The Effects of Soybean–Tea Intercropping on the Photosynthesis Activity of Tea Seedlings Based on Canopy Spectral, Transcriptome and Metabolome Analyses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experiment Field and Setup
2.2. Unmanned Aerial Systems and Image Acquisition
2.3. Determination of SPAD and Fv/Fm
2.4. RNA Extraction and RNA-Seq
2.5. Transcriptome Data Analysis
2.6. Metabolite Extraction and Analysis
2.7. Quantitative Real-Time PCR Analysis
2.8. Statistical Analysis
3. Results
3.1. Effects of STS on Spectral Reflectance and VIs of Tea Seedlings Canopy
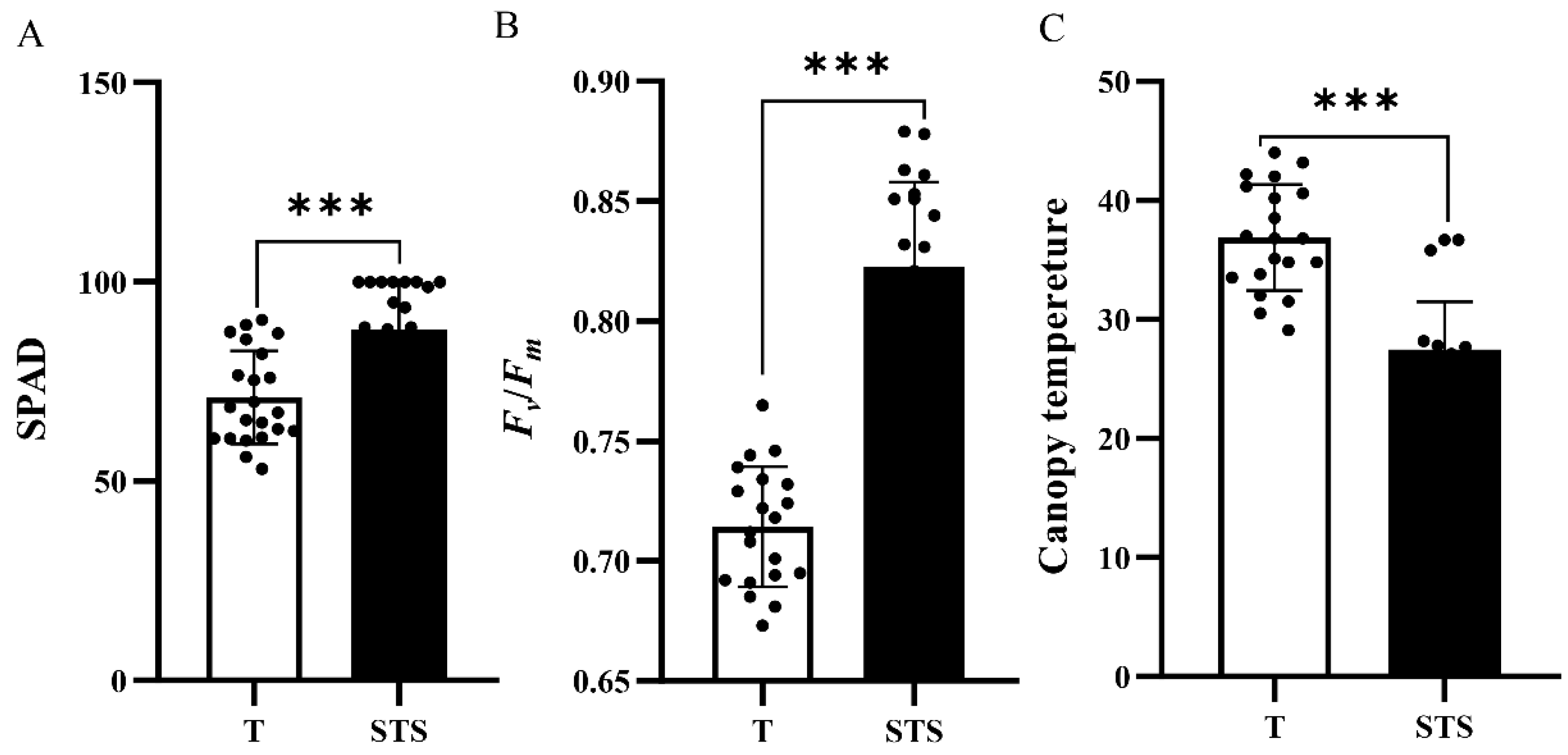
3.2. Effects of STS on the SPAD, Fv/Fm and Canopy Temperature of Tea Seedlings
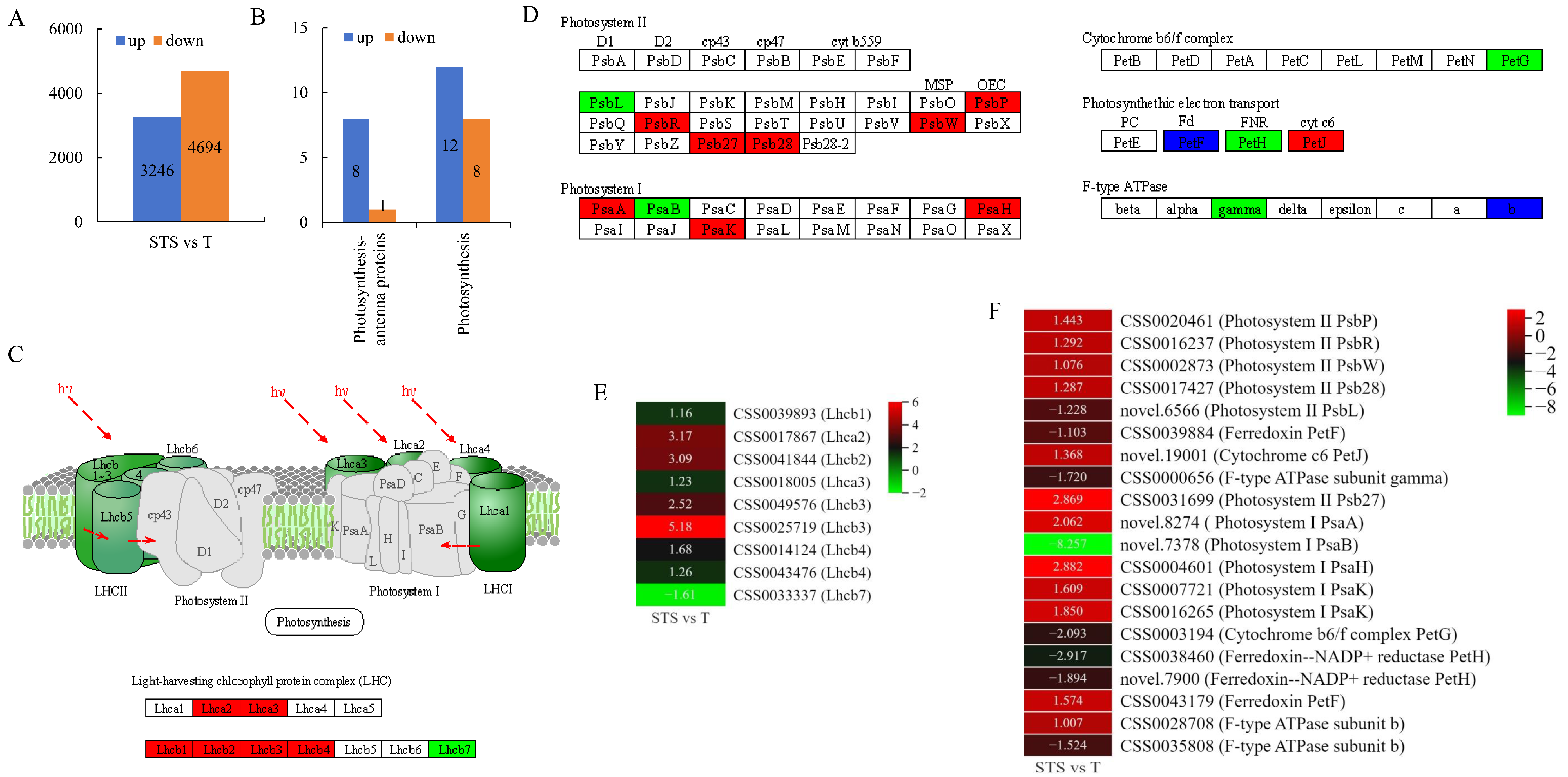
3.3. RNA Sequencing Data Analysis
3.4. Analysis of Photosynthesis-Antenna Proteins and Photosynthesis Pathways
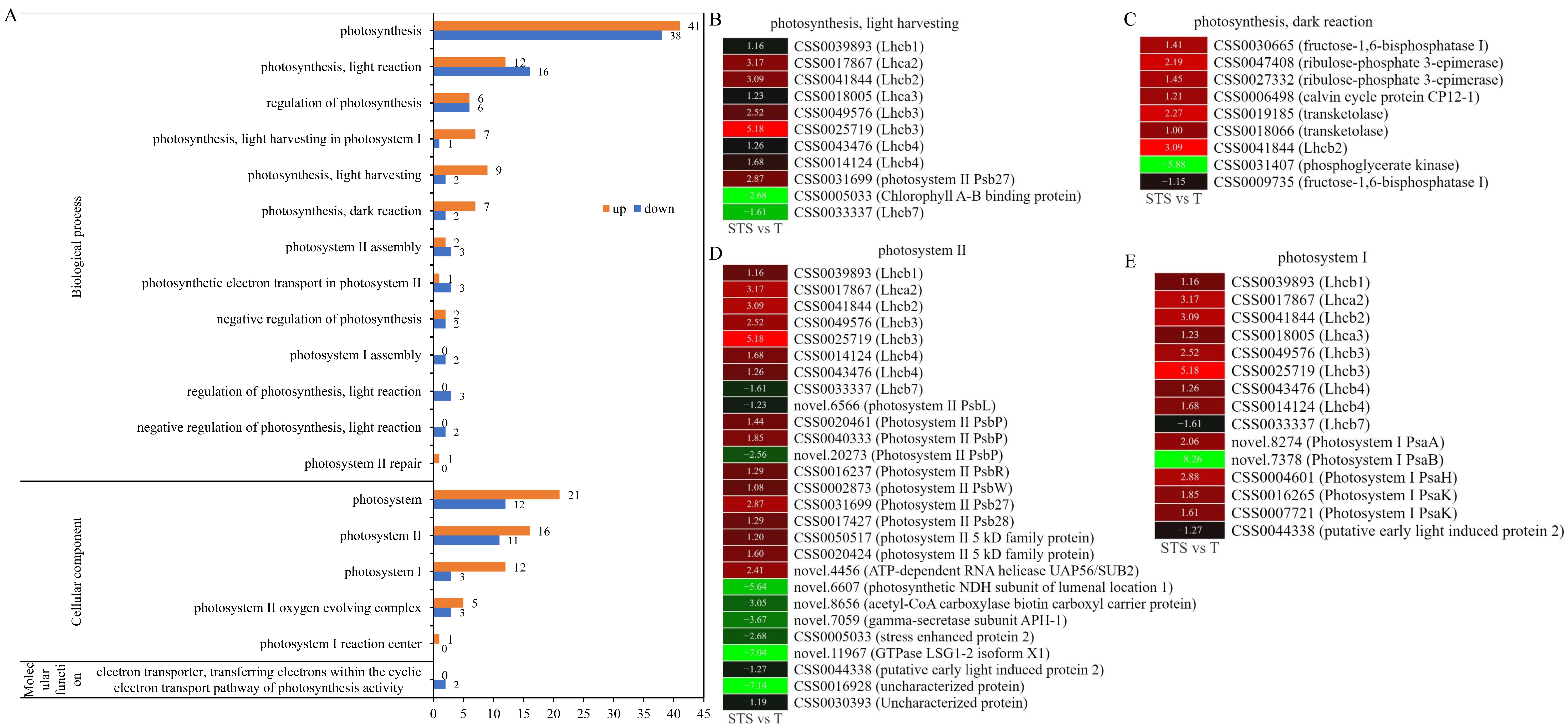
3.5. GO Analysis Related to Photosynthesis
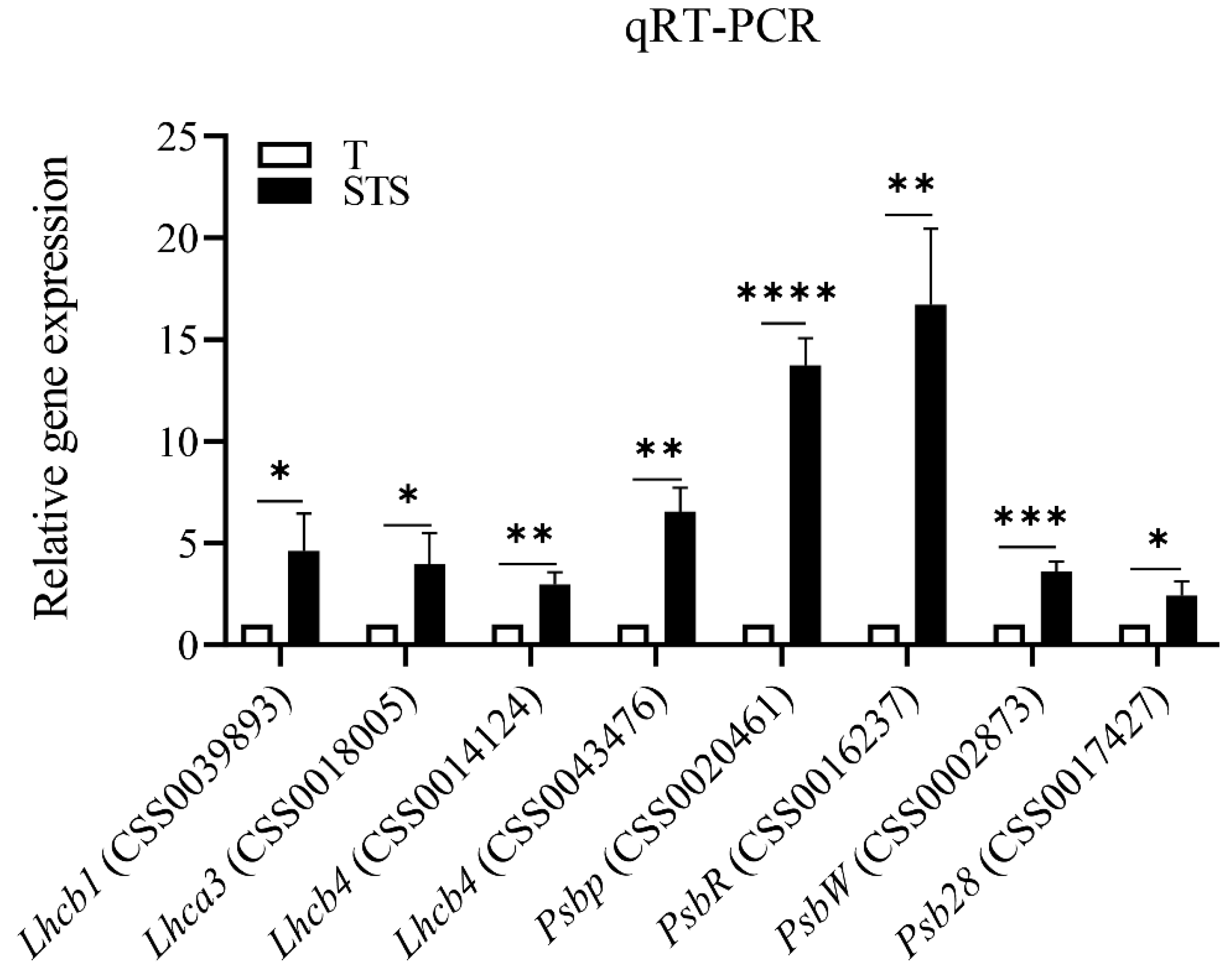
3.6. Validation of DEGs by qRT-PCR
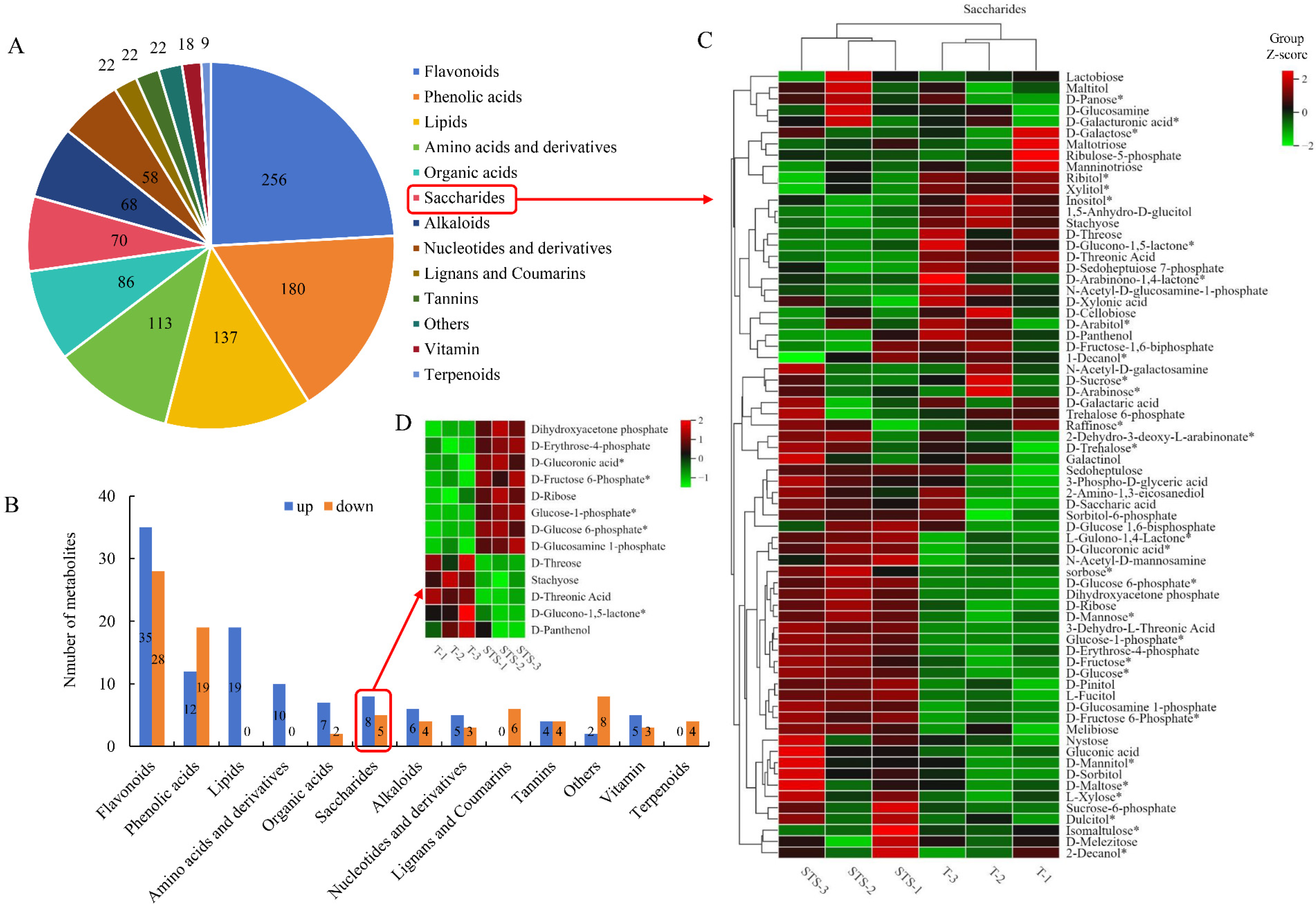
3.7. Differential Metabolite Identification
4. Discussion
4.1. Soybean–Tea Intercropping Changed the Spectral Characteristics of the Tea Seedlings Canopy
4.2. Soybean–Tea Intercropping Improved the Photosynthetic Characteristics of Tea Seedlings’ Leaves
4.3. Soybean–Tea Intercropping Enhanced Photosynthesis-Related Genes’ Expression Levels
4.4. Soybean–Tea Intercropping Promotes the Accumulation of Carbohydrate Substances
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xia, E.; Zhang, H.; Sheng, J.; Li, K.; Zhang, Q.; Kim, C.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The Tea Tree Genome Provides Insights into Tea Flavor and Independent Evolution of Caffeine Biosynthesis. Mol. Plant 2017, 10, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Han, M.H.; Teng, R.M.; Yang, Y.Z.; Wang, Y.H.; Xiong, A.S.; Zhuang, J. Exogenous Melatonin Enhances Photosynthetic Capacity and Related Gene Expression in A Dose-dependent Manner in the Tea Plant (Camellia sinensis (L.) Kuntze). Int. J. Mol. Sci. 2022, 23, 6694. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Z.; Wu, Z.; Wang, X.; Teng, R.; Zhuang, J. Differentially expressed protein and gene analysis revealed the effects of temperature on changes in ascorbic acid metabolism in harvested tea leaves. Hortic. Res. 2018, 5, 65. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.W.; Li, H.; Liu, J.X.; Wang, Y.; Zhuang, J. Integrative transcriptome, proteome, and microRNA analysis reveals the effects of nitrogen sufficiency and deficiency conditions on theanine metabolism in the tea plant (Camellia sinensis). Hortic. Res. 2020, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Shukla, S. Impact of various factors responsible for fluctuation in plant secondary metabolites. J. Appl. Res. Med. Aroma. 2015, 2, 105–113. [Google Scholar] [CrossRef]
- Croft, H.; Chen, J.M. Leaf Pigment Content. In Comprehensive Remote Sensing; Elsevier: Amsterdam, The Netherlands, 2018; pp. 117–142. [Google Scholar]
- Gitelson, A.A.; Peng, Y.; Arkebauer, T.J.; Schepers, J. Relationships between gross primary production, green LAI, and canopy chlorophyll content in maize: Implications for remote sensing of primary production. Remote Sens. Environ. 2014, 144, 65–72. [Google Scholar] [CrossRef]
- Féret, J.B.; François, C.; Gitelson, A.; Asner, G.P.; Barry, K.M.; Panigada, C.; Richardson, A.D.; Jacquemoud, S. Optimizing spectral indices and chemometric analysis of leaf chemical properties using radiative transfer modeling. Remote Sens. Environ. 2011, 115, 2742–2750. [Google Scholar] [CrossRef]
- Pan, W.J.; Wang, X.; Deng, Y.-R.; Li, J.H.; Chen, W.; Chiang, J.Y.; Yang, J.B.; Zheng, L. Nondestructive and intuitive determination of circadian chlorophyll rhythms in soybean leaves using multispectral imaging. Sci. Rep. 2015, 5, 11108. [Google Scholar] [CrossRef] [PubMed]
- Chungcharoen, T.; Donis-Gonzalez, I.; Phetpan, K.; Udompetaikul, V.; Sirisomboon, P.; Suwalak, R. Machine learning-based prediction of nutritional status in oil palm leaves using proximal multispectral images. Comput. Electron. Agric. 2022, 198, 107019. [Google Scholar] [CrossRef]
- Zhang, C.; Xue, Y. Estimation of Biochemical Pigment Content in Poplar Leaves Using Proximal Multispectral Imaging and Regression Modeling Combined with Feature Selection. Sensors 2024, 24, 217. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Koh, S.C. Photosystem II photochemical efficiency and photosynthetic capacity in leaves of tea plant (Camellia sinensis L.) under winter stress in the field. Hortic. Environ. Biotechnol. 2014, 55, 363–371. [Google Scholar] [CrossRef]
- Gunell, S.; Lempiäinen, T.; Rintamäki, E.; Aro, E.M.; Tikkanen, M. Enhanced function of non-photoinhibited photosystem II complexes upon PSII photoinhibition. BBA Bioenerg. 2023, 1864, 148978. [Google Scholar] [CrossRef] [PubMed]
- Van Bezouwen, L.S.; Caffarri, S.; Kale, R.S.; Kouřil, R.; Thunnissen, A.M.W.H.; Oostergetel, G.T.; Boekema, E.J. Subunit and chlorophyll organization of the plant photosystem II supercomplex. Nat. Plants 2017, 3, 17080. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Shang, X.; Liu, G.; Zou, Z.; Zhu, X.; Ma, Y.; Li, F.; Fang, W. The effects of tea plants-soybean intercropping on the secondary metabolites of tea plants by metabolomics analysis. BMC Plant Biol. 2021, 21, 482. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Dong, X.; Wang, Y.; Maker, G.; Agarwal, M.; Ding, Z. Tea-Soybean Intercropping Improves Tea Quality and Nutrition Uptake by Inducing Changes of Rhizosphere Bacterial Communities. Microorganisms 2022, 10, 2149. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Fu, S.; Zhang, X.; Zhao, K.; Chen, H. Intercropping improves soil nutrient availability, soil enzyme activity and tea quantity and quality. Appl. Soil Ecol. 2017, 119, 171–178. [Google Scholar] [CrossRef]
- Wu, T.; Zou, R.; Pu, D.; Lan, Z.; Zhao, B. Non-targeted and targeted metabolomics profiling of tea plants (Camellia sinensis) in response to its intercropping with Chinese chestnut. BMC Plant Biol. 2021, 21, 55. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Li, B.; Xu, C.; Raza, M.; Wang, Q.; Wang, Q.; Fu, Y.; Hu, J.; Imoulan, A.; Hussain, M.; et al. Intercropping Walnut and Tea: Effects on Soil Nutrients, Enzyme Activity, and Microbial Communities. Front. Microbiol. 2022, 13, 852342. [Google Scholar] [CrossRef]
- Wang, T.; Duan, Y.; Lei, X.G.; Cao, Y.; Liu, L.F.; Shang, X.W.; Wang, M.H.; Lv, C.J.; Ma, Y.C.; Fang, W.P.; et al. Tea Plantation Intercropping Legume Improves Soil Ecosystem Multifunctionality and Tea Quality by Regulating Rare Bacterial Taxa. Agronomy 2023, 13, 1110. [Google Scholar] [CrossRef]
- Huang, Z.; Cui, C.; Cao, Y.; Dai, J.; Cheng, X.; Hua, S.; Wang, W.; Duan, Y.; Petropoulos, E.; Wang, H. Tea plant-legume intercropping simultaneously improves soil fertility and tea quality by changing Bacillus species composition. Hortic. Res. 2022, 9, uhac046. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Wang, T.; Zhang, P.X.; Zhao, X.J.; Jiang, J.; Ma, Y.C.; Zhu, X.J.; Fang, W.P. The effect of intercropping leguminous green manure on theanine accumulation in the tea plant: A metagenomic analysis. Plant Cell Environ. 2024, 47, 1141–1159. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.J.; Liang, L.N.; Xu, R.N.; Xu, H.Y.; Sun, L.L.; Liao, H. Intercropping tea plantations with soybean and rapeseed enhances nitrogen fixation through shifts in soil microbial communities. Front. Agric. Sci. Eng. 2022, 9, 344–355. [Google Scholar]
- Mao, Y.; Li, H.; Wang, Y.; Wang, H.; Shen, J.; Xu, Y.; Ding, S.; Wang, H.; Ding, Z.; Fan, K. Rapid monitoring of tea plants under cold stress based on UAV multi-sensor data. Comput. Electron. Agric. 2023, 213, 108176. [Google Scholar] [CrossRef]
- Tian, F.; Jia, T.; Yu, B. Physiological regulation of seed soaking with soybean isoflavones on drought tolerance of Glycine max and Glycine soja. Plant Growth Regul. 2014, 74, 229–237. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.X.; Liu, L.L.; Tang, Q.H.; Sun, K.; Zeng, L.; Wu, Z.J. Evaluation and selection of suitable qRT-PCR reference genes for light responses in tea plant (Camellia sinensis). Sci. Hortic. 2021, 289, 110488. [Google Scholar] [CrossRef]
- Arocho, A.; Chen, B.Y.; Ladanyi, M.; Pan, Q.L. Validation of the 2−ΔΔCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn. Mol. Pathol. 2006, 15, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Neuwirthova, E.; Lhotakova, Z.; Cervena, L.; Lukes, P.; Campbell, P.; Albrechtova, J. Asymmetry of leaf internal structure affects PLSR modelling of anatomical traits using VIS-NIR leaf level spectra. Eur. J. Remote Sens. 2024, 57, 2292154. [Google Scholar] [CrossRef]
- Béland, M.; Kobayashi, H. Drivers of deciduous forest near-infrared reflectance: A 3D radiative transfer modeling exercise based on ground lidar. Remote Sens. Environ. 2024, 302, 113951. [Google Scholar] [CrossRef]
- Pappula-Reddy, S.P.; Kumar, S.; Pang, J.Y.; Chellapilla, B.; Pal, M.; Millar, A.H.; Siddique, K.H.M. High-throughput phenotyping for terminal drought stress in chickpea (Cicer arietinum L.). Plant Stress 2024, 11, 100386. [Google Scholar] [CrossRef]
- Huang, G.J.; Zeng, Y.J. Increased stomatal conductance and leaf biochemical capacity, not mesophyll conductance, contributing to the enhanced photosynthesis in Oryza plants during domestication. Planta 2024, 259, 28. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, H.; Takaragawa, H. Leaf Photosynthetic Reduction at High Temperatures in Various Genotypes of Passion Fruit (Passiflora spp.). Hortic. J. 2023, 92, 412–423. [Google Scholar] [CrossRef]
- Li, X.; Zhao, S.W.; Lin, A.Y.; Yang, Y.Y.; Zhang, G.Z.; Xu, P.; Wu, Y.J.; Yang, Z.C. Effect of Different Ratios of Red and Blue Light on Maximum Stomatal Conductance and Response Rate of Cucumber Seedling Leaves. Agronomy 2023, 13, 1941. [Google Scholar] [CrossRef]
- Yin, H.; Huang, W.L.; Li, F.; Yang, H.B.; Li, Y.; Hu, Y.C.; Yu, K. Multi-temporal UAV Imaging-Based Mapping of Chlorophyll Content in Potato Crop. PFG–J. Photogramm. Remote Sens. Geoinf. Sci. 2022, 91, 91–106. [Google Scholar] [CrossRef]
- Peng, Y.; Nguy-Robertson, A.; Arkebauer, T.; Gitelson, A.A. Assessment of Canopy Chlorophyll Content Retrieval in Maize and Soybean: Implications of Hysteresis on the Development of Generic Algorithms. Remote Sens. 2017, 9, 226. [Google Scholar] [CrossRef]
- Su, W.; Zhao, X.F.; Sun, Z.P.; Zhang, M.Z.; Zou, Z.C.; Wang, W.; Shi, Y.L. Estimating the Corn Canopy Chlorophyll Content Using the Sentinel-2A Image. Spectrosc. Spect. Anal. 2019, 39, 1535–1542. [Google Scholar]
- Schlemmer, M.; Gitelson, A.; Schepers, J.; Ferguson, R.; Peng, Y.; Shanahan, J.; Rundquist, D. Remote estimation of nitrogen and chlorophyll contents in maize at leaf and canopy levels. Int. J. Appl. Earth. Obs. 2013, 25, 47–54. [Google Scholar] [CrossRef]
- Greer, D.H.; Weedon, M.M. Modelling photosynthetic responses to temperature of grapevine (Vitis vinifera cv. Semillon) leaves on vines grown in a hot climate. Plant Cell Environ. 2012, 35, 1050–1064. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Meng, Q.; Brestic, M.; Olsovska, K.; Yang, X. Photosynthesis is improved by exogenous calcium in heat-stressed tobacco plants. J. Plant Physiol. 2011, 168, 2063–2071. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Zhang, Y.C.; Niu, S.C.; Hao, L.H.; Yu, W.B.; Chen, D.F.; Xiang, D.Y. Response of Dahlia Photosynthesis and Transpiration to High-Temperature Stress. Horticulturae 2023, 9, 1047. [Google Scholar] [CrossRef]
- Yang, Q.; Guo, Y.; Li, J.; Wang, L.; Wang, H.; Liu, G.; Fang, W.; Qiang, S.; Strasser, R.J.; Chen, S.G. Natural plant inducer 2-Amino-3-Methylhexanoic acid protects physiological activity against high-temperature damage to tea (Camellia sinensis). Sci. Hortic. 2023, 312, 111836. [Google Scholar] [CrossRef]
- Grieco, M.; Suorsa, M.; Jajoo, A.; Tikkanen, M.; Aro, E.M. Light-harvesting II antenna trimers connect energetically the entire photosynthetic machinery-including both photosystems II and I. BBA Bioenerg. 2015, 1847, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Rantala, S.; Tikkanen, M. Phosphorylation-induced lateral rearrangements of thylakoid protein complexes upon light acclimation. Plant Direct 2018, 2, 39. [Google Scholar] [CrossRef] [PubMed]
- Elias, E.; Liguori, N.; Croce, R. At the origin of the selectivity of the chlorophyll-binding sites in Light Harvesting Complex II (LHCII). Int. J. Biol. Macromol. 2023, 243, 125069. [Google Scholar] [CrossRef] [PubMed]
- Palm, D.M.; Agostini, A.; Averesch, V.; Girr, P.; Werwie, M.; Takahashi, S.; Satoh, H.; Jaenicke, E.; Paulsen, H. Chlorophyll a/b binding-specificity in water-soluble chlorophyll protein. Nat. Plants 2018, 4, 920–929. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.F.; Li, J.J.; Yu, Y.; Kou, X.B.; Periakaruppan, R.; Chen, X.; Li, X.H. STAY-GREEN and light-harvesting complex II chlorophyll a/b binding protein are involved in albinism of a novel albino tea germplasm ‘Huabai 1′. Sci. Hortic. 2022, 293, 110653. [Google Scholar] [CrossRef]
- Zaman, S.; Shen, J.; Wang, S.; Song, D.; Wang, H.; Ding, S.; Pang, X.; Wang, M.; Sabir, I.A.; Wang, Y.; et al. Effect of shading on physiological attributes and comparative transcriptome analysis of Camellia sinensis cultivar reveals tolerance mechanisms to low temperatures. Front. Plant Sci. 2023, 14, 1114988. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Zheng, X.; Liang, Y. High-Light-Induced Degradation of Photosystem II Subunits’ Involvement in the Albino Phenotype in Tea Plants. Int. J. Mol. Sci. 2022, 23, 8522. [Google Scholar] [CrossRef] [PubMed]
- Silveira, J.A.G.; Carvalho, F.E.L. Proteomics, photosynthesis and salt resistance in crops: An integrative view. J. Proteom. 2016, 143, 24–35. [Google Scholar] [CrossRef]
- Cai, W.; Ma, J.; Guo, J.; Zhang, L. Function of ROC4 in the Efficient Repair of Photodamaged Photosystem II in Arabidopsis. Photochem. Photobiol. 2008, 84, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Q.; Wang, Y.H.; Lin, S.X.; Chen, M.H.; Cheng, P.Y.; Du, M.R.; Jia, X.L.; Ye, J.H.; Wang, H.B. Study on the effect of magnesium on leaf metabolites, growth and quality of tea tree. Front. Plant Sci. 2023, 14, 1192151. [Google Scholar] [CrossRef] [PubMed]
- Zhi, Y.C.; Li, X.A.; Wang, X.W.; Jia, M.H.; Wang, Z.Y. Photosynthesis promotion mechanisms of artificial humic acid depend on plant types: A hydroponic study on C3 and C4 plants. Sci. Total Environ. 2024, 917, 170404. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.Y.; Wang, S.Y.; Zhang, L.P.; Xiao, X.D.; Wu, B.G.; Jaroniec, M.; Jiang, B.J. Unveiling enhanced dark photocatalysis: Electron storage-enabled hydrogen production in polymeric carbon nitride. Appl. Catal. B-Environ. 2024, 343, 123475. [Google Scholar] [CrossRef]
- Li, Y.Y.; Sun, W.P.; Yao, Y.H.; Zhang, L.; Xu, S.W.; Zhang, Q.; Huang, T. FRUCTOSE INSENSITIVE1 regulates stem cell function in Arabidopsis in response to fructose signalling. J. Exp. Bot. 2023, 74, 3060–3073. [Google Scholar] [CrossRef] [PubMed]
- Pack, M.; Gulde, T.N.; Völcker, M.V.; Boewe, A.S.; Wrublewsky, S.; Ampofo, E.; Montenarh, M.; Götz, C. Protein Kinase CK2 Contributes to Glucose Homeostasis by Targeting Fructose-1,6-Bisphosphatase 1. Int. J. Mol. Sci. 2023, 24, 428. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.X.; Zhang, X.Y.; Li, M.Y.; Li, T.T.; Duan, X.W.; Zhang, D.D.; Hu, L.M.; Huang, R.M. Label-Free Proteomic Analysis of Molecular Effects of 2-Methoxy-1,4-naphthoquinone on Penicillium italicum. Int. J. Mol. Sci. 2019, 20, 3459. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Kaloudas, D.; Raines, C.A. Expression analysis of the Arabidopsis CP12 gene family suggests novel roles for these proteins in roots and floral tissues. J. Exp. Bot. 2008, 59, 3975–3985. [Google Scholar] [CrossRef] [PubMed]
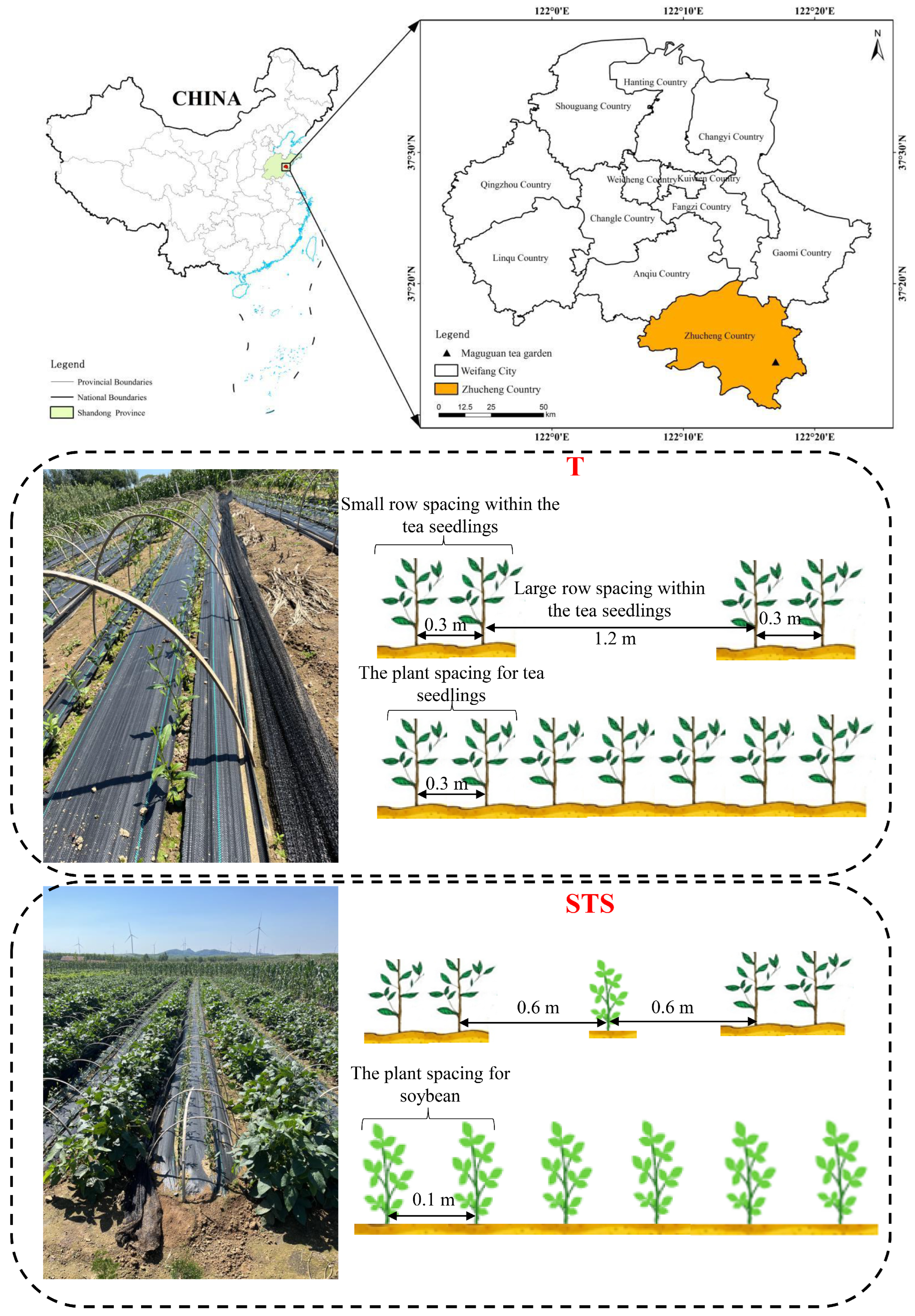

| Wavelength | Centre (nm) | Bandwidth (nm) |
|---|---|---|
| Blue | 452 | 35 |
| Green | 555 | 25 |
| Red | 660 | 20 |
| Red edge1 | 720 | 10 |
| Red edge2 | 750 | 15 |
| Near infrared | 840 | 35 |
| VIs | Formulation |
|---|---|
| LC | (NIR − Edge1)/(NIR + Red) |
| MTC | (Edge2 − Edge1)/(Edge1 − Red) |
| NDRE | (NIR − Edge1)/(NIR + Edge1) |
| NDVI | (Edge2 − Green)/(Edge2 + Green) |
| RENDVI | (Edge2 − Edge1)/(Edge2 + Edge1) |
| VOG1 | Edge2/Edge1 |
| CIgreen | (NIR/Green) − 1 |
| CIred-edge | (NIR/Edge1) − 1 |
| CV | NIR × (RED/Blue2) |
| GLI | (2 × Green − Red − Blue)/(2 × Green + Red + Blue) |
| GNDV | (NIR − Green)/(NIR + Green) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Xu, Y.; Mao, Y.; Wang, S.; Sun, L.; Shen, J.; Xu, X.; Wang, Y.; Ding, Z. The Effects of Soybean–Tea Intercropping on the Photosynthesis Activity of Tea Seedlings Based on Canopy Spectral, Transcriptome and Metabolome Analyses. Agronomy 2024, 14, 850. https://doi.org/10.3390/agronomy14040850
Li X, Xu Y, Mao Y, Wang S, Sun L, Shen J, Xu X, Wang Y, Ding Z. The Effects of Soybean–Tea Intercropping on the Photosynthesis Activity of Tea Seedlings Based on Canopy Spectral, Transcriptome and Metabolome Analyses. Agronomy. 2024; 14(4):850. https://doi.org/10.3390/agronomy14040850
Chicago/Turabian StyleLi, Xiaojiang, Yang Xu, Yilin Mao, Shuangshuang Wang, Litao Sun, Jiazhi Shen, Xiuxiu Xu, Yu Wang, and Zhaotang Ding. 2024. "The Effects of Soybean–Tea Intercropping on the Photosynthesis Activity of Tea Seedlings Based on Canopy Spectral, Transcriptome and Metabolome Analyses" Agronomy 14, no. 4: 850. https://doi.org/10.3390/agronomy14040850






