.org/10.3390/agronomy10091434
Chicago/Turabian StyleMoore, Virginia, Brian Davis, Megan Poskaitis, Jude E. Maul, Lisa Kissing Kucek, and Steven Mirsky. 2020. "Phenotypic and Nodule Microbial Diversity among Crimson Clover (Trifolium incarnatum L.) Accessions" Agronomy 10, no. 9: 1434. https://doi.org/10.3390/agronomy10091434






