Genetic Potential of New Maize Inbred Lines in Single-Cross Hybrid Combinations under Low-Nitrogen Stress and Optimal Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Germplasm and Test Sites
2.2. Experimental Design
2.3. Management
2.4. Data Collection
2.5. Statistical Analysis
3. Results
3.1. F1 Hybrid Performance and Combining Ability Effects
3.2. Potential of the New Inbred Line Parents for Grain-Yield Performance in Single-Cross Hybrid Combinations
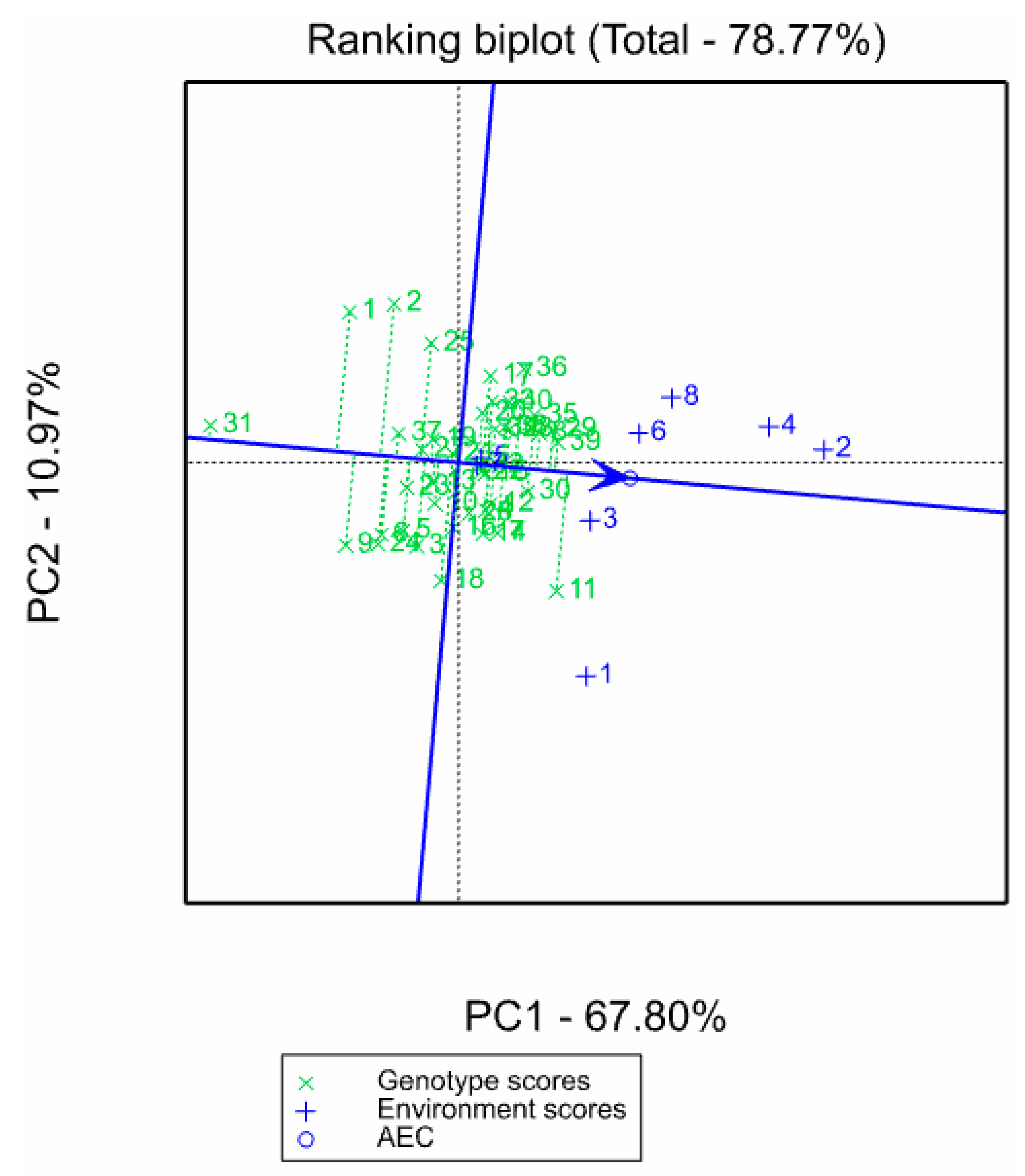
3.3. Stable High Yielding Single-Cross Hybrids under Low-Nitrogen Stress and Optimal Conditions
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aslam, M.; Cengiz, R. Effects of Drought on Maize. In Drought Stress in Maize (Zea mays L.); Springer International Publishing: Cham, Switzerland, 2015; pp. 5–17. [Google Scholar]
- Doswell, C. Maize in the Third World; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Ekpa, O.; Palacios-Rojas, N. Sub-Saharan African maize-based foods: Technological perspectives to increase the food and nutrition security impacts of maize breeding programmes. Glob. Food Secur. 2018, 17, 48–56. [Google Scholar] [CrossRef]
- Lydonn. Available online: https://www.world-grain.com/ (accessed on 28 August 2018).
- Nelimor, C.; Badu-Apraku, B. Phenotypic Characterization of Maize Landraces from Sahel and Coastal West Africa Reveals Marked Diversity and Potential for Genetic Improvement. J. Crop Improv. 2019, 34, 122–138. [Google Scholar] [CrossRef]
- Bradshaw, C.D.; Pope, E.; Kay, G.; Davie, J.C.; Cottrell, A.; Bacon, J.; Cosse, A.; Dunstone, N.; Jennings, S.; Challinor, A.; et al. Unprecedented climate extremes in South Africa and implications for maize production. Environ. Res. Lett. 2022, 17, 084028. [Google Scholar] [CrossRef]
- Ortiz-Bobea, A.; Ault, T.R.; Carrillo, C.M.; Chambers, R.G.; Lobell, D.B. Anthropogenic climate change has slowed global agricultural productivity growth. Nat. Clim. Chang. 2021, 11, 306–312. [Google Scholar] [CrossRef]
- Badu-Apraku, B.; Fakorede, M.A.B. Maize in Sub-Saharan Africa: Importance and Production Constraints. In Advances in Genetic Enhancement of Early and Extra-Early Maize for Sub-Saharan Africa; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Dakora, F.D.; Keya, S.O. Contribution of legume nitrogen fixation to sustainable agriculture in Sub-Saharan Africa. Soil Biol. Biochem. 1997, 29, 809–817. [Google Scholar] [CrossRef]
- Ricker-Gilbert, J. Inorganic fertilizer use among smallholder farmers in Sub-Saharan Africa: Implications for input subsidy policies. In The Role of Smallholder Farms in Food and Nutrition Security; Springer: Cham, Switzerland, 2020. [Google Scholar] [CrossRef]
- Masuka, B.; Magorokosho, C. Gains in Maize Genetic Improvement in Eastern and Southern Africa: II. CIMMYT Open-Pollinated Variety Breeding Pipeline. Crop Sci. 2017, 57, 180. [Google Scholar] [CrossRef]
- Grafton, R.; Williams, Q.J.; Jiang, Q. Food and Water Gaps to 2050: Preliminary Results from the Global Food and Water System (GFWS) Platform. Food Secur. 2015, 7, 209–210. [Google Scholar] [CrossRef]
- MacRoberts, J.; Setimela, P.S. Maize Hybrid Seed Production Manual. Handbook. 2014, pp. 27–31. Available online: https://excellenceinbreeding.org/sites/default/files/manual/98078.pdf (accessed on 19 August 2021).
- Eberhart, S.A. Theoretical relations among single, three-way and double-cross hybrids. Int. Biom. Soc. 1964, 20, 522–539. [Google Scholar] [CrossRef]
- Troyer, A.F. Background of U.S hybrid corn II: Breeding, climate, and food. Crop Sci. 2004, 44, 370–380. [Google Scholar] [CrossRef]
- Dari, S.; Macrobert, J. Effect of the fewbranched-1 (Fbr1) tassel mutation on performance of maize inbred lines and hybrids evaluated under stress and optimum environments. Maydica 2017, 12, 62. [Google Scholar]
- Griffing, B. Concept of General and Specific Combining Ability in Relation to Diallel Crossing Systems. Austrialian J. Biol. Sci. 1956, 9, 463–493. [Google Scholar] [CrossRef]
- Wu, Y.; San Vicente, F.; Huang, K.; Dhliwayo, T.; Costich, D.E.; Semagn, K.; Sudha, N.; Olsen, M.; Prasanna, B.M.; Zhang, X.; et al. Molecular characterization of CIMMYT maize inbred lines with genotyping-by-sequencing SNPs. Theor. Appl. Genet. 2016, 129, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Semagn, K.; Magorokosho, C. Molecular characterization of diverse CIMMYT maize inbred lines from eastern and Southern Africa using single nucleotide polymorphic markers. BMC Genom. 2012, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Patterson, H.D.; Williams, E.R.; Hunter, E.A. Block Designs for Variety Trials. J. Agric. Sci. (Camb.) 1978, 90, 395–400. [Google Scholar] [CrossRef]
- Arisede, C.; Mainassara, Z.A.; Jill, C.; Amsal, T.; Cosmos, M.; Bish, D.; Benhildah, M.; Mike, O.; Maruthi, P.B. Low-N stress tolerant maize hybrids have higher fertilizer N recovery efficiency and reduced N-dilution in the grain compared to susceptible hybrids under low N conditions. Plant Prod. Sci. 2020, 23, 417–426. [Google Scholar] [CrossRef]
- Tapera, T. Expression of Tolerance to Drought and Low Nitrogen Levels in Maize Inbred Lines and Hybrids in Southern Africa. Ph.D. Thesis, University of the Free State, Bloemfontein, South Africa, 2017; p. 36. [Google Scholar]
- Banziger, M.; Setimela, P. Breeding for Improved Abiotic Stress Tolerance in Maize Adapted to Southern Africa. Agric. Water Manag. 2006, 8, 212–214. [Google Scholar] [CrossRef]
- VSN International. GenStat for Windows, 14th ed.; VSN International: Hemel Hempstead, UK, 2011. [Google Scholar]
- Barreto, H.J.; Edmeades, G.O. The Alpha Lattice Design in Plant Breeding and Agronomy: Generation and Analysis. CIMMYT, Mexico DF (Mexico) [Corporate Author, Physics]. 1997. Available online: https://www.semanticscholar.org/paper/The-alpha-lattice-design-in-plant-breeding-and-and-Barreto-Edmeades/65d3efaaaee5396eef88d580c2799c5b448df6d2 (accessed on 19 August 2021).
- Singh, R.K.; Chaudhary, B.D. Biometrical Methods in Quantitative Genetic Analysis; Kalyani Publishers: New Delhi, India, 1979. [Google Scholar]
- Hallauer, A.R.; Miranda, J.B. Quantitative Genetics in Maize Breeding, 2nd ed.; Iowa State University Press: Ames, IA, USA, 1988. [Google Scholar]
- Rodriguez, F.; Alvarado, G. AGD-R (Analysis of Genetic Designs with R for Windows) (version 3.0). CIMMYT Res. Data Softw. Repos. Netw. 2015, 12, 53. [Google Scholar]
- Yan, W.; Kang, M.S. GGE biplot vs. AMMI analysis of genotype-by-environment data. Crop Sci. 2007, 47, 643–655. [Google Scholar] [CrossRef]
- Meseka, S.K.; Menkir, A.; Ibrahim, A.E.S. Yield potential and yield stability of maize hybrids selected for drought tolerance. J. Appl. Biosci. 2008, 3, 82–90. [Google Scholar]
- Dehghanpour, Z.; Ehdaie, B. Stability of general and specific combining ability effects for grain yield in elite Iranian maize inbred lines. J. Crop Improv. 2013, 27, 137–152. [Google Scholar] [CrossRef]
- Dabholkar, A.R. Elements of Biometrical Genetics; Concept Publishing Company: New Delhi, India, 1992; p. 166. [Google Scholar]
- Ertiro, B.T.; Beyene, Y. Combining ability and testcross performance of drought-tolerant maize inbred lines under stress and non-stress environments in Kenya. Plant Breed. 2017, 136, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Talukder, M.Z.A.; Karim, A.S. Combining ability and heterosis on yield and its component traits in maize (Zea mays L.). Bangladesh J. Agric. Res. 2016, 41, 565–577. [Google Scholar] [CrossRef] [Green Version]
- Makumbi, D.; Betrán, J.F. Combining ability, heterosis and genetic diversity in tropical maize (Zea mays L.) under stress and non-stress conditions. Euphytica 2011, 180, 143–162. [Google Scholar] [CrossRef]
- San Vicente, F.; Bejarano, A. Analysis of diallel crosses among improved tropical white endosperm maize populations. Maydica 1998, 43, 147–153. [Google Scholar]

| Parent | Name | Description | Heterotic Group |
|---|---|---|---|
| P1 | DJL173833 | New inbred line | B |
| P2 | DJL173527 | New inbred line | A |
| P3 | DJL173887 | New inbred line | B |
| P4 | CL1211559 | New inbred line | B |
| P5 | CL1212902 | New inbred line | A |
| P6 | CML311 | Elite | A |
| P7 | CML312 | Elite | A |
| P8 | CML543 | Elite | B |
| P9 | CML566 | Elite | B |
| No. | Sites | Management | Year | Longitude | Latitude | Altitude (masl) | Annual Rainfall (mm) | Annual Temp Range (°C) |
|---|---|---|---|---|---|---|---|---|
| 1 | ART | Optimal | 2018 | 31°03′ E | 17°49′ S | 1480 | 830 | 13–28.5 |
| 2 | CIMMYT-Harare | Optimal | 2018 | 31°2′ E | 17°5′ S | 1483 | 1000 | 10–37 |
| 3 | CIMMYT Harare | Low Nitrogen | 2018 | 31°2′ E | 17°5′ S | 1483 | 1000 | 11–37 |
| 4 | RARS | Optimal | 2018 | 31°14′ E | 17°14′ S | 1300 | 918 | 12.8–28.6 |
| 5 | RARS | Low Nitrogen | 2019 | 31°14′ E | 17°14′ S | 1300 | 918 | 12.8–28.6 |
| 6 | RARS | Optimal | 2019 | 31°14′ E | 17°14′ S | 1300 | 918 | 12.8–28.6 |
| 7 | Lusaka West | Low Nitrogen | 2019 | 28°04′ E | 15°24′ S | 1216 | 1000 | 14.2–28.9 |
| 8 | Mpongwe South | Optimal | 2019 | 28°03′ E | 13°32′ S | 1206 | 1200 | 20–25.3 |
| Optimal Management | Managed Low Nitrogen | Across | ||||
|---|---|---|---|---|---|---|
| DF | MS | DF | MS | DF | MS | |
| Site | 4 | 128.28 *** | 2 | 108.50 *** | 7 | 803.91 *** |
| Replication (Site) | 5 | 2.26 | 3 | 5.31 *** | 8 | 4.17 *** |
| Cross | 35 | 26.41 *** | 35 | 1.75 | 35 | 20.92 *** |
| GCA | 8 | 64.19 ** | 8 | 0.90 | 8 | 44.75 * |
| SCA | 27 | 15.22 *** | 27 | 2.06 | 27 | 14.14 *** |
| Cross × Site | 140 | 3.73 ** | 70 | 1.21 * | 245 | 3.45 *** |
| GCA × Site | 32 | 5.89 ** | 16 | 0.71 | 56 | 6.48 *** |
| SCA × Site | 108 | 3.08 * | 54 | 1.431 * | 189 | 2.61 ** |
| Residual | 104 | 2.17 | 63 | 0.74 | 167 | 1.64 |
| GCA variance | 4.43 | 0.01 | 3.08 | |||
| SCA variance | 6.53 | 0.66 | 6.25 | |||
| GCA-SCA ratio | 0.68 | 0.02 | 0.49 | |||
| Phenotypic variance | 17.55 | 1.42 | 14.05 | |||
| Narrow-sense heritability | 0.51 | 0.02 | 0.44 | |||
| Broad-sense heritability | 0.88 | 0.48 | 0.88 | |||
| Optimal Management | Managed Low Nitrogen | Across | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parent | Name | Description | Heterotic Group | GCA (tha−1) | p-Value | Rank | GCA (tha−1) | p-Value | Rank | GCA (tha−1) | p-Value | Rank |
| P1 | DJL173833 | New | B | −0.44 | *** | 5 | −0.28 | ** | 9 | −0.39 | *** | 6 |
| P2 | DJL173527 | New | A | −0.11 | ns | 4 | 0.17 | ns | 1 | 0.00 | ns | 4 |
| P3 | DJL173887 | New | B | −0.6 | *** | 7 | −0.04 | ns | 7 | −0.39 | *** | 7 |
| P4 | CL1211559 | New | B | −0.51 | *** | 6 | −0.13 | ns | 8 | −0.37 | *** | 5 |
| P5 | CL1212902 | New | A | 1.40 | *** | 1 | 0.15 | ns | 2 | 0.93 | *** | 1 |
| P6 | CML311 | Elite | A | −1.08 | *** | 9 | −0.03 | ns | 6 | −0.68 | *** | 9 |
| P7 | CML312 | Elite | A | −0.90 | *** | 8 | −0.03 | ns | 5 | −0.58 | *** | 8 |
| P8 | CML543 | Elite | B | 0.93 | *** | 3 | 0.10 | ns | 3 | 0.62 | *** | 3 |
| P9 | CML566 | Elite | B | 1.31 | *** | 2 | 0.09 | ns | 4 | 0.85 | *** | 2 |
| Optimal Management | Low-Nitrogen Management | Across | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Cross | SCA (tha−1) | Prob_T | Rank | SCA (tha−1) | Prob_T | Rank | SCA (tha−1) | Prob_T | Rank |
| P2 × P1 | −1.95 | *** | 35 | −0.57 | ns | 32 | −1.43 | *** | 35 |
| P3 × P1 | −0.68 | ns | 28 | −0.08 | ns | 22 | −0.45 | ns | 29 |
| P4 × P1 | 0.35 | ns | 15 | 0.02 | ns | 19 | 0.21 | ns | 16 |
| P5 × P1 | 0.23 | ns | 17 | −0.19 | ns | 24 | 0.08 | ns | 17 |
| P6 × P1 | 0.66 | ns | 10 | −0.23 | ns | 26 | 0.32 | ns | 14 |
| P7 × P1 | −0.34 | ns | 27 | 0.35 | ns | 11 | −0.09 | ns | 22 |
| P8 × P1 | 0.65 | ns | 11 | 0.55 | ns | 4 | 0.64 | * | 8 |
| P9 × P1 | 1.08 | ** | 5 | 0.16 | ns | 14 | 0.73 | ** | 6 |
| P3 × P2 | −1.42 | *** | 34 | −1.13 | *** | 35 | −1.31 | *** | 34 |
| P4 × P2 | 0.25 | ns | 16 | 0.33 | ns | 12 | 0.28 | ns | 15 |
| P5 × P2 | 1.26 | ** | 3 | 0.06 | ns | 17 | 0.81 | ** | 5 |
| P6 × P2 | 1.86 | *** | 1 | 0.60 | ns | 3 | 1.41 | *** | 1 |
| P7 × P2 | 0.53 | ns | 13 | 0.52 | ns | 7 | 0.53 | ns | 10 |
| P8 × P2 | −0.31 | ns | 26 | 0.38 | ns | 10 | −0.07 | ns | 20 |
| P9 × P2 | −0.21 | ns | 25 | −0.20 | ns | 25 | −0.22 | ns | 25 |
| P4 × P3 | 0.85 | * | 7 | 1.07 | *** | 1 | 0.93 | *** | 3 |
| P5 × P3 | −0.20 | ns | 24 | 0.16 | ns | 13 | −0.08 | ns | 21 |
| P6 × P3 | 1.25 | ** | 4 | 0.54 | ns | 6 | 1.00 | *** | 2 |
| P7 × P3 | 0.75 | ns | 8 | 0.63 | * | 2 | 0.70 | * | 7 |
| P8 × P3 | 0.13 | ns | 18 | −0.68 | * | 34 | −0.18 | ns | 24 |
| P9 × P3 | −0.68 | ns | 29 | −0.51 | ns | 31 | −0.62 | * | 31 |
| P5 × P4 | −0.18 | ns | 23 | −0.28 | ns | 28 | −0.22 | ns | 26 |
| P6 × P4 | 0.07 | ns | 20 | 0.09 | ns | 16 | 0.07 | ns | 18 |
| P7 × P4 | −0.03 | ns | 21 | −0.25 | ns | 27 | −0.10 | ns | 23 |
| P8 × P4 | −1.17 | ** | 33 | −0.61 | * | 33 | −0.95 | *** | 33 |
| P9 × P4 | −0.15 | ns | 22 | −0.37 | ns | 29 | −0.22 | ns | 27 |
| P6 × P5 | −0.89 | * | 30 | −0.45 | ns | 30 | −0.74 | ** | 32 |
| P7 × P5 | 0.70 | ns | 9 | 0.14 | ns | 15 | 0.50 | ns | 12 |
| P8 × P5 | 0.07 | ns | 19 | 0.01 | ns | 20 | 0.05 | ns | 19 |
| P9 × P5 | −0.99 | * | 32 | 0.55 | ns | 5 | −0.41 | ns | 28 |
| P7 × P6 | −3.96 | *** | 36 | −1.42 | *** | 36 | −3.01 | *** | 36 |
| P8 × P6 | 0.62 | ns | 12 | 0.41 | ns | 9 | 0.53 | ns | 11 |
| P9 × P6 | 0.40 | ns | 14 | 0.46 | ns | 8 | 0.44 | ns | 13 |
| P8 × P7 | 0.90 | * | 6 | 0.03 | ns | 18 | 0.57 | * | 9 |
| P9 × P7 | 1.44 | *** | 2 | 0.00 | ns | 21 | 0.89 | ** | 4 |
| P9 × P8 | −0.89 | * | 31 | −0.09 | ns | 23 | −0.60 | * | 30 |
| Genotype | Cross | Optimal Management (tha−1) | Managed Low Nitrogen (tha−1) | Across (tha−1) |
|---|---|---|---|---|
| G1 | P2 × P1 | 5.27 | 0.83 | 3.61 |
| G2 | P3 × P1 | 6.18 | 1.16 | 4.28 |
| G3 | P4 × P1 | 6.93 | 1.10 | 4.76 |
| G4 | P5 × P1 | 8.75 | 1.24 | 5.94 |
| G5 | P6 × P1 | 6.91 | 0.96 | 4.68 |
| G6 | P7 × P1 | 5.97 | 1.54 | 4.34 |
| G7 | P8 × P1 | 8.82 | 2.01 | 6.25 |
| G8 | P9 × P1 | 9.71 | 1.45 | 6.65 |
| G9 | P3 × P2 | 5.54 | 0.59 | 3.67 |
| G10 | P4 × P2 | 7.43 | 1.92 | 5.37 |
| G11 | P5 × P2 | 10.21 | 1.95 | 7.10 |
| G12 | P6 × P2 | 8.41 | 2.35 | 6.13 |
| G13 | P7 × P2 | 7.22 | 2.24 | 5.34 |
| G14 | P8 × P2 | 8.23 | 2.16 | 5.95 |
| G15 | P9 × P2 | 8.68 | 1.58 | 6.03 |
| G16 | P4 × P3 | 7.45 | 2.44 | 5.56 |
| G17 | P5 × P3 | 8.27 | 1.79 | 5.83 |
| G18 | P6 × P3 | 7.20 | 2.07 | 5.27 |
| G19 | P7 × P3 | 6.96 | 2.11 | 5.13 |
| G20 | P8 × P3 | 8.12 | 0.93 | 5.42 |
| G21 | P9 × P3 | 7.65 | 1.11 | 5.17 |
| G22 | P5 × P4 | 8.36 | 1.28 | 5.70 |
| G23 | P6 × P4 | 6.15 | 1.49 | 4.41 |
| G24 | P7 × P4 | 6.25 | 1.17 | 4.34 |
| G25 | P8 × P4 | 6.95 | 0.93 | 4.68 |
| G26 | P9 × P4 | 8.34 | 1.18 | 5.64 |
| G27 | P6 × P5 | 7.13 | 1.15 | 4.91 |
| G28 | P7 × P5 | 8.90 | 1.83 | 6.25 |
| G29 | P8 × P5 | 10.15 | 1.79 | 7.02 |
| G30 | P9 × P5 | 9.43 | 2.34 | 6.78 |
| G31 | P7 × P6 | 1.80 | 0.07 | 1.15 |
| G32 | P8 × P6 | 8.12 | 2.01 | 5.82 |
| G33 | P9 × P6 | 8.34 | 2.11 | 6.01 |
| G34 | P8 × P7 | 8.57 | 1.65 | 5.97 |
| G35 | P9 × P7 | 9.55 | 1.57 | 6.58 |
| G36 | P9 × P8 | 9.05 | 1.63 | 6.29 |
| G37 | Check 1 | 6.65 | 0.65 | 4.39 |
| G38 | Check 2 | 8.75 | 1.95 | 6.22 |
| G39 | Check 3 | 10.06 | 2.64 | 7.28 |
| G40 | Check 4 | 8.88 | 1.75 | 6.20 |
| Heritability | 0.87 | 0.36 | 0.85 | |
| Genotype variance | 2.21 | 0.12 | 1.12 | |
| Genotype × Location variance | 0.67 | 0.26 | 0.82 | |
| Environment variance | 1.61 | 2.19 | 11.87 | |
| Residual variance | 2.11 | 0.74 | 1.58 | |
| Grand mean | 7.78 | 1.57 | 5.45 | |
| Least significance difference (5% probability level) | 1.16 | 0.55 | 0.87 | |
| Coefficient of variation | 18.66 | 54.98 | 23.08 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makore, F.; Magorokosho, C.; Dari, S.; Gasura, E.; Mazarura, U.; Kamutando, C.N. Genetic Potential of New Maize Inbred Lines in Single-Cross Hybrid Combinations under Low-Nitrogen Stress and Optimal Conditions. Agronomy 2022, 12, 2205. https://doi.org/10.3390/agronomy12092205
Makore F, Magorokosho C, Dari S, Gasura E, Mazarura U, Kamutando CN. Genetic Potential of New Maize Inbred Lines in Single-Cross Hybrid Combinations under Low-Nitrogen Stress and Optimal Conditions. Agronomy. 2022; 12(9):2205. https://doi.org/10.3390/agronomy12092205
Chicago/Turabian StyleMakore, Fortunate, Cosmos Magorokosho, Shorai Dari, Edmore Gasura, Upenyu Mazarura, and Casper Nyaradzai Kamutando. 2022. "Genetic Potential of New Maize Inbred Lines in Single-Cross Hybrid Combinations under Low-Nitrogen Stress and Optimal Conditions" Agronomy 12, no. 9: 2205. https://doi.org/10.3390/agronomy12092205
APA StyleMakore, F., Magorokosho, C., Dari, S., Gasura, E., Mazarura, U., & Kamutando, C. N. (2022). Genetic Potential of New Maize Inbred Lines in Single-Cross Hybrid Combinations under Low-Nitrogen Stress and Optimal Conditions. Agronomy, 12(9), 2205. https://doi.org/10.3390/agronomy12092205







