Variations in Protein and Gene Expression Involved in the Pathways of Carbohydrate, Abscisic Acid, and ATP-Binding Cassette Transporter in Soybean Roots under Drought Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Proteomics Analysis and Functional Enrichment Analysis of Differentially Expressed Proteins Identified in Soybean Roots
2.3. Determination of Physiological Indices of Soybean Roots
2.4. RNA Extraction and Quantitative Real-Time PCR Analysis
2.5. Statistical Analysis
3. Results
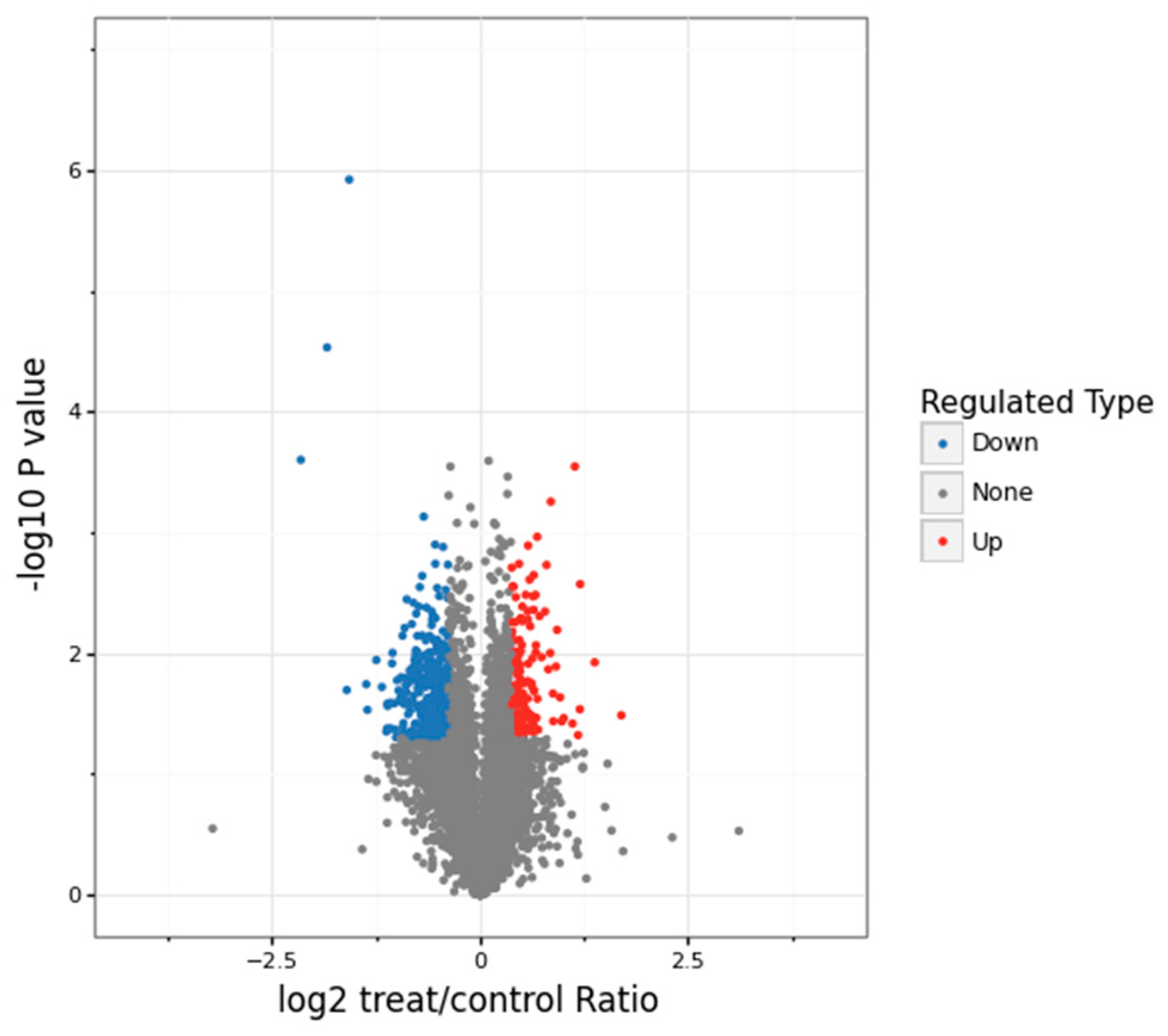
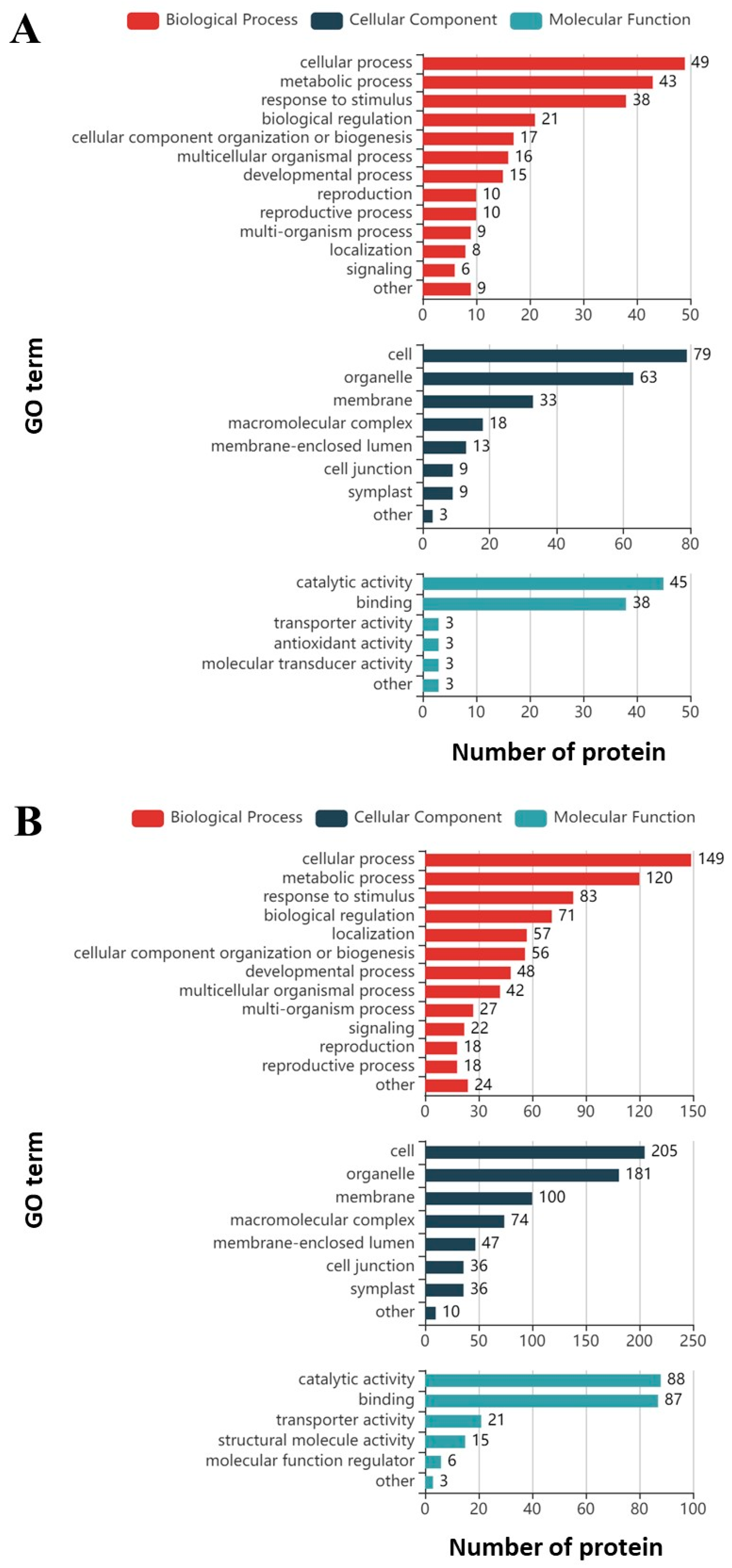
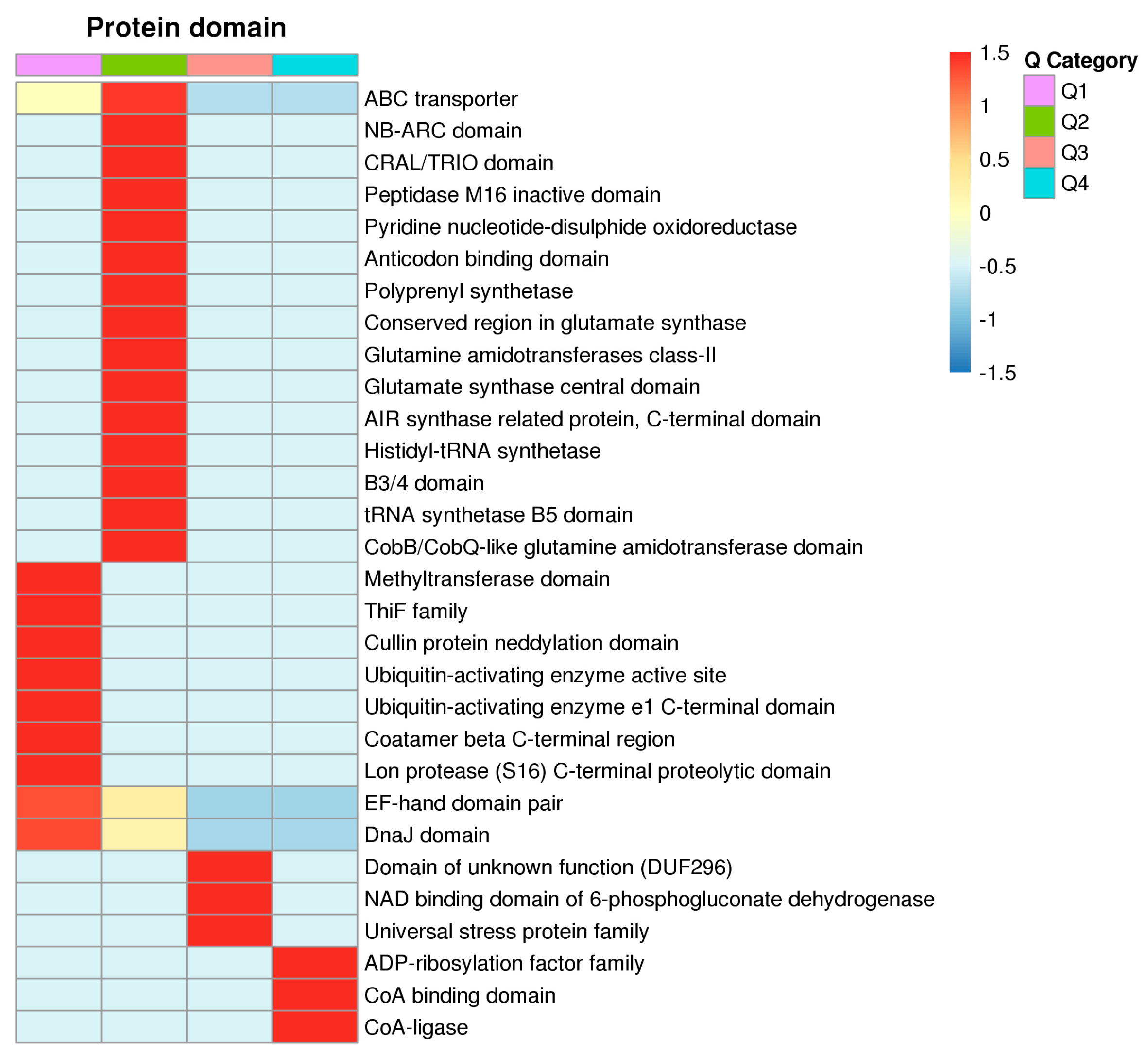
3.1. Proteomics Analysis and Functional Enrichment Analysis of Differentially Expressed Proteins in Soybean Roots under Drought Stress
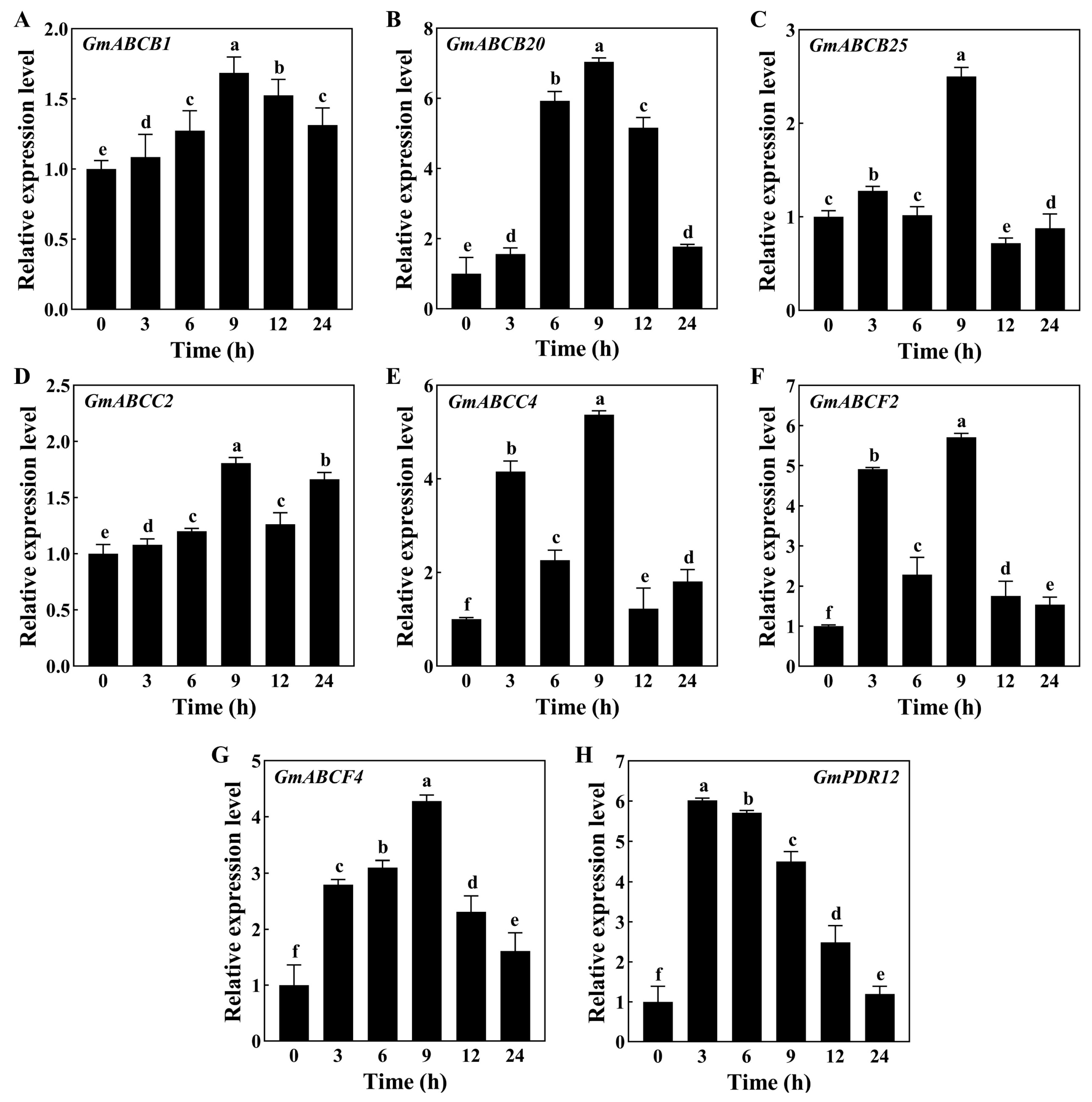
3.2. Variations in Expression Pattern of Genes Encoding ABC Transporters in Soybean Roots under Drought Stress
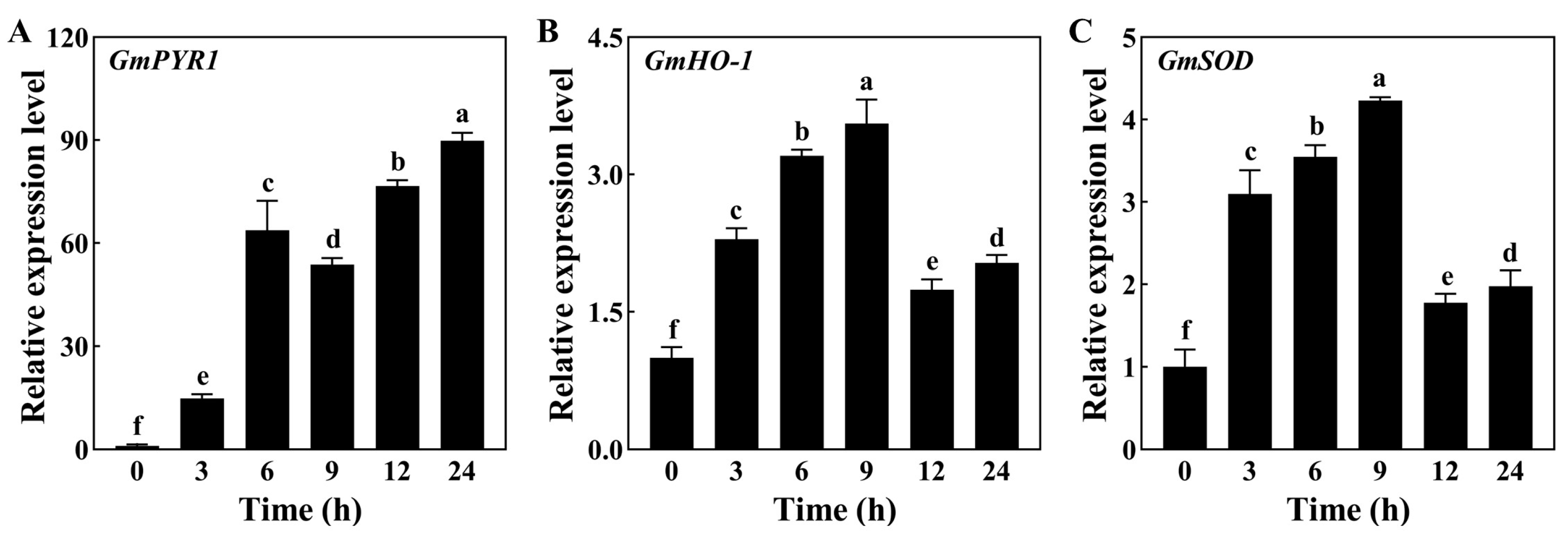
3.3. Evaluation of the Expression Levels of Genes Associated with the ABA Signaling Pathway in Soybean Root Response to Drought Stress
3.4. Evaluation of Expression Patterns of Genes Associated with Sucrose and Betaine Biosynthesis in Soybean Root Response to Drought Stress
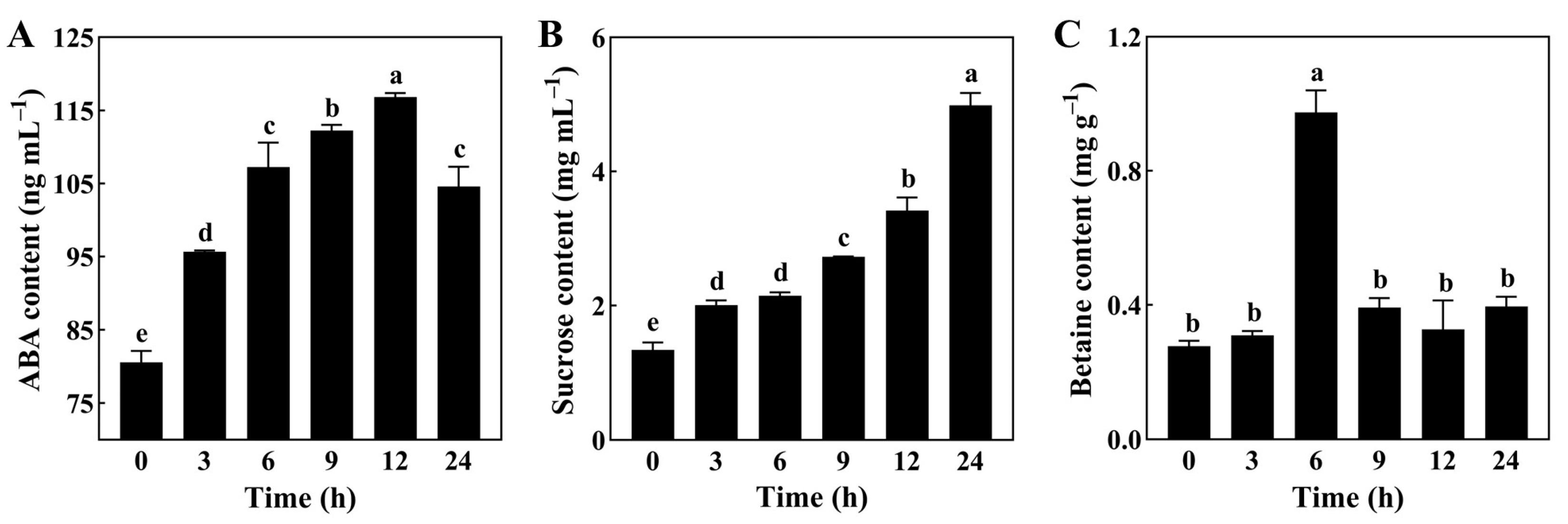
3.5. Determination of the Contents of Osmoregulatory Substances and ABA in Soybean Roots under Drought Stress
4. Discussion
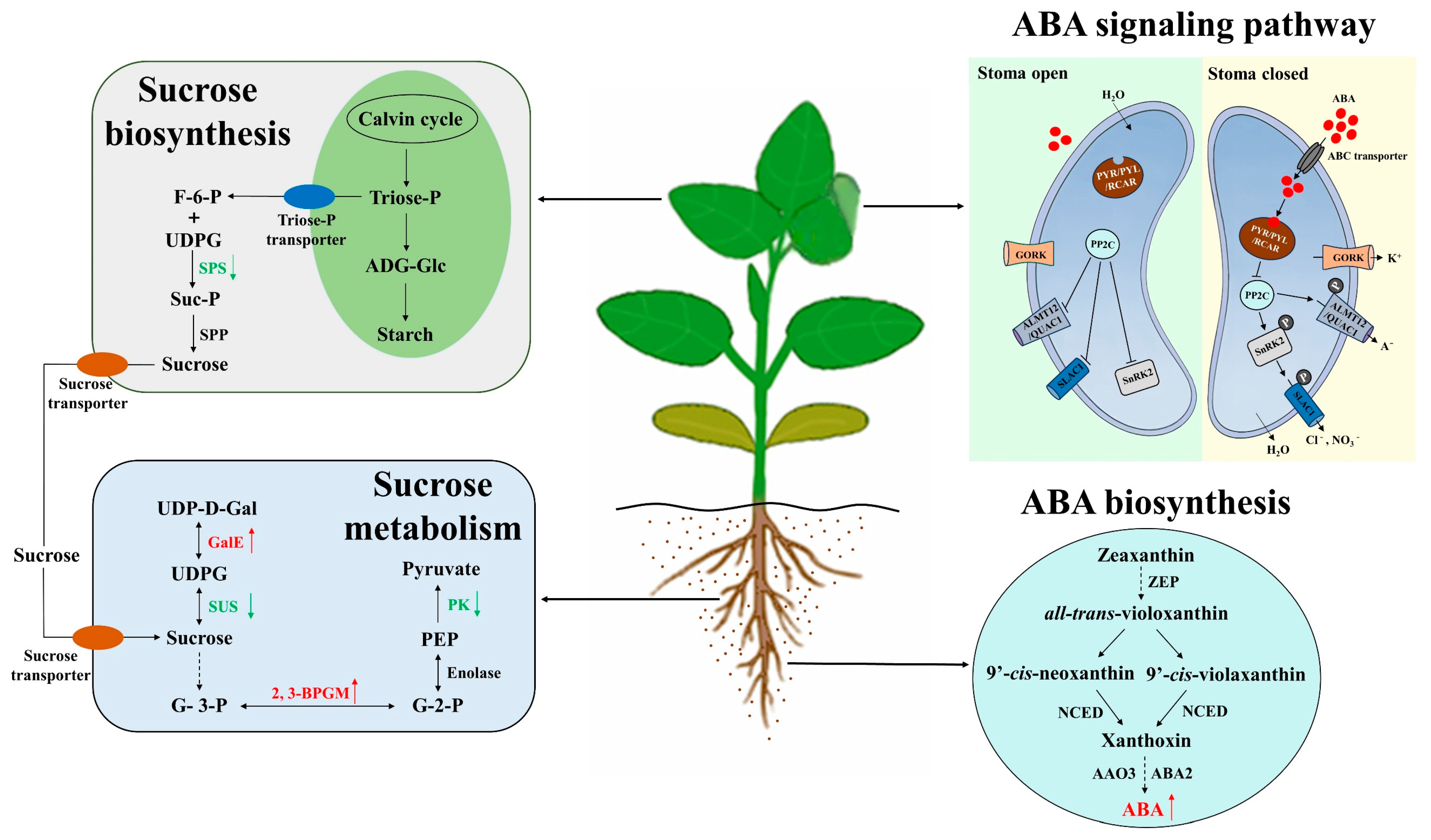
4.1. DEGs Involved in ABC Biosynthesis and Signaling Pathways in Soybean Roots under Drought Stress
4.2. DEGs Involved in ABA Biosynthesis and Signaling Pathways in Soybean Roots under Drought Stress
4.3. DEGs Involved in Sucrose and Betaine Biosynthesis and Signaling Pathways in Soybean Roots under Drought Stress
4.4. A Proposed Model and the Effect of Drought Stress on Soybean Stomatal Movement
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tian, T.; Wang, S.; Yang, S.; Yang, Z.; Liu, S.; Wang, Y.; Gao, H.; Zhang, S.; Yang, X.; Jiang, C.; et al. Genome assembly and genetic dissection of a prominent drought-resistant maize germplasm. Nat. Genet. 2023, 55, 496–506. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Du, X.; Zhang, X.; Chen, X.; Jin, W.; Huang, Z.; Kong, L. Drought stress reduces the photosynthetic source of subtending leaves and the transit sink function of podshells, leading to reduced seed weight in soybean plants. Front. Plant Sci. 2024, 15, 1337544. [Google Scholar] [CrossRef]
- Xu, C.; Shan, J.; Liu, T.; Wang, Q.; Ji, Y.; Zhang, Y.; Wang, M.; Xia, N.; Zhao, L. CONSTANS-LIKE 1a positively regulates salt and drought tolerance in soybean. Plant Physiol. 2023, 191, 2427–2446. [Google Scholar] [CrossRef]
- Wei, Y.; Jin, J.; Jiang, S.; Ning, S.; Liu, L. Quantitative response of soybean development and yield to drought stress during different growth stages in the Huaibei Plain China. Agronomy 2018, 8, 97. [Google Scholar] [CrossRef]
- Zhang, Z.; Tong, T.; Fang, Y.; Zheng, J.; Zhang, X.; Niu, C.; Li, J.; Zhang, X.; Xue, D. Genome-Wide Identification of Barley ABC Genes and Their Expression in Response to Abiotic Stress Treatment. Plants 2020, 9, 1281. [Google Scholar] [CrossRef]
- Abid, M.; Ali, S.; Qi, L.K.; Zahoor, R.; Tian, Z.; Jiang, D.; Snider, J.L.; Dai, T. Physiological and biochemical changes during drought and recovery periods at tillering and jointing stages in wheat (Triticum aestivum L.). Sci. Rep. 2018, 8, 4615. [Google Scholar] [CrossRef]
- Wang, B.; Yang, L.; Zhang, Y.; Chen, S.; Gao, X.; Wan, C. Investigation of the dynamical expression of Nostoc flagelliforme proteome in response to rehydration. J. Proteom. 2019, 192, 160–168. [Google Scholar] [CrossRef]
- Wang, X.; Song, S.; Wang, X.; Liu, J.; Dong, S. Transcriptomic and Metabolomic Analysis of Seedling-Stage Soybean Responses to PEG-Simulated Drought Stress. Int. J. Mol. Sci. 2022, 23, 6869. [Google Scholar] [CrossRef]
- Roychowdhury, R.; Das, S.P.; Gupta, A.; Parihar, P.; Chandrasekhar, K.; Sarker, U.; Kumar, A.; Ramrao, D.P.; Sudhakar, C. Multi-omics pipeline and omics-integration approach to decipher plant’s abiotic stress tolerance response. Genes 2023, 14, 1281. [Google Scholar] [CrossRef]
- Feng, Z.; Ding, C.; Li, W.; Wang, D.; Cui, D. Applications of metabolomics in the research of soybean plant under abiotic stress. Food Chem. 2020, 310, 125914. [Google Scholar] [CrossRef] [PubMed]
- Higgins, C.F. ABC transporters: From microorganisms to man. Annu. Rev. Cell Biol. 1992, 8, 67–113. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.U.; Song, W.Y.; Hong, D.; Ko, D.; Yamaoka, Y.; Jang, S.; Yim, S.; Lee, E.; Khare, D.; Kim, K.; et al. Plant ABC Transporters Enable Many Unique Aspects of a Terrestrial Plant’s Lifestyle. Mol. Plant 2016, 9, 338–355. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC Transporters. Arab. Book 2011, 9, e0153. [Google Scholar] [CrossRef] [PubMed]
- Rea, P.A. Plant ATP-binding cassette transporters. Annu. Rev. Plant Biol. 2007, 58, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Dawson, R.J.; Hollenstein, K.; Locher, K.P. Uptake or extrusion: Crystal structures of full ABC transporters suggest a common mechanism. Mol. Microbiol. 2007, 65, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Lefèvre, F.; Boutry, M. Towards identification of the substrates of ATP-Binding cassette transporters. Plant Physiol. 2018, 178, 18–39. [Google Scholar] [CrossRef] [PubMed]
- Theodoulou, F.L.; Kerr, L.D. ABC transporter research: Going strong 40 years on. Biochem. Soc. Trans. 2015, 43, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Jin, J.Y.; Alejandro, S.; Martinoia, E.; Lee, Y. Overexpression of AtABCG36 improves drought and salt stress resistance in Arabidopsis. Physiol. Plant. 2010, 139, 170–180. [Google Scholar] [CrossRef]
- Long, H.; Zheng, Z.; Zhang, Y.; Xing, P.; Wan, X.; Zheng, Y.; Li, L. An abscisic acid (ABA) homeostasis regulated by its production, catabolism and transport in peanut leaves in response to drought stress. PLoS ONE 2019, 14, e0213963. [Google Scholar] [CrossRef]
- Thomas, C.; Tampé, R. Structural and Mechanistic Principles of ABC Transporters. Annu. Rev. Biochem. 2020, 89, 605–636. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Zhuang, Y.; Cai, Y.; Agüero, C.B.; Liu, S.; Wu, J.; Deng, S.; Walker, M.A.; Lu, J.; Zhang, Y. Overexpression of 9-cis-Epoxycarotenoid Dioxygenase Cisgene in Grapevine Increases Drought Tolerance and Results in Pleiotropic Effects. Front. Plant Sci. 2018, 9, 970. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Qiu, C.; Zeng, Y.; Lin, Q. Molecular and Enzymatic Characterization of 9-Cis-epoxycarotenoid Dioxygenases from Mulberry. Protein J. 2022, 41, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.; Susila, H.; Nasim, Z.; Jung, J.Y.; Ahn, J.H. Arabidopsis ABF3 and ABF4 Transcription Factors Act with the NF-YC Complex to Regulate SOC1 Expression and Mediate Drought-Accelerated Flowering. Mol. Plant 2019, 12, 489–505. [Google Scholar] [CrossRef]
- Mora, L.; Bramley, P.M.; Fraser, P.D. Development and optimisation of a label-free quantitative proteomic procedure and its application in the assessment of genetically modified tomato fruit. Proteomics 2013, 13, 2016–2030. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, H.; Chen, H.; Yang, X.; Yu, T.; Wang, Y.; Wang, Y.; Jiang, K.; Wang, Y.; Chen, Z.; et al. Proteomic Investigation of Molecular Mechanisms in Response to PEG-Induced Drought Stress in Soybean Roots. Plants 2022, 11, 1173. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tang, K.; Zhang, Y.; Zhang, H.; Xu, P.; Liu, J.; Ma, J.; Lv, M.; Li, D.; Katirai, F.; Shen, G.X.; et al. Delivery of chemotherapeutic drugs in tumour cell-derived microparticles. Nat. Commun. 2012, 3, 1282. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Khan, T.; Farooq, I.; Shah, L.R.; Sharma, V.; Sonne, C.; Rinklebe, J.; Ahmad, P. Drought and global hunger: Biotechnological interventions in sustainability and management. Planta 2022, 256, 97. [Google Scholar] [CrossRef] [PubMed]
- Thao, N.P.; Tran, L.S. Potentials toward genetic engineering of drought-tolerant soybean. Crit. Rev. Biotechnol. 2012, 32, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Miao, R.; Khanna, M. Maladaptation of U.S. corn and soybeans to a changing climate. Sci. Rep. 2021, 11, 12351. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Lu, Y.; Zhang, X.; Yang, G.; Chao, D.; Wang, Z.; Shi, M.; Chen, J.; Chao, D.Y.; Li, R.; et al. The ABC transporter ABCG36 is required for cadmium tolerance in rice. J. Exp. Bot. 2019, 70, 5909–5918. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Yu, Q.; Wang, J.; Han, H.; Mao, L.; Nyporko, A.; Maguza, A.; Fan, L.; Bai, L.; Powles, S. An ABCC-type transporter endowing glyphosate resistance in plants. Proc. Natl. Acad. Sci. USA 2021, 118, e2100136118. [Google Scholar] [CrossRef] [PubMed]
- Song, W.Y.; Park, J.; Mendoza-Cózatl, D.G.; Suter-Grotemeyer, M.; Shim, D.; Hörtensteiner, S.; Geisler, M.; Weder, B.; Rea, P.A.; Rentsch, D.; et al. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. USA 2010, 107, 21187–21192. [Google Scholar] [CrossRef]
- Dhara, A.; Raichaudhuri, A. ABCG transporter proteins with beneficial activity on plants. Phytochemistry 2021, 184, 112663. [Google Scholar] [CrossRef]
- Martignago, D.; Rico-Medina, A.; Blasco-Escámez, D.; Fontanet-Manzaneque, J.B.; Caño-Delgado, A.I. Drought Resistance by Engineering Plant Tissue-Specific Responses. Front. Plant Sci. 2020, 10, 1676. [Google Scholar] [CrossRef]
- Daszkowska-Golec, A.; Szarejko, I. Open or close the gate-stomata action under the control of phytohormones in drought stress conditions. Front. Plant Sci. 2013, 4, 138. [Google Scholar] [CrossRef]
- Gulzar, F.; Fu, J.; Zhu, C.; Yan, J.; Li, X.; Meraj, T.A.; Shen, Q.; Hassan, B.; Wang, Q. Maize WRKY Transcription Factor ZmWRKY79 Positively Regulates Drought Tolerance through Elevating ABA Biosynthesis. Int. J. Mol. Sci. 2021, 22, 10080. [Google Scholar] [CrossRef]
- Yan, J.; Ninkuu, V.; Fu, Z.; Yang, T.; Ren, J.; Li, G.; Yang, X.; Zeng, H. OsOLP1 contributes to drought tolerance in rice by regulating ABA biosynthesis and lignin accumulation. Front. Plant Sci. 2023, 14, 1163939. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Mao, Y.; Duan, X.; Zhou, H.; Lai, D.; Zhang, Y.; Shen, W. Arabidopsis HY1-Modulated Stomatal Movement: An Integrative Hub Is Functionally Associated with ABI4 in Dehydration-Induced ABA Responsiveness. Plant Physiol. 2016, 170, 1699–1713. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.K.; Jin, Q.J.; Xie, Y.J.; Liu, Y.H.; Lin, Y.T.; Shen, W.B.; Zhou, Y.J. Characterization of a wheat heme oxygenase-1 gene and its responses to different abiotic stresses. Int. J. Mol. Sci. 2011, 12, 7692–7707. [Google Scholar] [CrossRef]
- Tounsi, S.; Jemli, S.; Feki, K.; Brini, F.; Najib Saïdi, M. Superoxide dismutase (SOD) family in durum wheat: Promising candidates for improving crop resilience. Protoplasma 2023, 260, 145–158. [Google Scholar] [CrossRef]
- Kuromori, T.; Sugimoto, E.; Shinozaki, K. Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J. 2011, 67, 885–894. [Google Scholar] [CrossRef]
- Hong, Y.; Wang, Z.; Liu, X.; Yao, J.; Kong, X.; Shi, H.; Zhu, J.K. Two Chloroplast Proteins Negatively Regulate Plant Drought Resistance Through Separate Pathways. Plant Physiol. 2020, 182, 1007–1021. [Google Scholar] [CrossRef]
- Du, Y.; Zhao, Q.; Chen, L.; Yao, X.; Zhang, W.; Zhang, B.; Xie, F. Effect of drought stress on sugar metabolism in leaves and roots of soybean seedlings. Plant Physiol. Biochem. 2020, 146, 1–12. [Google Scholar] [CrossRef]
- Schäfer, W.E.; Rohwer, J.M.; Botha, F.C. Partial purification and characterisation of sucrose synthase in sugarcane. J. Plant Physiol. 2005, 162, 11–20. [Google Scholar] [CrossRef]
- Xiao, N.; Ma, H.; Wang, W.; Sun, Z.; Li, P.; Xia, T. Overexpression of ZmSUS1 increased drought resistance of maize (Zea mays L.) by regulating sucrose metabolism and soluble sugar content. Planta 2024, 259, 43. [Google Scholar] [CrossRef]
- Barrero-Sicilia, C.; Hernando-Amado, S.; González-Melendi, P.; Carbonero, P. Structure, expression profile and subcellular localisation of four different sucrose synthase genes from barley. Planta 2011, 234, 391–403. [Google Scholar] [CrossRef]
- Xiao, X.; Tang, C.; Fang, Y.; Yang, M.; Zhou, B.; Qi, J.; Zhang, Y. Structure and expression profile of the sucrose synthase gene family in the rubber tree: Indicative of roles in stress response and sucrose utilization in the laticifers. FEBS J. 2014, 281, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Vaultier, M.N.; Rochat, C. Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J. Exp. Bot. 2004, 55, 397–409. [Google Scholar] [CrossRef]
- Annunziata, M.G.; Ciarmiello, L.F.; Woodrow, P.; Dell’Aversana, E.; Carillo, P. Spatial and Temporal Profile of Glycine Betaine Accumulation in Plants Under Abiotic Stresses. Front. Plant Sci. 2019, 10, 230. [Google Scholar] [CrossRef]
- You, L.; Song, Q.; Wu, Y.; Li, S.; Jiang, C.; Chang, L.; Yang, X.; Zhang, J. Accumulation of glycine betaine in transplastomic potato plants expressing choline oxidase confers improved drought tolerance. Planta 2019, 249, 1963–1975. [Google Scholar] [CrossRef]
- Park, E.J.; Jeknić, Z.; Sakamoto, A.; DeNoma, J.; Yuwansiri, R.; Murata, N.; Chen, T.H. Genetic engineering of glycine betaine synthesis in tomato protects seeds, plants, and flowers from chilling damage. Plant J. 2004, 40, 474–487. [Google Scholar] [CrossRef]
- Lee, S.C.; Lan, W.; Buchanan, B.B.; Luan, S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA 2009, 106, 21419–21424. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Hwang, J.U.; Lee, M.; Kim, Y.Y.; Assmann, S.M.; Martinoia, E.; Lee, Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 2355–2360. [Google Scholar] [CrossRef]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Lim, C.W.; Lan, W.; He, K.; Luan, S. ABA signaling in guard cells entails a dynamic protein-protein interaction relay from the PYL-RCAR family receptors to ion channels. Mol. Plant 2013, 6, 528–538. [Google Scholar] [CrossRef] [PubMed]

| Primer | Sequence (5′→3′) |
| GmABCB1 | F: AGCAGTGGAAATGGTCCGAA |
| R: TTCTCCCCACCACCGTTACT | |
| GmABCB20 | F: TGATGGTTTCTCGGGGACTG |
| R: CTCCATGGGCTGAGACGTG | |
| GmABCB25 | F: GAACATGGTGATGCAGTGCC |
| R: GTGGCTGCAATCAACAGAGC | |
| GmABCC2 | F: CGAGGAATCTTTGAACCAGCG |
| R: TGATGGAGCCTGTTCTTGGC | |
| GmABCC4 | F: AACTCAGAGCTCGACAAGCC |
| R: AGTTTGCTTCCACGTCCCAT | |
| GmABCF2 | F: CCAAGAGAGGAGGCAAAGCA |
| R: GGAAAGAGGATGCGAGCAGA | |
| GmABCF4 | F: CAAGCGGCTAAGAGGAGTGG |
| R: ATCTTCCCGGTTTGGGTAGC | |
| GmPDR12 | F: AGAACCAATGACGTCTGAAGGT |
| R: CAATGCAAAGAGAACGGCGA | |
| GmPYR1 | F: TGGACAAAACGCACAGCG |
| R: GTCCTCGAACTCTTCCGGG | |
| GmHO-1 | F: TCCCAGTCCCAATCGCTCTA |
| R: CATGACAGCAGCACGAACAC | |
| GmSOD | F: GAGCCCTGTTGACCAGAAAAAC |
| R: GTACACGTGCAACCCACGA | |
| GmUGTA1 | F: CACAACTCCTCAGAACAAC |
| R: GAGATAGGGAATGGGAAGAA | |
| GmUGTA2 | F: CTAAGATGTTGGTGGAAGA |
| R: CTCAGCACTGTCTCTAAC | |
| GmSUS6 | F: GAGCTTGGTGAACCTTGTGG |
| R: GTTGCGGTAGCGATCGGTTT | |
| GmSUS7 | F: GACAGCCTTAACTCTGCTGC |
| R: TTCATCGGGTCTGGTGCTTG | |
| GmUDP141 | F: GCTTGTGTTCTCATCTTC |
| R: AACTGTCCAATCTGAATCT | |
| GmUDP189 | F: CTATGGTGTGGAGGAGAT |
| R: CATTCGTAGATGAGCAGTAT | |
| Tubulin | F: GGAAGGCTTTCTTGCATTGGTA |
| R: AGTGGCATCCTGGTACTGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Cui, X.; Chang, J.; Wang, J.; Wang, Y.; Liu, H.; Wang, Y.; Chen, Y.; Yang, Y.; Yao, D.; et al. Variations in Protein and Gene Expression Involved in the Pathways of Carbohydrate, Abscisic Acid, and ATP-Binding Cassette Transporter in Soybean Roots under Drought Stress. Agronomy 2024, 14, 843. https://doi.org/10.3390/agronomy14040843
Yang X, Cui X, Chang J, Wang J, Wang Y, Liu H, Wang Y, Chen Y, Yang Y, Yao D, et al. Variations in Protein and Gene Expression Involved in the Pathways of Carbohydrate, Abscisic Acid, and ATP-Binding Cassette Transporter in Soybean Roots under Drought Stress. Agronomy. 2024; 14(4):843. https://doi.org/10.3390/agronomy14040843
Chicago/Turabian StyleYang, Xiaoqin, Xiyan Cui, Jiageng Chang, Jianan Wang, Yujue Wang, Haoye Liu, Yan Wang, Yanbo Chen, Yuhan Yang, Dan Yao, and et al. 2024. "Variations in Protein and Gene Expression Involved in the Pathways of Carbohydrate, Abscisic Acid, and ATP-Binding Cassette Transporter in Soybean Roots under Drought Stress" Agronomy 14, no. 4: 843. https://doi.org/10.3390/agronomy14040843






