Abstract
Understanding the cytological pattern of genome size and ploidy level of the bermudagrass (Cynodon dactylon) is vital to explore the evolution pattern and breeding of the species. To study the diversification of the cytological pattern of bermudagrass along the longitudinal gradient, the genome size and ploidy level were measured and explored with the relationship to climate factors. The corresponding ploidy level was verified through the mitotic chromosome counts method. Bermudagrass accessions ploidy level included diploids, triploid, tetraploid, pentaploid and hexaploid with a basic chromosome number of x = 9. The major ploidy level was tetraploid (45%) and aneuploidy was commonly discovered in collected regions. Mean genome size of bermudagrasswas was estimated to be 1.31 pg/1Cx along longitudinal gradient. The 1Cx values of diploid were higher than that of triploid and tetraploid, while the tetraploid had minimum basic genome size. In the current study, we observed that genome downsizing exists in tetraploids of Cynodon dactylon. Tetraploids have a wider distribution than other ploidy levels, especially in arid areas, occupying a relatively high proportion. In addition, at the same ploidy level, genome size was remarkably variable in the current study. The coefficient of determination analysis showed that longitude and mean annual rainfall were significantly correlated to genome size rather than ploidy level. This cytological study will be helpful for further genetic mechanisms and molecular characteristics to landscape adaptation of bermudagrass.
Keywords:
Cynodon dactylon; flow cytometry; genome size; polyploidy; genome downsizing; longitude; climate 1. Introduction
The grass family (Gramineae or Poaceae) includes about 789 genera and 11,738 species; the genus Cynodon belongs to one of them [1]. Cynodon genera include nine species and several varieties [2], widely used for turfgrass, forage, soil stabilization and remediation, and phytoremediation [3]. Bermudagrass (Cynodon dactylon (L.) Pers.) is native to Africa and widely distributed in tropical and subtropical regions worldwide [4,5]. In China, bermudagrass is distributed in the reaches of the Yellow River and its south and also sparsely distributed in Xingjiang, Tibet, and Yunnan province [6,7]. The extensive geographic distribution indicated that huge genetic variation potential existed in the bermudagrass resource [8]. Bermudagrass accessions were considered autoploid and included diploids, triploids, tetraploids, pentaploids, and hexaploids, with a basic chromosome number of x = 9 [9,10].
Studies of plant genome size and ploidy level play an important role in the progress of genomics. The Feulgen microspectrophotometry of root tip or shoot tip mitotic cells and the primordial digestion maceration technique have been applied for estimating the genome size and ploidy level [11,12,13]. Flow cytometry is used to analyze cells and any estimated change in ploidy level and genome size that has been applied to determine it in several turfgrasses [14,15,16,17]. The method of measuring the genome size and ploidy level of bermudagrass was first observed in the late 20th century [18]. The ploidy determination provides a cytological basis for genetic diversity studies [16].
Bermudagrass is worldwide in distribution and has huge genetic variation. There are quite a large number of research on the genus Cynodon. There are relevant experimental studies on variation in ploidy level and genome size of Cynodon dactylon along a latitudinal gradient [10]. However, latitude and longitude have different climates, such as temperature, rainfall, and precipitation, that affect plant traits and evolution differently. The mean annual precipitation decreases along longitude from east to west with the increase in altitude. Longitude is the main factor for studying the distribution of species. The distribution of Carex physodes was controlled by longitude. Carex physodes is basically distributed in the longitude range of 84° E~91° E of the Gurbantunggut Desert. [19]. Genetic isolation of longitude gradient induced by mountain environment has also been found in other widespread Eurasian plants [20]. In addition, studies have found that the number of species decreases with increasing latitude. Before 86° E, richness decreased sharply with the increase of longitude, but after 86°E, the number of species increased with elevation (from west to east) in the Gurbantunggut Desert [19]. Meanwhile, the phenomenon of genome decreasing and increasing ploidy levels is often observed in polyploids [21]. Genome downsizing is defined as the mean DNA amount per basic genome decreased along with increasing ploidy level. This phenomenon is common in angiosperms. For example, it occurs in six monophyletic subdivisions of angiosperms (i.e., monocots, all eudicots, Caryophyllales, all core eudicots, asterids, and rosids [21,22,23]. A considerable change in nuclear DNA amount could be induced by environmental stress such as sudden heat, frost, and drought [24,25]. In many polyploids, natural selection on smaller c-values may simply be the main evolutionary force driving genome downsizing. Genome downsizing may result from the elimination of some specific DNA sequences and loss of chromosomal fragments in some species following polyploid formation. The occurrence of this phenomenon was observed after the formation of polyploid triticale [26,27,28]. It is also possible that genome downsizing is associated with the activation of retrotransposons. The extent to which polyploids lose DNA through this mechanism may depend on the species, as the degree of homologous recombination may vary among different polyploids [22]. The examples provided by the wheat model showed that genome downsizing may be a mechanism facilitating the success of polyploid speciation events. Such genetic modifications may favor polyploids to gain stability at the cytological and genetic levels [29].
There are several pieces of research on Cynodon dactylon (L.). However, the longitudinal factors driving the evolution of genome size and the genome downsizing phenomenon in Cynodon dactylon (L.) have never been investigated before. Here, we propose the hypothesis that the genome downsizing phenomenon exists in Cynodon dactylon (L.), and it would have a characteristic of adaptation to the mean annual rainfall by changing not ploidy level but only genome size at the cellular level. Therefore, this study has two objectives: (1) Evaluate the cytological pattern (ploidy level and genome size) of bermudagrass collected from different longitudes. (2) Explore the relationship between ploidy level, genome size, climate, and longitude in contrast with the latitude. Moreover, this cytological study could be beneficial to enhance our knowledge of the evolutionary biology of bermudagrass and will provide further development of more genetic resources for the breeding of bermudagrass.
2. Materials and Methods
2.1. Plant Materials Collection
During the season of vigorous growth in July 2016, 260 bermudagrass individuals were collected from the east to the west in 13 different regions along a continuous longitude gradient in China. Bermudagrass was collected in the regions from 105 to 119° E of longitude and 34° N of latitude (Figure 1). The experimental fields were in the range of 7 °C to 13 °C for mean annual temperature (MAT). Meteorological data from about 30 years (1981–2010), including mean average rainfall (MAR) and MAT, were recorded for the 13 sampled regions (Table 1). Mean annual precipitation and temperature at the collection sites were provided by the China Meteorological Administration. These sampled individuals were taken from each region, and the distance between each individual was about 5 m. All bermudagrass individuals were sampled and transplanted to the experimental field on Zhoukou farm. However, some of them did not survive because of weather or other environmental factors. Finally, 227 plants were analyzed with 15–19 individuals per population by flow cytometry at the Henan Academy of Agricultural Sciences soon afterward (Table 1).
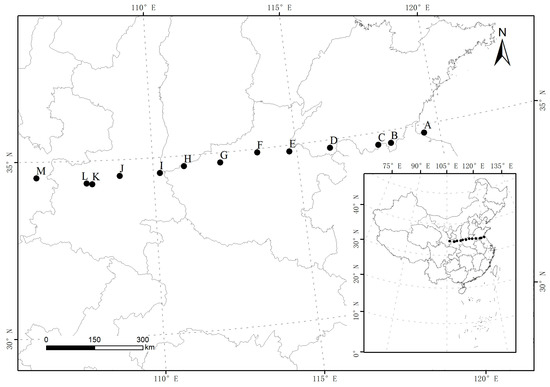
Figure 1.
Distribution of sampling sites. A, Lianyungang; B, Tancheng; C, Zaozhuang; D, Shanxian; E, Lankao; F, Zhengzhou; G, Luoyang; H, Sanmenxia; I, Tongguan; J, Jingyang; K, Fufeng; L, Baoji; M, Tianshui.

Table 1.
Geography and climate information of bermudagrass populations collected from 13 different longitude regions. Formatting of mathematical components.
2.2. Flow Cytometry Analysis
Flow cytometry (Cube8, Partec, Münster, Germany) was used to estimate genome size and ploidy level of bermudagrass at the China Henan Academy of Agricultural Sciences. Fresh leaf samples were taken from the experimental field to the lab for flow cytometry analysis. Preparation for samples was followed using a reagent kit of CyStain® PI to determine genome size (Sysmex Partec gmbH, Goerlitz, Germany), as mentioned below. Firstly, approximately 0.5 cm2 (or less) of fresh and tender leaves were put in a plastic petri dish. Secondly, a 500 uL nuclei extraction buffer was added. Thirdly, a sharp razor blade was used to cut up the leaf tissues for 30–60 s, and then incubated for 30–90 s and filtered through a 50 uM CellTrics® filter into a sample tube. Moreover, 2 mL of propidium iodide was added. Finally, the solution was incubated for 30–60 min, protected from light at room temperature, and then filtered through a 30 uM nylon mesh into a 5 mL test tube for the next analyses. In this study, we selected leaves of Pisum sativum L. as the internal standard because the genome size of Pisum sativum L. 4.397 pg/1Cx was reported in a previous study and suitable for the investigation of bermudagrass or grass family (Plant DNA C-values database accessed on 18 March 2017 http://www.kew.org/cval/) [30,31]. At least 5000–10,000 cell nuclei were analyzed per sample. We analyzed the data of genome size by CyFlow Cube 13 software. Three measurements (three replications) were obtained for each sample, and the sample genome size was calculated by using the formula: sample genome size (pg/1Cx) = [(mean value of the sample peak)/(mean value of the internal standard)] × known genome size of internal standard. The genome size values: base-pair numbers was 1 pg: 978 Mbp [31]. Ploidy levels of bermudagrass were assessed by comparing genome size with thresholds reported in [18]. The coefficient of variation (CV) of the G0/G1 peak for all samples was measured, which can be used to assess the integrity of nuclei and the variability of the DNA staining (Supplementary Table S1). The CV < 4 indicates the reliability of the experimental trial to show the ploidy level [30].
2.3. Root Fixation and Chromosome Preparation
In order to confirm the results of ploidy level based on the flow cytometry estimation, root tip samples of bermudagrass were used for chromosome counting. The sandy soil was used to cultivate the stolon for further production of adventitious roots. Roots were sampled in the early morning and counted by chromosome tablet method when growing up to 0.5–0.8 cm. First, apical tissues were incubated with 0.05% colchicine for 4 h. Next, apical tissues were rinsed with water for 10 min. After that, apexes were fixed for 24 h using Carnoy’s fixative (anhydrous alcohol: glacial acetic acid = 3:1), and then samples were washed three times using sterile water. Finally, apexes were put into 70% ethanol for about 10 min. If longer storage was required, then soaked samples would need to stay at a constant temperature of 4 °C. For chromosome preparation, we first dissociated the cell walls of the apical root in HCL (1 mol/L) for 9 min at 60 °C and then flushed for 30 min with flowing water. The slides were counter-stained with modified carbonate fuchsin and examined under a Zeiss Scope.A1 fluorescence microscope (made in Germany). More than 10 cells at metaphase were observed per operation using an optical microscope. There were representative figures of chromosome counts for each ploidy level.
2.4. Data Analysis
Maximum, minimum, mean standard deviation (SD), and coefficient of variation (CV) for genome size at each longitude site were estimated by using SPSS 22.0. ANOVA was used to calculate the significant differences among populations and within populations for genome size and ploidy level of bermudagrass. Pearson’s correlation coefficients were used to analyze genome size, ploidy level, longitude, and climate shown by heat map using HemI 1.0.3.3 software. Moreover, regression analysis was applied to estimate the relationship between genome size, longitude, and environmental factors and the optimal fitted standard regression equation. All statistical analyses were conducted using Excel and SPSS 22.0 for Windows (SPSS Inc. Chicago, IL, USA).
3. Results
3.1. Genome Size of Bermudagrass along Longitude Gradient
Genome size estimations of bermudagrass individuals ranged from 0.68 pg/1Cx in Baoji to 2.24 pg/1 Cx in Tancheng (Figure 2). The mean genome size of all bermudagrass individuals was 1.31 pg/1Cx (Table 2). Genome size was significantly correlated to longitude. The genome size would be smaller in regions at lower longitude, which indicated that longitude was an extremely important factor in genome size variation. The relationship between longitude and genome size was interpreted well by a quadratic curve (r = 0.267, p = 0.000) (Figure 3). In addition, genome size was significantly influenced by MAR (mean average rainfall). Bermudagrass individuals with a smaller genome size are more frequent in dry habitats. Regression analysis showed that the quadratic could explain the relationship between genome size and MAR (r = 0.350, p = 0.000) (Figure 4).

Figure 2.
Maximum, minimum, median, and interquartile range of genome size of bermudagrass in different longitudes were shown in Box-plot. “●” represented outliers greater than 1.5 times the interquartile range, while “*” represented outliers greater than 3 times the interquartile range.

Table 2.
Genome size of bermudagrass populations along longitudinal gradient.
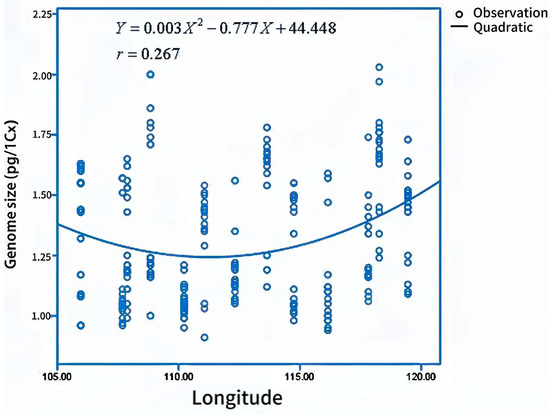
Figure 3.
Regression analysis between genome size and longitude.

Figure 4.
Regression analysis between genome size and MAR.
3.2. Ploidy Level of Bermudagrass in Different Sites
The method of chromosome counts was applied to confirm the accuracy of the ploidy level of bermudagrass. The reliable ploidy of bermudagrass was determined by flow cytometry in this study. The observed chromosome in bermudagrass was x = 9. The number of chromosomes observed under light microscopy is consistent with the ploidy level inferred by previous flow cytometry (Figure 5). Based on the position of referred Pisum sativum L. as the internal standard and the value of genome size, six different ploidy levels of bermudagrass were detected such as diploid, triploid, tetraploid, pentaploid, hexaploid, and aneuploid (Figure 5). Tetraploids (45%) were the most common cytotype across all the bermudagrass individuals. The second was pentaploid (18.3%), followed by hexaploid (17.1%), while the aneuploid was ubiquitous in most regions (9.2%). The ploidy level with the least number of individuals was diploid (5%), which was slightly lower than that of triploid (5.4%) (Table 3 and Figure 6). As regards the distribution pattern of ploidy levels along the longitudinal gradient, tetraploids have a wider distribution than other ploidy levels, especially in arid areas, occupying a relatively high proportion (40.7%). There are also many tetraploid individualities distributed in low elevations (Table 3). Unlike genome size, ploidy level was not highly correlated with longitude, although ploidy level was significantly associated with genome size. The same phenomenon was also found between MAR and ploidy level along the longitude gradient.
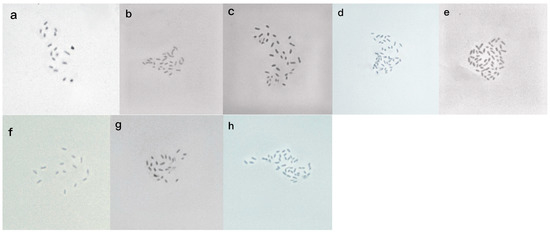
Figure 5.
Mioic metaphase chromosomes of bermudagrass individuals with different ploidy levels as follows: (a) diploid (2n = 18); (b) triploid (2n = 27); (c) tetraploid (2n = 36); (d) pentaploid (2n = 45); (e) hexaploid (2n = 54); (f) aneuploidy (2n = 14); (g) aneuploidy(2n = 23); (h) aneuploidy (2n = 43).

Table 3.
The distribution of polyploids of bermudagrass in different longitudes.

Figure 6.
Flow cytometric histogram of representative ploidy levels of bermudagrass individuals: (a) diploid; (b) triploid; (c) tetraploid; (d) pentaploid; (e) hexaploid.
3.3. Relationship between Ploidy and Genome Size
At the same ploidy level, the genome size of bermudagrass individuals was remarkably variable. In the meantime, the mean 1Cx genome size of different ploidy levels showed that hexaploid had a maximum value of genome size, while aneuploid had higher genome size than diploid, triploid, and tetraploid (Table 4). In addition, 1Cx values of triploid and tetraploid were lower than that of diploid, while the minimum 1Cx values existed in tetraploid. ANOVA analysis showed that significant statistical differences existed in genome size and ploidy level along the longitude gradient (Table 5).

Table 4.
The variation of genome size in different ploidy levels.

Table 5.
ANOVA analysis of genome size and ploidy level for each population.
4. Discussion
4.1. Genome Size Variation of Bermudagrass along the Longitudinal Gradient
In plants, the change in genome size was considered to be correlated with some geographical and environmental factors [32,33,34]. It was clearly indicated that significant differences in genome size existed in different longitude sites. The coefficient of determination analysis indicated that genome size was significantly associated with longitude. Previous reports have confirmed this conclusion [35,36], while genome size was not significantly correlated to the geographical distance matrix along the latitude gradient by Mantel tests. The quadratic curve can properly highlight the relationship between genome size and latitude, which was consistent with the tendency between genome size and longitude [10]. In addition, genome size was also significantly correlated with MAR and genome size as well. The areas with less rainfall have smaller genome sizes, but their variation could not be interpreted by MAR because the rainfall gradient is usually deemed to be the proxy for a significant correlation between genome size and longitude [37]. The large genome constraint hypothesis predicts that populations with large genomes are restricted to narrower environmental ranges because they do not thrive in extreme environments. The result of this research revealed that smaller genome sizes appeared at the low longitude regions, which supports the large genome constraint hypothesis that species with small genome sizes are able to adapt to environmental variations owing to being short of evolutionary limitations linked with a great deal of repetitive DNA [34,38]. Plants of smaller genome sizes usually have shorter cell cycles in order to enable rapid plant growth during the period of harsh climates. There was a temporary link between higher genome size and reduced capability of adaption for the extreme and swift environment [39,40]. Bermudagrass with a smaller genome size was believed to be more successful under stressful conditions such as drought at lower longitudes in the current study. Survival and reproduction of species with higher genome size could demand more energy supply and longer growth cycles, which may put it at a selective disadvantage in source shortage environments [41]. Species with large genomes tend to be excluded from extreme environments with a short growing season [32]. Environment (altitude, temperature, and drought) appears to play a key role in shaping genome size by imposing selective pressure on physiological and life-history traits associated with genome size. In different environments, plants may control gene dosage by changing genome size so that their gene expression, adaptation mechanism, physiological mechanism, and traits change to adapt to different environments [37,38].
4.2. Polyploidy of Bermudagrass along a Longitudinal Gradient
Polyploidy is necessary for the speciation process in plants and is mainly energy-driving to biodiversity in angiosperms [42,43]. Comparative genomics is a further step to confirm that one or more polyploidization events are experienced for their ancestry [44,45]. A certain percentage of diploid, triploid, tetraploid, pentaploid, and hexaploid was observed, and the most common ploidy level was tetraploid. The probability of polyploidy was as high as 85% in this study. The ploidy levels of bermudagrass have a similar distribution in latitude [10]. After polyploidization, a series of changes in genome structure and gene expression will cause the polyploid to exhibit a new phenotype different from its diploid ancestor. This change will facilitate the polyploid to enter a new ecological niche [46,47]. Therefore, polyploids may be an evolutionary mechanism to adapt to various environments, and tetraploids could be one of the most adaptable ecotypes in bermudagrass. Tetraploids have a wider distribution than other ploidy levels in our study. In addition, due to intergenomic recombination and multi-genome inheritance, plant polyploidy can accelerate epigenetic inheritance and thus rapidly adapt to the ecological environment [48,49,50]. In latitude, ploidy levels of mid-latitudes are lower than that of high and low latitudes [10]. Longitude has no regular effect on ploidy level, which supports previous studies showing that plant ploidy level did nothing to their geographical conditions, such as little bluestem and buffalograss [51,52]. The complex ploidy level composition of each site and narrow range of temperature may be contributed to the unnoticeable effect between ploidy level and longitude variation in this study. Temperature appears to be the most important factor affecting the polyploid distribution [53]. It has been shown to produce changes in cell division, cell expansion, and DNA endoreduplication, with effects on cell size and the number of cells as well [54]. In the study of cabbage flowers, it is found that there is a correlation between ploidy levels and cell size. The formation of large differentiated cells is accompanied by increased ploidy levels [55]. At the sampling point involved in our experiment, MAT ranged from 6.9 °C in Tianshui to 10.6 °C in Lianyungang. The temperature variation amplitude is too small. Ploidy level was not highly correlated with longitude in our study. It seems that the latitude significantly affected the change of ploidy level rather than the longitude in bermudagrass. By comparing the temperature of longitude and latitude, it is clear that the range of temperature in latitude is wider than in longitude. Cold weather at higher latitudes can increase percentages of polyploidization [56]. A warm environment at low latitude would be more suitable for survival and growth. The environmental pressure is smaller than other collected regions, which can cause the phenomenon that higher ploidy levels happen in bermudagrass. Polyploids have the ability to grow faster and produce more seeds than diploids [57,58]. From the above facts, C. dactylon in different latitudes and longitudes suggested that polyploidization might be an adaptation mechanism.
4.3. The Relationship between Genome Size and Ploidy Level in Bermudagrass along the Longitudinal Gradient
At the same ploidy level, genome size was remarkably variable in this study, possibly owing to the variation of chromosomal rearrangements and the occurrence of supernumerary chromosomes [41,59,60]. For example, the amplification and insertion of transposable elements or the evolution and amplification of satellite repeats [61,62]. Changes in genome size play an important role in speciation [21]. On the distribution pattern of ploidy levels along the longitude gradient, we found that tetraploids had lower 1Cx values than diploids. The phenomenon is called genome downsizing [22]. Genome downsizing is widespread in angiosperms [21,22,23]. Some studies have shown that this phenomenon can occur by various recombinational mechanisms, such as unequal homologous recombination between homologous chromosomes, sister chromatids, interchromatin [63,64,65,66], and illegitimate recombination [66]. Species with smaller genomes have more small stomata and higher leaf vein densities [67]. A high density of small stomata can better absorb CO2 while also effectively improving water use efficiency [68,69]. In a nutrient-poor environment, natural selection may put pressure on the organism to eliminate unnecessary repetitive DNA sequences, thus reducing DNA amount and reducing the biochemical costs associated with this extra DNA [22]. Under these circumstances, individuals with smaller genome sizes may be more dominant when competing with individuals with larger genome sizes [69]. In the distribution pattern of ploidy levels along the longitudinal gradient, tetraploids have a wider distribution than other ploidy levels. Especially in arid areas, tetraploids occupy a relatively high proportion. Genome downsizing expands the range of final cell size. Tetraploids may reduce the minimum cell size by shrinking the genome size to optimize the trade-off between photosynthesis and water use efficiency to adapt to a wider range of habitats [32,67,69,70]. Such genetic modification may be beneficial for polyploids to gain stability at the cytological and genetic levels [65]. The findings of this study will provide further knowledge of more genetic resources for bermudagrass breeding.
4.4. Aneuploidy of Bermudagrass Is Ubiquitous
Aneuploidy is put down to segregation errors and non-disjunction during cell division via meiosis or mitosis. Aneuploidy was not only found in bermudagrass [71] but also in many plant species discovered, such as sugarcane and wild switchgrass [72,73]. The appearance of a fragile site could further interpret the occurrence of aneuploidy in plants, which leads to an increase in the number of chromosomes. In plants, the existence of fragile sites has been first studied in Lolium spp. Studies have found that the 45S rDNA regions are chromosome-fragile sites in Lolium spp., expressed as gaps in vitro on metaphase chromosomes of Root-Tip meristematic cells [74]. Bermudagrass may have fragile sites with chromosome numbers that may be lower than the aneuploidy numbers. There may also be other reasons to cause the phenomenon of aneuploidy in the bermudagrass, which needs further research. It is a stable genome that plants need in order to live, and higher instability than polyploidy existence in aneuploidy because of adding or subtracting the number of chromosomes [75,76]. The presence of aneuploidy is harmful to species in most circumstances, but it can enhance phenotypes sometimes when it occurs without serious harm to growth [74,77,78]. Mostly bermudagrass is persistent by vegetative propagation, which allows bermudagrass to bypass the “meiotic filter” and keep growing, just like the growth pattern of sugarcane [79].
5. Conclusions
The mean genome sizes of different ploidy levels showed that hexaploids had a maximum value of genome size, while the minimum genome size existed in tetraploids. Genome downsizing was found in tetraploids of bermudagrass. Tetraploids are widely distributed in arid regions. The individuals of bermudagrass with smaller genome sizes could have the ability to adapt to dry and complex geographic conditions. In cytological patterns, bermudagrass transformed the genome size rather than the ploidy level to adapt to the longitudinal environment variation. Polyploidization and existing aneuploidy of bermudagrass were specific phenomena to respond to the different geographic regions. Longitude had little effect on the ploidy level; no regular pattern was observed in the bermudagrass. It was observed that the plant ploidy level had no link to their geographical conditions. The results of the cytological landscape study will provide a base for further understanding of the genetic mechanism and molecular characteristics for the landscape adaptation of bermudagrass.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy13081984/s1, Table S1: The values of particles, mean, median and CV of each sample in Cynodon dactylon and Pisum sativum L. were measured by flow cytometry.
Author Contributions
Conceptualization, M.L., M.W. and J.Z.; data curation, M.L. and M.W.; formal analysis, M.L., M.W., J.Z., G.F., Z.G., Y.G. (Yuxia Guo) and Y.G. (Yongzhuo Guan); investigation, Z.G., Y.G. (Yuxia Guo) and Y.G. (Yongzhuo Guan); methodology, X.Y., M.L., G.F., Z.G., Y.G. (Yuxia Guo) and Y.G. (Yongzhuo Guan); resources, M.L., M.W. and J.Z.; software, X.Y., M.L., M.W., J.Z., G.F., Z.G., Y.G. (Yuxia Guo) and Y.G. (Yongzhuo Guan); supervision, X.Y., G.F., Y.G. (Yuxia Guo) and Y.G. (Yongzhuo Guan); validation, Z.G.; visualization, X.Y., M.L., G.F., Y.G. (Yuxia Guo) and Y.G. (Yongzhuo Guan); writing—original draft, M.L., M.W. and J.Z.; writing—review and editing, X.Y., M.N., M.L., M.W., J.Z., Z.G., Y.G. (Yuxia Guo) and Y.G. (Yongzhuo Guan). All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Grant No. 32171672), the Project of Forestry Science and Technology Innovation and Promotion of Jiangsu (Grant No. LYKJ [2021]09), and the Jiangsu Students’ Innovation and Entrepreneurship Training Program (No.202211117005Z).
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank Ming-Zhang for his help in the sample collection. The help and guidance provided by Yi-Xiang during the determination of genome size and ploidy level of this study is much appreciated.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Soreng, R.J.; Peterson, P.M.; Zuloaga, F.O.; Romaschenko, K.; Clark, L.G.; Teisher, J.K.; Gillespie, L.J.; Barberá, P.; Welker, C.A.; Kellogg, E.A.; et al. worldwide phylogenetic classification of the Poaceae (Gramineae) III: An update. J. Syst. Evol. 2022, 60, 476–521. [Google Scholar] [CrossRef]
- Zheng, Y.; Xu, S.; Liu, J.; Zhao, Y.; Liu, J. Genetic diversity and population structure of Chinese natural bermudagrass [Cynodon dactylon (L.) Pers.] germplasm based on SRAP markers. PLoS ONE 2017, 12, e0177508. [Google Scholar] [CrossRef] [PubMed]
- Taliaferro, C.M.; Rouquette, F.M.; Mislevy, P. Bermudagrass and stargrass. In Warm-Season (C4) Grasses; Moser, L.E., Burson, B.L., Sollenberger, L.E., Eds.; Wiley Online Library: Hoboken, NJ, USA, 2004; pp. 417–475. [Google Scholar]
- Harlan, J.R.; De Wet, J.M.J. Sources of variation in Cynodon dactylon (L). Pers. 1. Crop Sci. 1969, 9, 774–778. [Google Scholar] [CrossRef]
- Baxter, L.L.; Anderson, W.F.; Gates, R.N.; Rios, E.F.; Hancock, D.W. Moving warm-season forage bermudagrass (Cynodon spp.) into temperate regions of North America. Grass Forage Sci. 2022, 77, 141–150. [Google Scholar] [CrossRef]
- Wu, Y.Q.; Taliaferro, C.M.; Martin, D.L.; Anderson, J.A.; Anderson, M.P. Genetic variability and relationships for adaptive, morphological, and biomass traits in Chinese bermudagrass accessions. Crop Sci. 2007, 47, 1985–1994. [Google Scholar] [CrossRef]
- Huang, C.; Liu, G.; Bai, C.; Wang, W. Genetic analysis of 430 Chinese Cynodon dactylon accessions using sequence-related amplified polymorphism markers. Int. J. Mol. Sci. 2014, 15, 19134–19146. [Google Scholar] [CrossRef]
- Taliaferro, C.M. Diversity and vulnerability of bermuda turfgrass species. Crop Sci. 1995, 35, 327–332. [Google Scholar] [CrossRef]
- Yu, S.; Dong, H.; Fang, T.; Wu, Y. Comparative analysis reveals chromosome number reductions in the evolution of African bermudagrass (Cynodon transvaalensis Burtt-Davy). Genome 2022, 65, 341–348. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, M.; Guo, Z.; Guan, Y.; Guo, Y.; Yan, X. Variation in ploidy level and genome size of Cynodon dactylon (L.) Pers. Along A Latitudinal Gradient. Folia Geobot. 2019, 54, 267–278. [Google Scholar] [CrossRef]
- Temsch, E.M.; Koutecký, P.; Urfus, T.; Šmarda, P.; Doležel, J. Reference standards for flow cytometric estimation of absolute nuclear DNA content in plants. Cytom. Part A 2022, 101, 710–724. [Google Scholar] [CrossRef]
- Galbraith, D.W. Flow cytometric analysis of plant genomes. Method Cell Biol. 1990, 33, 549–562. [Google Scholar]
- Heller, F.O. DNA measurement of Vicia faba L. with the pulse cytophotometry. Berichte Dtsch. Bot. Ges. 1973, 86, 437–441. [Google Scholar] [CrossRef]
- Loureiro, J.; Rodriguez, E.; Dolezel, J.; Santos, C. Comparison of four nuclear isolation buffers for plant DNA flow cytometry. Ann. Bot. 2006, 98, 679–689. [Google Scholar] [PubMed]
- Carović-Stanko, K.; Liber, Z.; Besendorfer, V.; Javornik, B.; Bohanec, B.; Kolak, I.; Satovic, Z. Genetic relations among basil taxa (Ocimum L.) based on molecular markers, nuclear DNA content, and chromosome number. Plant Syst. Evol. 2010, 285, 13–22. [Google Scholar] [CrossRef]
- Wu, Y.Q.; Taliaferro, C.M.; Bai, G.H.; Martin, D.L.; Anderson, J.A.; Anderson, M.P.; Edwards, R.M. Genetic analyses of Chinese Cynodon accessions by flow cytometry and AFLP markers. Crop Sci. 2006, 46, 917–926. [Google Scholar] [CrossRef]
- Sakhanokho, H.F.; Islam-Faridi, M.N.; Rajasekaran, K.; Pounders, C.T. Diversity in nuclear DNA content and ploidy level of Hedychium species and hybrids. J. Crop Improv. 2018, 32, 431–439. [Google Scholar] [CrossRef]
- Taliaferro, C.M.; Hopkins, A.A.; Henthorn, J.C.; Murphy, C.D.; Edwards, R.M. Use of flow cytometry to estimate ploidy level in Cynodon species. Int. Turfgrass Soc. Res. J. 1997, 8, 385–392. [Google Scholar]
- Zhang, R.; Liu, T.; Zhang, J.L.; Sun, Q.M. Spatial and environmental determinants of plant species diversity in a temperate desert. J. Plant Ecol. 2016, 9, 124–131. [Google Scholar] [CrossRef]
- Bartha, L.; Sramkó, G.; Volkova, P.A.; Surina, B.; Ivanov, A.L.; Banciu, H.L. Patterns of plastid DNA differentiation in Erythronium (Liliaceae) are consistent with allopatric lineage divergence in Europe across longitude and latitude. Plant Syst. Evol. 2015, 301, 1747–1758. [Google Scholar]
- Eilam, T.; Anikster, Y.; Millet, E.; Manisterski, J.; Feldman, M. Genome size in diploids, allopolyploids, and autopolyploids of mediterranean triticeae. J. Bot. 2010, 2010, 34138. [Google Scholar]
- Leitch, I.J.; Bennett, M.D. Genome downsizing in polyploid plants. Biol. J. Linn. Soc. 2004, 82, 651–663. [Google Scholar]
- Lidia, P.; Florencia, R.M.; Florencia, F.M.; María, G.A.; Esther, G.G. Genome downsizing and karyotype constancy in diploid and polyploid congeners: A model of genome size variation. AoB PLANTS 2014, 6, plu029. [Google Scholar]
- Walbot, V.; Cullis, C.A. Rapid genomic change in higher plants. Annu. Rev. Plant Biol. 1985, 36, 367–396. [Google Scholar]
- Grandbastien, M.-A. Activation of plant retrotransposons under stress conditions. Trends Plant Science. 1998, 3, 181–187. [Google Scholar]
- Ozkan, H.; Levy, A.A.; Feldman, M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops–Triticum) group. Plant Cell 2001, 13, 1735–1747. [Google Scholar]
- Shaked, H.; Kashkush, K.; Ozkan, H.; Feldman, M.; Levy, A.A. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 2001, 13, 1749–1759. [Google Scholar]
- Gustafson, J.P.; Bennett, M.D. The effect of telomericheterochromatin from Secale cereale on Triticale (× Triticosecale). I. The influence of the loss of several blocks of telo-meric heterochromatin on early endosperm development and kernel characteristics at maturity. Can. J. Genet. Cytol. 1982, 24, 83–92. [Google Scholar]
- Ozkan, H.; Tuna, M.; Arumuganathan, K. Nonadditive changes in genome size during allopolyploidization in the wheat (Aegilops–Triticum) group). J. Hered. 2003, 94, 260–264. [Google Scholar]
- Bennett, M.D.; Smith, J.B. Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976, 274, 227–274. [Google Scholar] [PubMed]
- Dolezel, J. Nuclear DNA content and genome size of trout and human. Cytom. Part A 2003, 51, 127–128.33. [Google Scholar]
- Knight, C.A.; Ackerly, D.D. Variation in nuclear DNA content across environmental gradients: A quantile regression analysis. Ecol. Lett. 2002, 5, 66–76. [Google Scholar]
- Akbudak, M.A.; Şakiroğlu, M.; Tuna, M. Estimation of nuclear DNA content and determination of relationship between altitude and genome size of USDA Turkish oat (Avena spp.) collection. Gesunde Pflanz. 2018, 70, 171–178. [Google Scholar]
- Carta, A.; Peruzzi, L. Testing the large genome constraint hypothesis: Plant traits, habitat and climate seasonality in L iliaceae. New Phytol. 2016, 210, 709–716. [Google Scholar] [PubMed]
- Bottini, M.C.J.; Greizerstein, E.J.; Aulicino, M.B.; Poggio, L. Relationships among genome size, environmental conditions and geographical distribution in natural populations of NW Patagonian species of Berberis L. (Berberidaceae). Ann. Bot. 2000, 86, 565–573. [Google Scholar]
- Bogunic, F.; Muratovic, E.; Ballian, D.; Siljak-Yakovlev, S.; Brown, S. Genome size stability among five subspecies of Pinus nigra Arnold sl. Environ. Exp. Bot. 2007, 59, 354–360. [Google Scholar]
- Basak, S.; Sun, X.; Wang, G.; Yang, Y. Genome size unaffected by variation in morphological traits, temperature, and precipitation in turnip. Appl. Sci. 2019, 9, 253. [Google Scholar]
- Knight, C.A.; Molinari, N.A.; Petrov, D.A. The large genome constraint hypothesis: Evolution, ecology and phenotype. Ann. Bot. 2005, 95, 177–190. [Google Scholar]
- Grime, J.P.; Mowforth, M.A. Variation in genome size—An ecological interpretation. Nature 1982, 299, 151–153. [Google Scholar]
- Grime, J.P.; Shacklock, J.M.L.; Band, S.R. Nuclear DNA contents, shoot phenology and species co-existence in a limestone grassland community. New Phytol. 1985, 100, 435–445. [Google Scholar]
- Šmarda, P.; Bureš, P. Understanding intraspecific variation in genome size in plants. Preslia 2010, 82, 41–61. [Google Scholar]
- Leitch, I.J.; Bennett, M.D. Polyploidy in angiosperms. Trends Plant Sci. 1997, 2, 470–476. [Google Scholar]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [PubMed]
- Wendel, J.F. Genome evolution in polyploids. Plant Mol. Evol. 2000, 42, 225–249.50. [Google Scholar]
- Doyle, J.J.; Flagel, L.E.; Paterson, A.H.; Rapp, R.A.; Soltis, D.E.; Soltis, P.S.; Wendel, J.F. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 2008, 42, 443–461. [Google Scholar] [PubMed]
- Dhawan, O.P.; Lavania, U.C. Enhancing the productivity of secondary metabolites via induced polyploidy: A review. Euphytica 1996, 87, 81–89. [Google Scholar]
- Ramsey, J.; Schemske, D.W. Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 2002, 33, 589–639. [Google Scholar]
- Parisod, C.; Holderegger, R.; Brochmann, C. Evolutionary consequences of autopolyploidy. New Phytol. 2010, 186, 5–17. [Google Scholar]
- Soltis, P.S.; Soltis, D.E. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 2009, 60, 561–588. [Google Scholar]
- Te Beest, M.; Le Roux, J.J.; Richardson, D.M.; Brysting, A.K.; Suda, J.; Kubešová, M.; Pyšek, P. The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot. 2012, 109, 19–45. [Google Scholar]
- Huff, D.R.; Quinn, J.A.; Higgins, B.; Palazzo, A.J. Random amplified polymorphic DNA (RAPD) variation among native little bluestem [Schizachyrium scoparium (Michx.) Nash] populations from sites of high and low fertility in forest and grassland biomes. Mol. Ecol. 1998, 7, 1591–1597. [Google Scholar]
- Gulsen, O.; Shearman, R.C.; Vogel, K.P.; Lee, D.J.; Baenziger, P.S.; Heng-Moss, T.M.; Budak, H. Nuclear genome diversity and relationships among naturally occurring buffalograss genotypes determined by sequence-related amplified polymorphism markers. HortScience 2005, 40, 537–541. [Google Scholar]
- Rice, A.; Šmarda, P.; Novosolov, M.; Drori, M.; Glick, L.; Sabath, N.; Meiri, S.; Belmaker, J.; Mayrose, I. The global biogeography of polyploid plants. Nat. Ecol. Evol. 2019, 3, 265–273. [Google Scholar] [PubMed]
- Bertin, N. Analysis of the tomato fruit growth response to temperature and plant fruit load in relation to cell division, cell expansion and DNA endoreduplication. Ann. Bot. 2005, 95, 439–447. [Google Scholar] [PubMed]
- Kudo, N.; Kimura, Y. Cell and Molecular Biology, Biochemistry and Molecular Physiology. Nuclear DNA endoreduplication during petal development in cabbage: Relationship between ploidy levels and cell size. J. Exp. Bot. 2002, 53, 1017–1023. [Google Scholar]
- Ehrendorfer, F.; Lewis, W.H. Polyploidy: Biological relevance. In Polyploidy and Distribution; Springer: Berlin/Heidelberg, Germany, 1980; pp. 45–60. [Google Scholar]
- Maceira, N.O.; Jacquard, P.; Lumaret, R. Competition between diploid and derivative autotetraploid Dactylis glomerata L. from Galicia. Implications for the establishment of novel polyploid populations. New Phytol. 1993, 124, 321–328. [Google Scholar]
- Petit, C.; Bretagnolle, F.; Felber, F. Evolutionary consequences of diploid–polyploid hybrid zones in wild species. Trends Ecol. Evol. 1999, 14, 306–311. [Google Scholar]
- Bureš, P.; Wang, Y.F.; Horová, L.; Suda, J. Genome size variation in Central European species of Cirsium (Compositae) and their natural hybrids. Ann. Bot. 2004, 94, 353–363. [Google Scholar]
- Rosato, M.; Chiavarino, A.M.; Naranjo, C.A.; Hernandez, J.C.; Poggio, L. Genome size and numerical polymorphism for the B chromosome in races of maize (Zea mays ssp. mays, Poaceae). Am. J. Bot. 1998, 85, 168–174. [Google Scholar]
- Vitte, C.; Bennetzen, J.L. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl. Acad. Sci. USA 2006, 103, 17638–17643. [Google Scholar]
- Yoong Lim, K.; Kovarik, A.; Matyasek, R.; Chase, M.W.; Knapp, S.; McCarthy, E.; Clarkson, J.J.; Leitch, A.R. Comparative genomics and repetitive sequence divergence in the species of diploid Nicotiana section Alatae. Plant J. 2006, 48, 907–919. [Google Scholar]
- Bennetzen, J.L. Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica 2002, 115, 29–36. [Google Scholar] [PubMed]
- Vicient, C.M.; Suoniemi, A.; Anamthawat-Jonsson, K.; Tanskanen, J.; Beharav, A.; Nevo, E.; Schulman, A.H. Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 1999, 11, 1769–1784. [Google Scholar] [PubMed]
- Shirasu, K.; Schulman, A.H.; Lahaye, T.; Schulze-Lefert, P. A contiguous 66-kb barley DNA sequence provides evidence for r eversible genome expansion. Genome Res. 2000, 10, 908–915. [Google Scholar]
- Devos, K.M.; Brown, J.K.M.; Bennetzen, J.L. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002, 12, 1075–1079. [Google Scholar] [PubMed]
- Simonin, K.A.; Roddy, A.B. Genome downsizing, physiological novelty, and the global dominance of flowering plants. PLoS Biol. 2018, 16, e2003706. [Google Scholar]
- Roddy, A.B.; Theroux-Rancourt, G.; Abbo, T.; Benedetti, J.W.; Brodersen, C.R.; Castro, M.; Castro, S.; Gilbride, A.B.; Jensen, B.; Jiang, G.-F.; et al. The scaling of genome size and cell size limits maximum rates of photosynthesis with implications for ecological strategies. Int. J. Plant Sci. 2019, 181, 75–87. [Google Scholar]
- Wang, X.; Morton, J.; Pellicer, J.; Leitch, I.J.; Leitch, A.R. Genome downsizing after polyploidy: Mechanisms, rates and selection pressures. Plant J. 2021, 107, 1003–1015. [Google Scholar]
- Hao, G.Y.; Lucero, M.E.; Sanderson, S.C.; Zacharias, E.H.; Holbrook, N.M. Polyploidy enhances the occupation of heterogeneous environments through hydraulic related trade-offs in Atriplex canescens (Chenopodiaceae). New Phytol. 2013, 197, 970–978. [Google Scholar] [CrossRef]
- Silva, D.M.; Santos, Y.D.; Benites, F.R.G.; Techio, V.H. Microsporogenesis, viability and morphology of pollen grain in accessions of Cynodon L. C. Rich. (Poaceae). S. Afr. J. Bot. 2018, 118, 260–267. [Google Scholar]
- Grivet, L.; Arruda, P. Sugarcane genomics: Depicting the complex genome of an important tropical crop. Curr. Opin. Plant Biol. 2002, 5, 122–127. [Google Scholar]
- Costich, D.E.; Friebe, B.; Sheehan, M.J.; Casler, M.D.; Buckler, E.S. Genome-size variation in switchgrass (Panicum virgatum): Flow cytometry and cytology reveal rampant aneuploidy. Plant Genome 2010, 3. [Google Scholar] [CrossRef]
- Huang, J.; Ma, L.; Yang, F.; Fei, S.Z.; Li, L. 45S rDNA regions are chromosome fragile sites expressed as gaps in vitro on metaphase chromosomes of root-tip meristematic cells in Lolium spp. PLoS ONE. 2008, 3, e2167. [Google Scholar]
- Birchler, J.A. Reflections on studies of gene expression in aneuploids. Biochem. J. 2010, 426, 119–123. [Google Scholar] [PubMed]
- Li, Z.Y.; Ge, X.H. Unique chromosome behavior and genetic control in Brassica × Orychophragmus wide hybrids: A review. Plant Cell Rep. 2007, 26, 701–710. [Google Scholar]
- Henry, I.M.; Dilkes, B.P.; Young, K.; Watson, B.; Wu, H.; Comai, L. Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics 2005, 170, 1979–1988. [Google Scholar] [PubMed]
- Henry, I.M.; Dilkes, B.P.; Comai, L. Genetic basis for dosage sensitivity in Arabidopsis thaliana. PLoS Genet. 2007, 3, e70. [Google Scholar]
- Cuadrado, A.; Acevedo, R.; de la Espina, S.M.D.; Jouve, N.; De La Torre, C. Genome remodelling in three modern S. officinarum × S. spontaneum sugarcane cultivars. J. Exp. Bot. 2004, 55, 847–854. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).