Responses of Soil Microbial Communities in Soybean–Maize Rotation to Different Fertilization Treatments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site and Design
2.2. Soil Sample Collection
2.3. Measurement of Soil Properties
2.4. Agronomic Indexes and Yield
2.5. Microbiological Analysis
2.6. Data Analysis
3. Results
3.1. Effect of Soybean–Maize Rotation and Fertilization Treatments on Soil Properties
3.2. Effects of Soybean–Maize Rotation and Fertilization Treatments on Soybean Growth
3.3. Effects of Soybean–Maize Rotation and Fertilization Treatments on Soybean Yield and Nitrogen Utilization
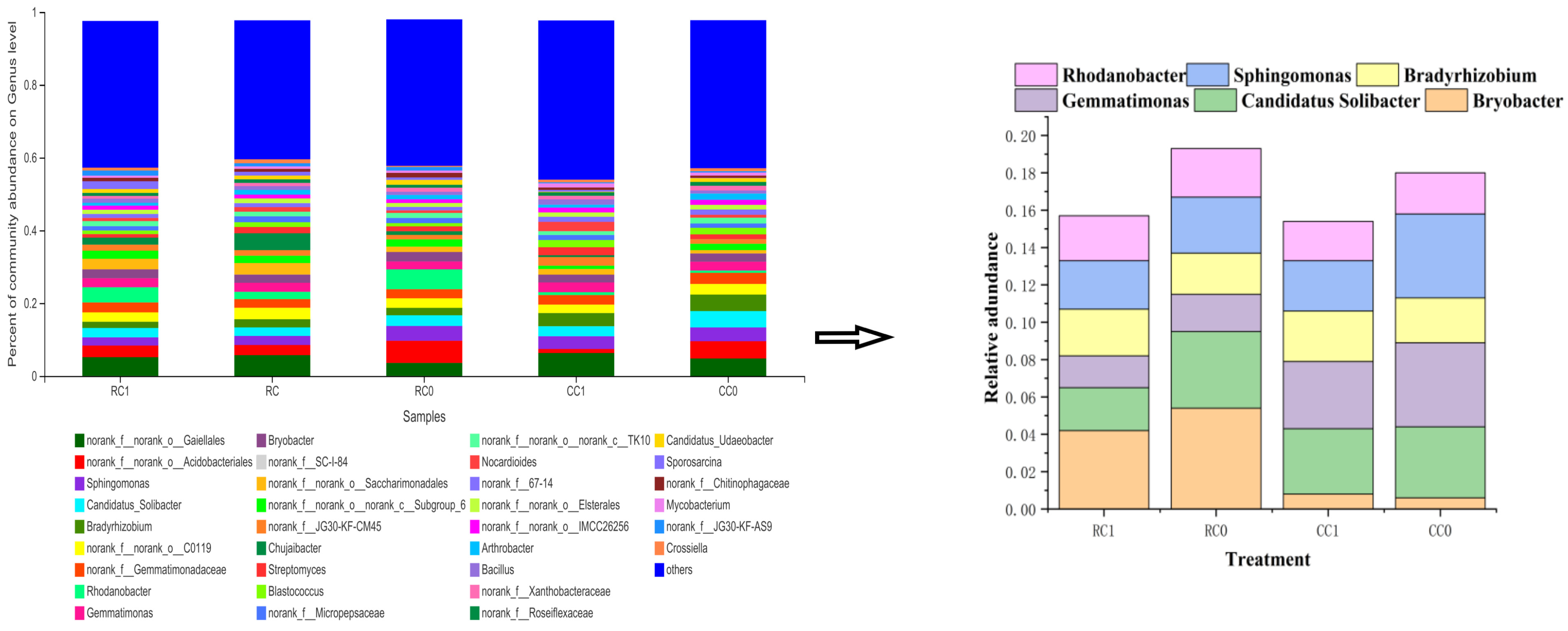
3.4. Response of Soil Bacterial Community to Soybean–Maize Rotation and Fertilization Treatments
3.4.1. Effects on Bacterial Diversity
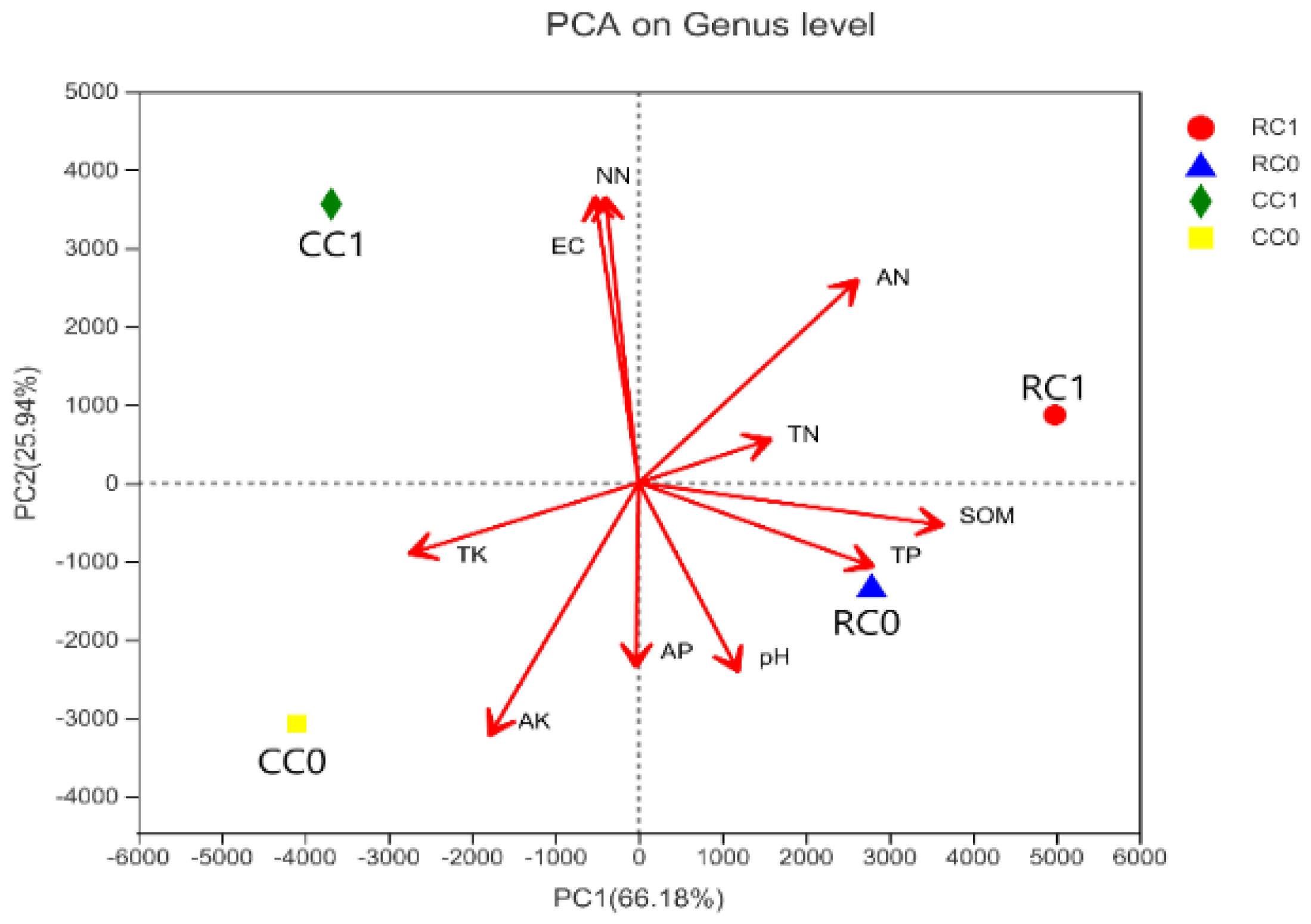
3.4.2. Soil-Bacterial-Community-Associated Environmental Factors
3.4.3. Correlation between Soil Bacterial Community Structure and Soil Environmental Factors
3.4.4. Molecular Ecological Network Structure
3.4.5. Molecular Ecological Network Structure
3.5. Effects of Soybean–Maize Rotation and Fertilization on Soil Fungal Community
3.5.1. Fungal Community Diversity in Soil
3.5.2. Fungal Species Composition
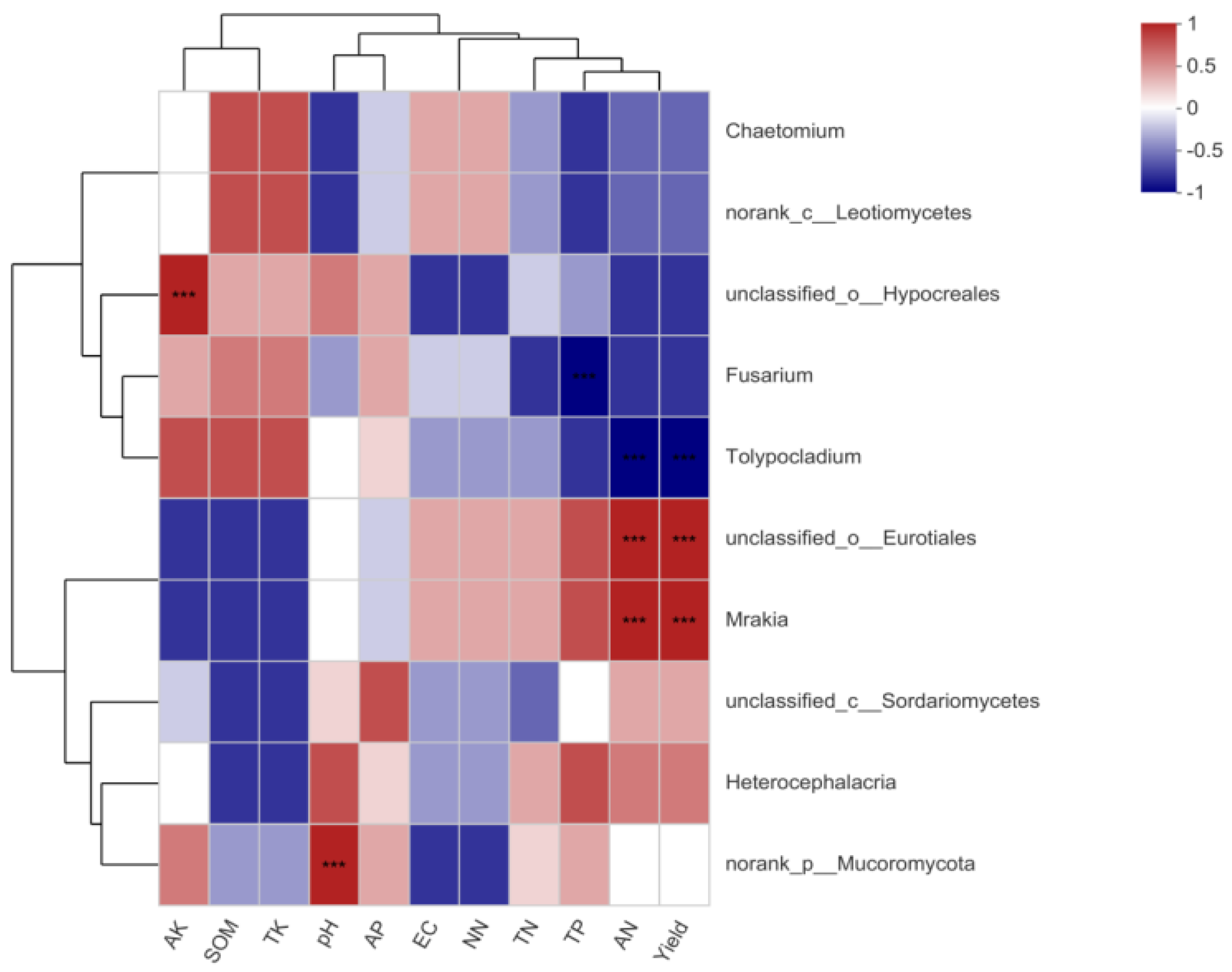
3.5.3. Correlation between Soil Fungal Community Diversity and Soil Environmental Factors
3.5.4. Molecular Ecological Network Structure of Fungal Community Diversity
3.5.5. FUNGuild Prediction of Soil Fungal Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hooper, D.U.; Chapin, F.S., III; Ewel, J.J.; Hector, A.; Inchausti, P.; Lavorel, S.; Lawton, J.H.; Lodge, D.M.; Loreau, M.; Naeem, S. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol. Monogr. 2005, 75, 3–35. [Google Scholar] [CrossRef]
- Ghimire, R.; Thapa, V.R.; Cano, A.; Acosta-Martinez, V. Soil organic matter and microbial community responses to semiarid croplands and grasslands management. Appl. Soil Ecol. 2019, 141, 30–37. [Google Scholar] [CrossRef]
- Meng, L.; Xia, Z.; Lv, J.; Liu, G.; Tan, Y.; Li, Q. Extraction and GC-MS analysis of phenolic acids in rhizosphere soil of Pinellia ternate. J. Radiat. Res. Appl. Sci. 2022, 15, 40–45. [Google Scholar] [CrossRef]
- Zeng, Z. Progress and perspective of legume-gramineae rotations. Zhongguo Shengtai Nongye Xuebao/Chin. J. Eco-Agric. 2018, 26, 57–61. [Google Scholar]
- Laberge, G.; IG Haussmann, B.; Ambus, P.; Høgh-Jensen, H. Cowpea N rhizodeposition and its below-ground transfer to a co-existing and to a subsequent millet crop on a sandy soil of the Sudano-Sahelian eco-zone. Plant Soil 2011, 340, 369–382. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.; Yi, L.; Hu, Y.; Zeng, Z.; Lin, C.; Tang, H.; Yang, G.; Xiao, X. Nitrogen release from incorporated 15 N-labelled Chinese milk vetch (Astragalus sinicus L.) residue and its dynamics in a double rice cropping system. Plant Soil 2014, 374, 331–344. [Google Scholar] [CrossRef]
- Vesterager, J.M.; Nielsen, N.E.; Høgh-Jensen, H. Nitrogen budgets in crop sequences with or without phosphorus-fertilised cowpea in the maize-based cropping systems of semi-arid eastern Africa. Afr. J. Agric. Res. 2007, 2, 261–268. [Google Scholar]
- Larkin, R.P.; Griffin, T.S.; Honeycutt, C.W. Rotation and cover crop effects on soilborne potato diseases, tuber yield, and soil microbial communities. Plant Dis. 2010, 94, 1491–1502. [Google Scholar] [CrossRef] [Green Version]
- Geisseler, D.; Linquist, B.A.; Lazicki, P.A. Effect of fertilization on soil microorganisms in paddy rice systems—A meta-analysis. Soil Biol. Biochem. 2017, 115, 452–460. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, N.; Chang, K.-F.; Hwang, S.-F.; Strelkov, S.E.; Conner, R.L.; McLaren, D.L.; Fu, H.; Harding, M.W.; Turnbull, G.D. Genetic diversity and aggressiveness of Fusarium species isolated from soybean in Alberta, Canada. Crop Prot. 2018, 105, 49–58. [Google Scholar] [CrossRef]
- Melero, S.; Vanderlinden, K.; Ruiz, J.C.; Madejon, E. Long-term effect on soil biochemical status of a Vertisol under conservation tillage system in semi-arid Mediterranean conditions. Eur. J. Soil Biol. 2008, 44, 437–442. [Google Scholar] [CrossRef]
- Zhang, X.; Duan, H.; Lu, A.; Li, H.; Wang, Y. Variation of rhizosphere soil microbial flora in healthy and black shank-susceptible tobacco fields at different growth stages. J. Henan Agric. Sci. 2018, 47, 63–69. [Google Scholar]
- Aschi, A.; Aubert, M.; Riah-Anglet, W.; Nélieu, S.; Dubois, C.; Akpa-Vinceslas, M.; Trinsoutrot-Gattin, I. Introduction of Faba bean in crop rotation: Impacts on soil chemical and biological characteristics. Appl. Soil Ecol. 2017, 120, 219–228. [Google Scholar] [CrossRef]
- Fanin, N.; Kardol, P.; Farrell, M.; Nilsson, M.-C.; Gundale, M.J.; Wardle, D.A. The ratio of Gram-positive to Gram-negative bacterial PLFA markers as an indicator of carbon availability in organic soils. Soil Biol. Biochem. 2019, 128, 111–114. [Google Scholar] [CrossRef]
- Liu, Z.; Ying, H.; Chen, M.; Bai, J.; Xue, Y.; Yin, Y.; Batchelor, W.D.; Yang, Y.; Bai, Z.; Du, M. Optimization of China’s maize and soy production can ensure feed sufficiency at lower nitrogen and carbon footprints. Nat. Food 2021, 2, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Simpson, M.J.; Otto, A.; Feng, X. Comparison of solid-state carbon-13 nuclear magnetic resonance and organic matter biomarkers for assessing soil organic matter degradation. Soil Sci. Soc. Am. J. 2008, 72, 268–276. [Google Scholar] [CrossRef]
- Bardgett, R.D.; Hobbs, P.J.; Frostegård, Å. Changes in soil fungal: Bacterial biomass ratios following reductions in the intensity of management of an upland grassland. Biol. Fertil. Soils 1996, 22, 261–264. [Google Scholar] [CrossRef]
- Carney, K.M.; Matson, P.A. Plant communities, soil microorganisms, and soil carbon cycling: Does altering the world belowground matter to ecosystem functioning? Ecosystems 2005, 8, 928–940. [Google Scholar] [CrossRef]
- Eilers, K.G.; Lauber, C.L.; Knight, R.; Fierer, N. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol. Biochem. 2010, 42, 896–903. [Google Scholar] [CrossRef]
- Meriles, J.M.; Gil, S.V.; Conforto, C.; Figoni, G.; Lovera, E.; March, G.J.; Guzman, C.A. Soil microbial communities under different soybean cropping systems: Characterization of microbial population dynamics, soil microbial activity, microbial biomass, and fatty acid profiles. Soil Tillage Res. 2009, 103, 271–281. [Google Scholar] [CrossRef]
- Trivedi, P.; He, Z.; Van Nostrand, J.D.; Albrigo, G.; Zhou, J.; Wang, N. Huanglongbing alters the structure and functional diversity of microbial communities associated with citrus rhizosphere. ISME J. 2012, 6, 363–383. [Google Scholar] [CrossRef] [Green Version]
- Thomson, B.C.; Ostle, N.; McNamara, N.; Bailey, M.J.; Whiteley, A.S.; Griffiths, R.I. Vegetation affects the relative abundances of dominant soil bacterial taxa and soil respiration rates in an upland grassland soil. Microb. Ecol. 2010, 59, 335–343. [Google Scholar] [CrossRef]
- Yin, C.; Jones, K.L.; Peterson, D.E.; Garrett, K.A.; Hulbert, S.H.; Paulitz, T.C. Members of soil bacterial communities sensitive to tillage and crop rotation. Soil Biol. Biochem. 2010, 42, 2111–2118. [Google Scholar] [CrossRef]
- Perez Brandan, C.; Arzeno, J.L.; Huidobro, D.J.; Conforto, E.C.; Grumberg, B.C.; Hilton, S.; Bending, G.D.; Meriles, J.M.; Vargas Gil, S. The Effect of Crop Sequences on Soil Microbial, Chemical and Physical Indicators and Its Relationship with Soybean Sudden Death Syndrome (Complex of Fusarium Species); 1695-971X; Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA): Madrid, Spain, 2014. [Google Scholar]
- Zhang, C.; Shu, D.; Wei, G. Soybean cropping patterns affect trait-based microbial strategies by changing soil properties. Appl. Soil Ecol. 2021, 167, 104095. [Google Scholar] [CrossRef]
- Fang, Y.; Xun, F.; Bai, W.; Zhang, W.; Li, L. Long-term nitrogen addition leads to loss of species richness due to litter accumulation and soil acidification in a temperate steppe. PLoS ONE 2012, 7, e47369. [Google Scholar] [CrossRef] [PubMed]
- Lennon, R.J.; Peach, W.J.; Dunn, J.C.; Shore, R.F.; Pereira, M.G.; Sleep, D.; Dodd, S.; Wheatley, C.J.; Arnold, K.E.; Brown, C.D. From seeds to plasma: Confirmed exposure of multiple farmland bird species to clothianidin during sowing of winter cereals. Sci. Total Environ. 2020, 723, 138056. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.P.; Montes-Molina, J.A.; Govaerts, B.; Six, J.; van Kessel, C.; Fonte, S.J. Crop residue retention enhances soil properties and nitrogen cycling in smallholder maize systems of Chiapas, Mexico. Appl. Soil Ecol. 2016, 103, 110–116. [Google Scholar] [CrossRef]
- Huang, X.; Wang, L.; Mei, Y.; Jia, Z.; Li, T.; Yu, G.; Ran, W. Towards a mechanistic understanding of microbial and nonmicrobial mediated topsoil organic carbon sequestration efficiency in a rice-wheat cropping system. Appl. Soil Ecol. 2022, 170, 104259. [Google Scholar] [CrossRef]
- Degrune, F.; Dufrêne, M.; Colinet, G.; Massart, S.; Taminiau, B.; Bodson, B.; Hiel, M.-P.; Daube, G.; Nezer, C.; Vandenbol, M. A novel sub-phylum method discriminates better the impact of crop management on soil microbial community. Agron. Sustain. Dev. 2015, 35, 1157–1166. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Shi, C.; Wei, D.; Gu, X.; Wang, Y.; Sun, L.; Cai, S.; Hu, Y.; Jin, L.; Wang, W. Soybean continuous cropping affects yield by changing soil chemical properties and microbial community richness. Front. Microbiol. 2022, 13, 5046. [Google Scholar] [CrossRef]
- Raglin, S.S.; Soman, C.; Ma, Y.; Kent, A.D. Long Term Influence of Fertility and Rotation on Soil Nitrification Potential and Nitrifier Communities. Front. Soil Sci. 2022, 2, 10. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, Z. Distribution and function of yeast in eco-environments. Environ. Sci. Technol. 2009, 32, 86–91. [Google Scholar]
- Detranaltes, C.; Saldanha, M.; Scofield, S.; Cai, G. First report of Fusarium commune causing root rot of soybean seedlings in Indiana. Plant Dis. 2022, 106, 3216. [Google Scholar] [CrossRef]
- Xu, H.; Yan, L.; Zhang, M.; Chang, X.; Zhu, D.; Wei, D.; Naeem, M.; Song, C.; Wu, X.; Liu, T. Changes in the density and composition of rhizosphere pathogenic Fusarium and beneficial Trichoderma contributing to reduced root rot of intercropped soybean. Pathogens 2022, 11, 478. [Google Scholar] [CrossRef]
- Jang, I.-H.; Lee, S. A review and perspective on soybean (Glycine max L.) breeding for the resistance to Phytophthora sojae in Korea. Plant Breed. Biotechnol. 2020, 8, 114–130. [Google Scholar] [CrossRef]
- Webster, R.W.; Roth, M.G.; Mueller, B.D.; Mueller, D.S.; Chilvers, M.I.; Willbur, J.F.; Mourtzinis, S.; Conley, S.P.; Smith, D.L. Integration of row spacing, seeding rates, and fungicide applications for control of Sclerotinia stem rot in Glycine max. Plant Dis. 2022, 106, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, S.J.; Campbell, B.M.; Ingram, J.S. Climate change and food systems. Annu. Rev. Environ. Resour. 2012, 37, 195–222. [Google Scholar] [CrossRef] [Green Version]
- Hooper, D.U.; Adair, E.C.; Cardinale, B.J.; Byrnes, J.E.; Hungate, B.A.; Matulich, K.L.; Gonzalez, A.; Duffy, J.E.; Gamfeldt, L.; O’Connor, M.I. A global synthesis reveals biodiversity loss as a major driver of ecosystem change. Nature 2012, 486, 105–108. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, W.; Zhang, X.; Yong, T.; Liu, W.; Su, B. Effects of shading at different growth stages on different traits and yield of soybean. Acta Agron. Sin. 2013, 39, 1871–1879. [Google Scholar] [CrossRef]
- Shi, A.; Chen, P.; Zhang, B.; Hou, A. Genetic diversity and association analysis of protein and oil content in food-grade soybeans from Asia and the United States. Plant Breed. 2010, 129, 250–256. [Google Scholar] [CrossRef]
- Chen, Y.-X.; Chen, X.-H.; Tang, Y.-Q.; Zhang, F.-S.; Chen, X.-P.; Zhang, C.-C.; Liu, J.; Xu, K.-W. Effect of nitrogen fertilizer on dry matter accumulation and yield in wheat/maize/soybean intercropping systems. Acta Prataculturae Sin. 2014, 23, 73. [Google Scholar]
- Morell, F.; Lampurlanés, J.; Álvaro-Fuentes, J.; Cantero-Martínez, C. Yield and water use efficiency of barley in a semiarid Mediterranean agroecosystem: Long-term effects of tillage and N fertilization. Soil Tillage Res. 2011, 117, 76–84. [Google Scholar] [CrossRef] [Green Version]
- Fan, K.; Delgado-Baquerizo, M.; Guo, X.; Wang, D.; Zhu, Y.-G.; Chu, H. Microbial resistance promotes plant production in a four-decade nutrient fertilization experiment. Soil Biol. Biochem. 2020, 141, 107679. [Google Scholar] [CrossRef]
- Sanginga, N.; Dashiell, K.E.; Diels, J.; Vanlauwe, B.; Lyasse, O.; Carsky, R.; Tarawali, S.; Asafo-Adjei, B.; Menkir, A.; Schulz, S. Sustainable resource management coupled to resilient germplasm to provide new intensive cereal–grain–legume–livestock systems in the dry savanna. Agric. Ecosyst. Environ. 2003, 100, 305–314. [Google Scholar] [CrossRef]
- Song, X.; Liang, H.; Huang, R.; Ke, C.; Tao, B.; Zhang, W. Mechanism underlying the response of fungi and their Fusarium symbiotic networks to the rotations of soybean and corn. Fungal Biol. 2022, 126, 609–619. [Google Scholar] [CrossRef]
- Kim, W.-S.; Krishnan, H.B. A nopA deletion mutant of Sinorhizobium fredii USDA257, a soybean symbiont, is impaired in nodulation. Curr. Microbiol. 2014, 68, 239–246. [Google Scholar] [CrossRef]
- Contador, C.A.; Lo, S.-K.; Chan, S.H.; Lam, H.-M. Metabolic analyses of nitrogen fixation in the soybean microsymbiont Sinorhizobium fredii using constraint-based modeling. MSystems 2020, 5, e00516–e00519. [Google Scholar] [CrossRef] [Green Version]
- Paudel, S.R.; Choi, O.; Khanal, S.K.; Chandran, K.; Kim, S.; Lee, J.W. Effects of temperature on nitrous oxide (N2O) emission from intensive aquaculture system. Sci. Total Environ. 2015, 518, 16–23. [Google Scholar] [CrossRef]
- Yamazaki, T.; Hozuki, T.; Arai, K.; Toyoda, S.; Koba, K.; Fujiwara, T.; Yoshida, N. Isotopomeric characterization of nitrous oxide produced by reaction of enzymes extracted from nitrifying and denitrifying bacteria. Biogeosciences 2014, 11, 2679–2689. [Google Scholar] [CrossRef] [Green Version]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Maslov, S.; Sneppen, K. Specificity and stability in topology of protein networks. Science 2002, 296, 910–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimitriu, P.A.; Grayston, S.J. Relationship between soil properties and patterns of bacterial β-diversity across reclaimed and natural boreal forest soils. Microb. Ecol. 2010, 59, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liang, Y.; Li, C.; Wang, F.; Sui, Y.; Suvannang, N.; Zhou, J.; Sun, B. Crop rotations alter bacterial and fungal diversity in paddy soils across East Asia. Soil Biol. Biochem. 2016, 95, 250–261. [Google Scholar] [CrossRef] [Green Version]








| Analytical Software/Database | Version Number | Use |
|---|---|---|
| Uparse | 7.0.1090 | OTU clustering |
| RDP Classifier | 2.11 | Sequence classification annotation |
| Usearch | 7 | OTU statistics |
| Mothur | 1.30.2 | Analysis of alpha diversity |
| PICRUSt | 1.1.0 | KEGG, COG, and Pfam functional predictions of the 16S sequence |
| SILVA | 138 | rRNA database |
| UNITE | 8 | Fungal ITS database |
| FunGene | 9.6 | Functional gene database |
| MaarjAM | 81 | Fungal 18S rRNA database |
| HPB | -- | 16S rRNA database of human pathogens |
| Funguild | 1 | Database of fungal functional annotation |
| MAFFT | 7.2 | Multiple sequence alignment |
| Treatment | EC (µS/cm) | pH | TN (g/kg) | TP (g/kg) | TK (g/kg) | SOC (g/kg) | AN (mg/kg) | AP (mg/kg) | AK (mg/kg) | NN (mg/kg) |
|---|---|---|---|---|---|---|---|---|---|---|
| RC1 | 27.22 ± 1.44 b | 5.6 ± 0.06 c | 0.6 ± 0.002 b | 1.17 ± 0.25 a | 6.66 ± 0.02 c | 76.6 ± 2.3 c | 4.69 ± 0.54 a | 48.4 ± 2.13 c | 76.59 ± 2.5 b | 14.45 ± 0.2 b |
| RC0 | 14.32 ± 0.75 d | 5.98 ± 0.02 a | 0.87 ± 0.004 c | 1.37 ± 0.02 b | 7.16 ± 0.06 a | 78.16 ± 0.93 c | 4.04 ± 0.11 b | 27.82 ± 0.48 c | 109.32 ± 2.65 b | 13.46 ± 0.05 a |
| CC1 | 42.98 ± 4.01 a | 5.49 ± 0.05 d | 0.66 ± 0.02 b | 1 ± 0.12 a | 7.2 ± 0.02 c | 66.99 ± 0.55 c | 3.67 ± 0.05 c | 57.27 ± 0.34 a | 97.01 ± 1.3 b | 16.44 ± 0.2 d |
| CC0 | 12.4 ± 0.99 a | 5.63 ± 0.02 c | 0.51 ± 0.1 a | 0.98 ± 0.09 b | 7.18 ± 0.06 a | 67.41 ± 1.48 c | 2.33 ± 0.41 a | 23.77 ± 0.6 b | 176.29 ± 0.19 a | 13.11 ± 0.13 c |
| Treatment | Height/cm | Stem Diameter/mm | Number of Nodes | |
|---|---|---|---|---|
| Rotation system | RC1 | 91.70 ± 4.16 a | 7.75 ± 0.38 a | 17.17 ± 0.31 a |
| RC0 | 89.72 ± 1.60 a | 6.42 ± 0.21 b | 16.30 ± 1.00 b | |
| Continuous cropping | CC1 | 95.21 ± 2.75 a | 8.13 ± 0.26 a | 17.11 ± 0.91 a |
| CC0 | 75.67 ± 0.96 b | 6.74 ± 0.17 c | 16.97 ± 0.56 ab | |
| Two-factor variance analysis(F) | ||||
| Cropping pattern Mode of fertilization Cropping pattern × Mode of fertilization | 8.084 * | 6.208 * | 2.418 | |
| 13.970 ** | 47.391 ** | 2.149 | ||
| 21.311 ** | 0.059 | 2.938 | ||
| Treatment | Yield/(kg/ha) | N-Fertilizer Agronomic Efficiency/[kg/(kg·hm2)] | Total Grain Weight/g | 100-Grain Weight/g | |
|---|---|---|---|---|---|
| Rotation cropping | RC1 | 2941.18 ± 101.89 a | 10.32 ± 0.36 c | 3.15 ± 0.07 a | 22.00 ± 0.80 a |
| RC0 | 2725.5 ± 290.17 ab | 12.12 ± 1.29 b | 2.86 ± 0.34 ab | 22.64 ± 0.51 a | |
| Continuous cropping | CC1 | 2623.53 ± 92.45 b | 21.87 ± 0.78 a | 2.52 ± 0.31 bc | 22.20 ± 1.78 a |
| CC0 | 2215.69 ± 97.08 c | 23.57 | 2.34 ± 0.20 c | 19.34 ± 1.61 b | |
| Two-factor variance analysis (F) | |||||
| Cropping pattern Mode of fertilization Cropping pattern × Mode of fertilization | 49.838 ** | 2095.038 ** | 135.903 ** | 43.805 ** | |
| 35.397 ** | 135.611 ** | 14.916 ** | 25.396 ** | ||
| 8.477 * | 30.929 ** | 2.946 | 18.228 * | ||
| Sample | Shannon | Chao1 | Coverage | Simpson |
|---|---|---|---|---|
| RC1 | 6.339251 | 3118.223 | 0.968376 | 0.004401 |
| RC0 | 6.3424 | 3241.878 | 0.96706 | 0.005188 |
| CC1 | 6.360753 | 3220.631 | 0.968456 | 0.004995 |
| CC0 | 6.30414 | 3246.365 | 0.966342 | 0.00628 |
| Metabolic Pathway | Enzyme Number | Enzyme | RC1 | RC0 | CC1 | CC0 |
|---|---|---|---|---|---|---|
| 1.7.2.6 | Hydroxylamine reductase | 99.50 | 63.00 | 69.00 | 85.00 | |
| Nitrification | 1.14.18.3 | Methane monooxygenase | 73.00 | 70.00 | 59.00 | 58.00 |
| 1.14.99.39 | Ammonia monooxygenase | 73.00 | 70.00 | 59.00 | 58.00 | |
| 1.7.2.5 | Nitric oxide reductase | 1241.33 | 898.65 | 2195.47 | 2564.67 | |
| Denitrification | 1.7.2.1 | Nitrite reductase | 2421.03 | 2099.65 | 3157.49 | 3542.19 |
| 1.7.99.1 | Hydroxylamine reductase | 649.03 | 649.43 | 769.32 | 664.72 | |
| 1.7.2.4 | Nitrous oxide reductase | 1270.53 | 1195.15 | 1502.66 | 1662.69 | |
| 1.7.7.2 | Nitrate reductase | 706.65 | 597.82 | 695.66 | 790.99 | |
| 3.5.5.1 | Nitrilase | 2242.52 | 2212.55 | 1845.52 | 1847.66 | |
| Ammoniation | 1.4.1.4 | Glutamate dehydrogenase | 3701.32 | 3346.57 | 3703.46 | 3292.57 |
| 1.4.1.2 | Glutamate dehydrogenase | 11,380.13 | 8804.01 | 8569.94 | 9067.96 | |
| 2.7.2.2 | Carbamate kinase | 2353.5 | 1761.50 | 1325.72 | 1261.13 | |
| 3.5.1.49 | Formamidase | 3708.94 | 3123.03 | 2036.37 | 2043.31 | |
| 4.2.1.104 | Hydrogenase | 2813.80 | 2884.13 | 1729.50 | 1805.40 | |
| 1.18.6.1 | Nitrogenase | 2846.84 | 3032.04 | 1438.33 | 1660.12 | |
| Nitrogen fixation |
| Sample | Shannon | Chao1 | Coverage | Simpson |
|---|---|---|---|---|
| RC1 | 3.275589 | 252.5172 | 0.999179 | 0.077116 |
| RC0 | 3.528299 | 299.3871 | 0.998921 | 0.056446 |
| CC1 | 3.249732 | 233.3226 | 0.999155 | 0.075076 |
| CC0 | 3.495963 | 267.75 | 0.999179 | 0.057651 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhang, L.; Meng, F.; Lou, Z.; An, X.; Jiang, X.; Zhao, H.; Zhang, W. Responses of Soil Microbial Communities in Soybean–Maize Rotation to Different Fertilization Treatments. Agronomy 2023, 13, 1590. https://doi.org/10.3390/agronomy13061590
Wang Y, Zhang L, Meng F, Lou Z, An X, Jiang X, Zhao H, Zhang W. Responses of Soil Microbial Communities in Soybean–Maize Rotation to Different Fertilization Treatments. Agronomy. 2023; 13(6):1590. https://doi.org/10.3390/agronomy13061590
Chicago/Turabian StyleWang, Yunlong, Liqiang Zhang, Fangang Meng, Zixi Lou, Xiaoya An, Xinbo Jiang, Hongyan Zhao, and Wei Zhang. 2023. "Responses of Soil Microbial Communities in Soybean–Maize Rotation to Different Fertilization Treatments" Agronomy 13, no. 6: 1590. https://doi.org/10.3390/agronomy13061590






