The Myc Road to Hearing Restoration
Abstract
:1. Introduction
Hearing Loss Is a Worldwide Problem with No Perfect Solution
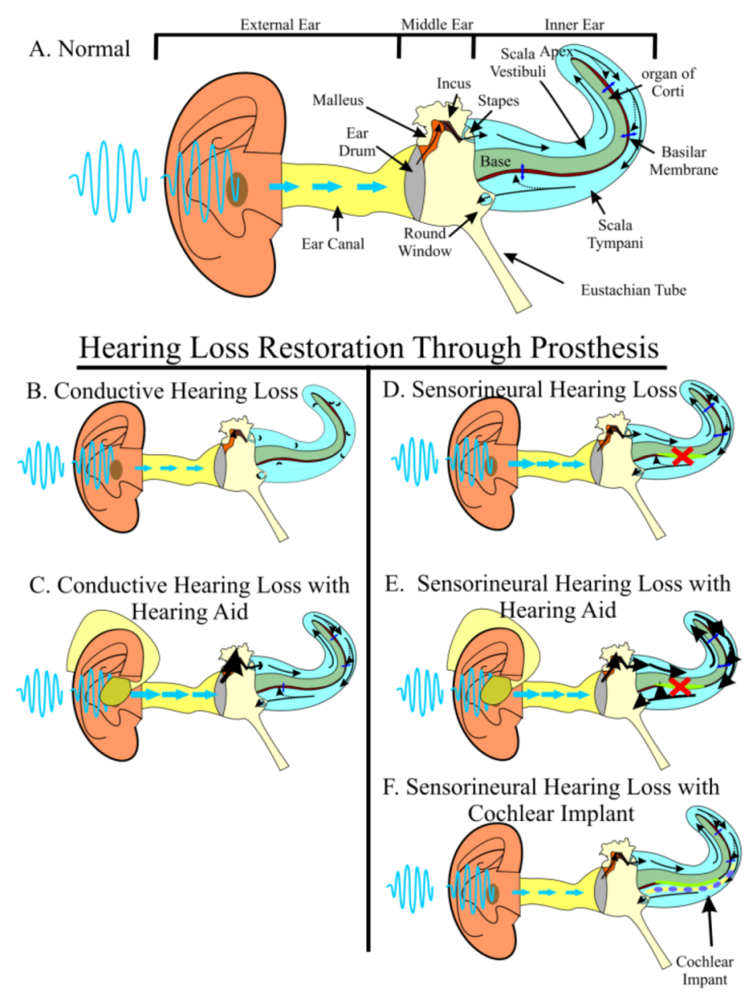
2. The Two-Pronged Attack of Hearing Loss Treatment
2.1. Prevention of Hearing Loss as a First-Line Treatment
2.2. Regeneration of Hair Cells when Prevention Fails
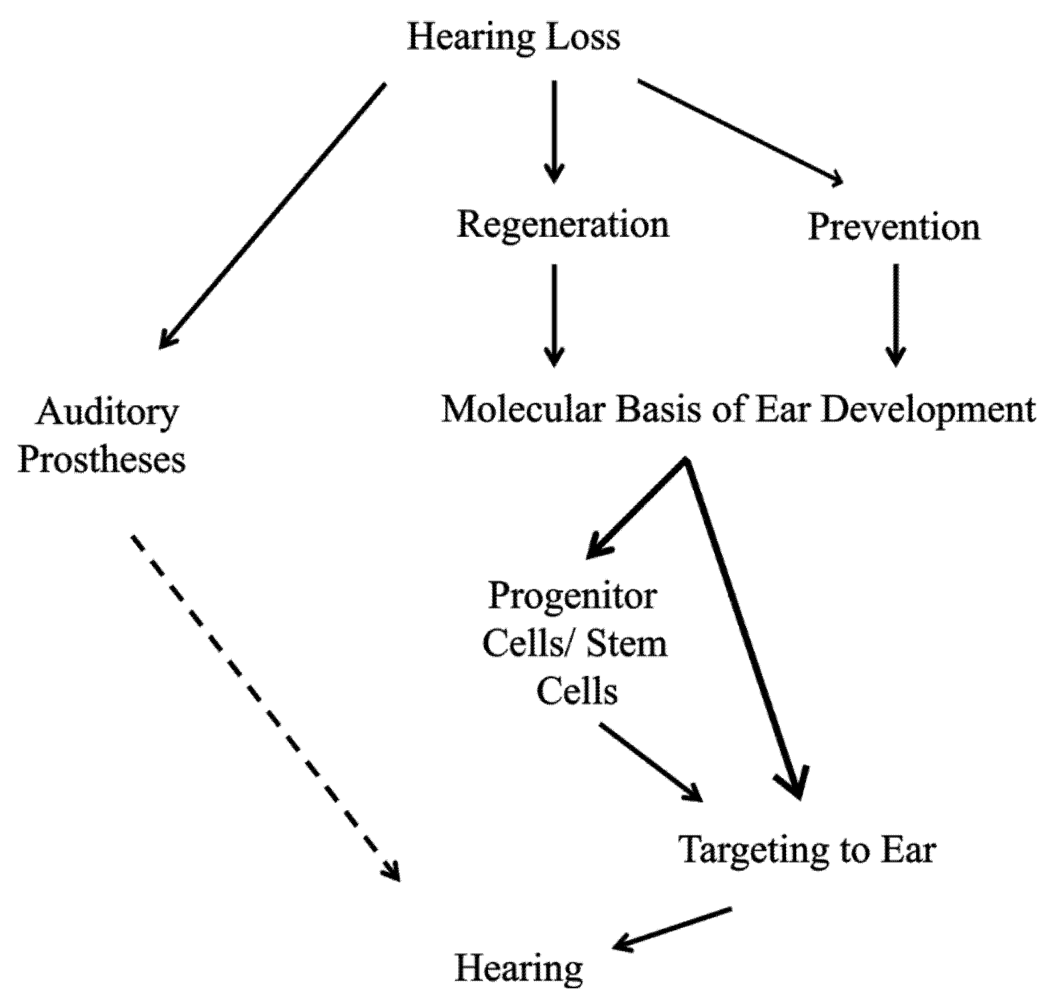
3. Proliferation Is Essential to Regeneration of Organ of Corti Hair Cells
3.1. Ear Development Is a Controlled Balance of Proliferation and Differentiation
3.2. Otic Placode Induction
3.3. Organ of Corti Specification
3.4. Neurosensory Formation
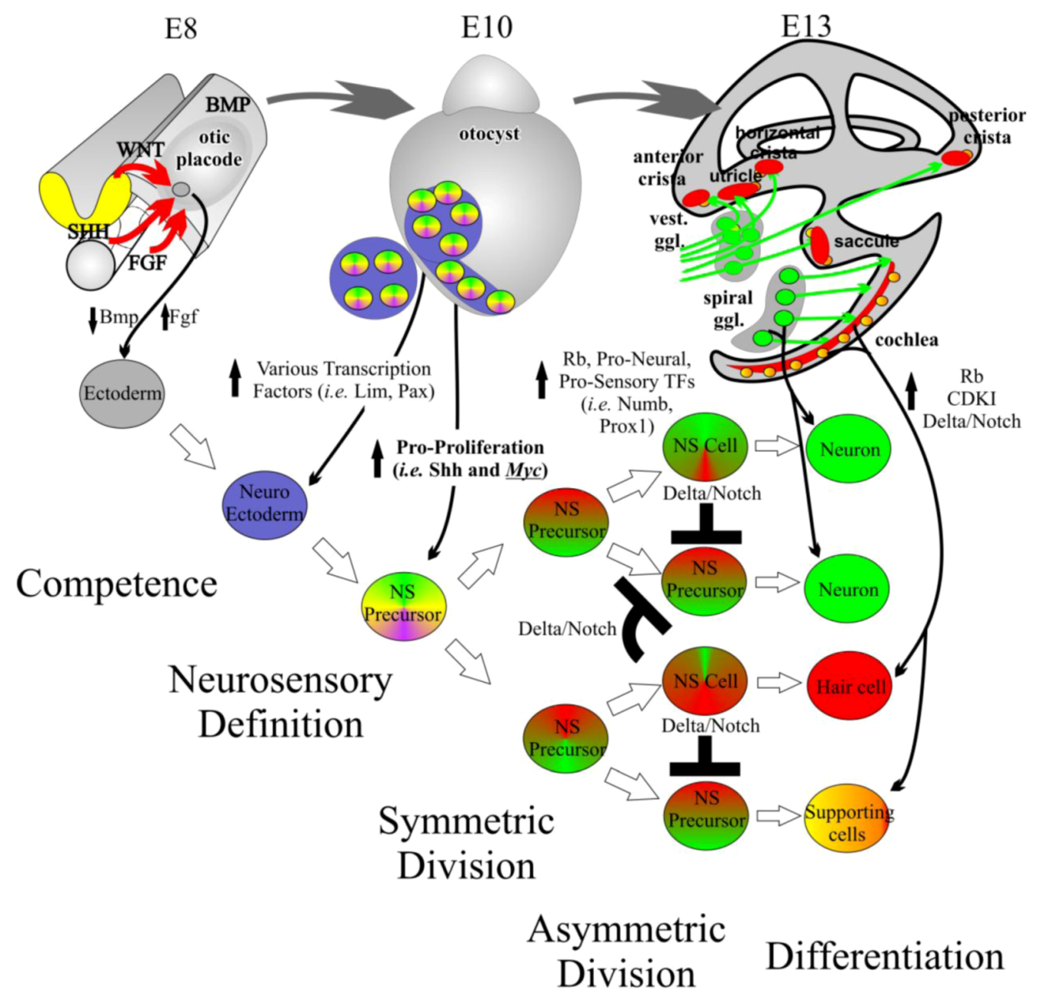
4. The Complex Balance of Proliferation and Differentiation Is Not Well Understood in the Mammalian Inner Ear
5. The Mycs Are an Important Node Integrating Proliferation with Cellular Differentiation
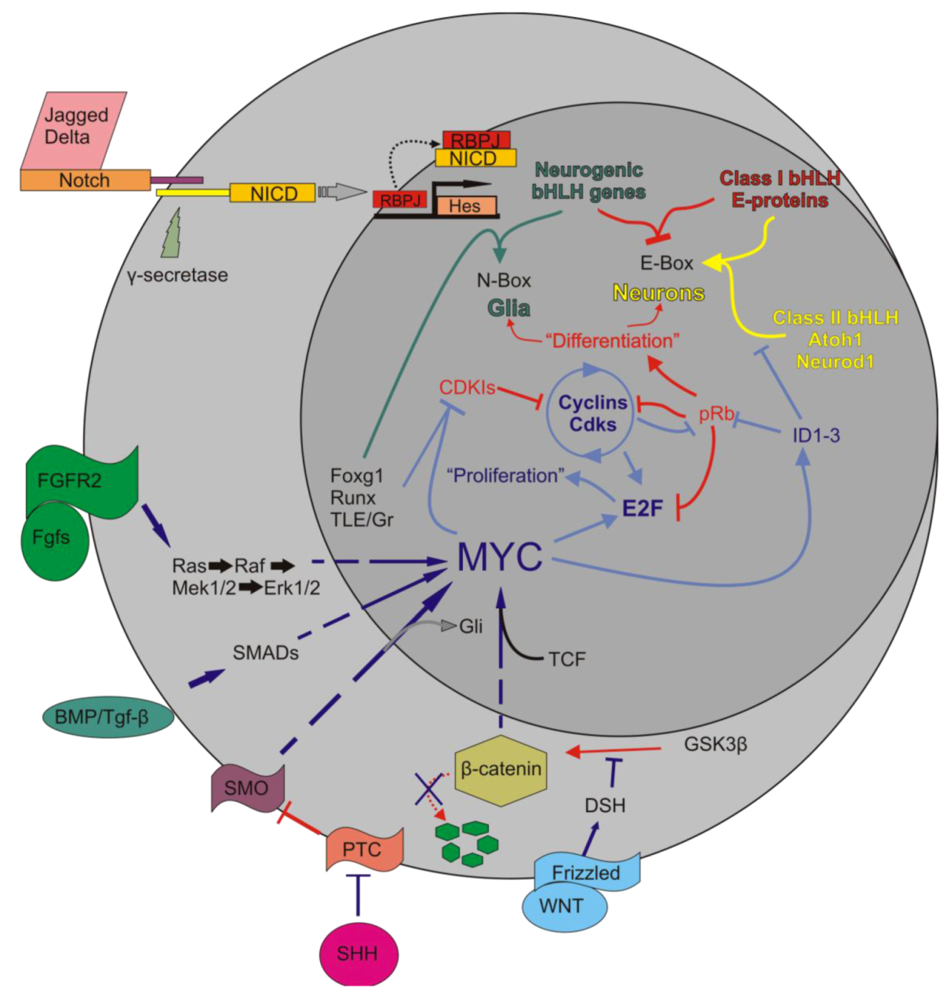
5.1. Myc Is Tightly Regulated to Ensure a Properly Synchronized State
5.2. Myc/Max/Mad Network Balances Proliferation in a Cell
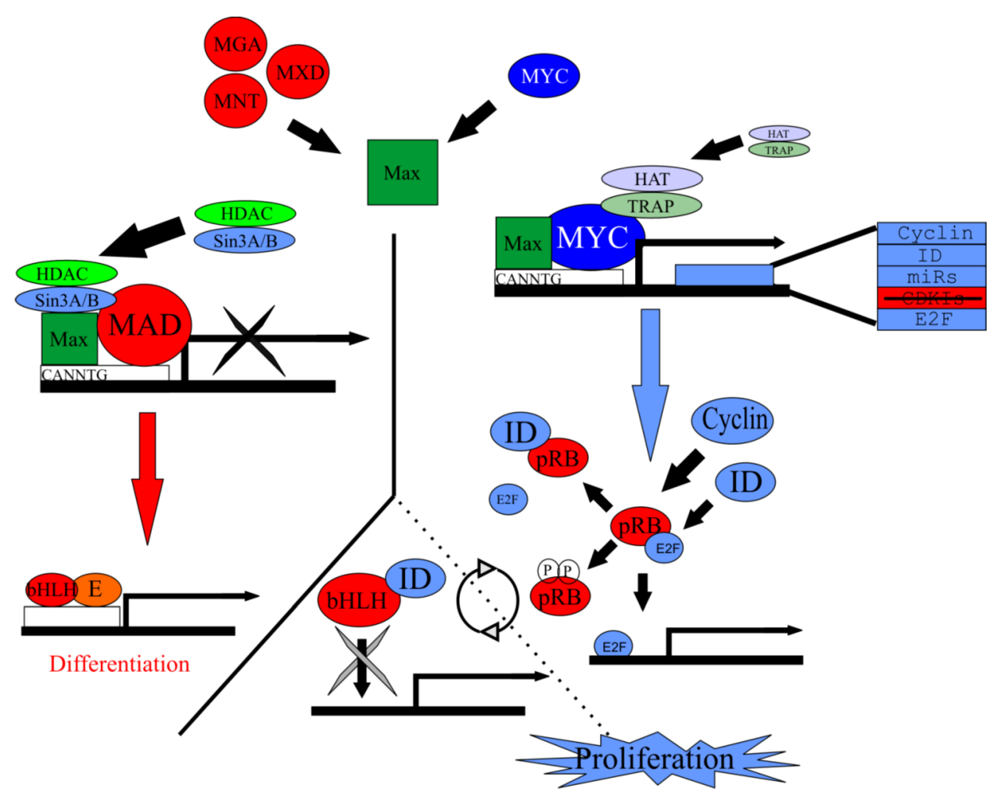
5.3. Myc/Max/Mad Is a Highly Conserved Regulator of Proliferation
6. Ear Specific Knockouts of N-Myc and L-Myc May Provide Translational Insights
7. Summary of Current Literature of N-Myc and L-Myc Function in the Inner Ear
7.1. N-Myc Is Essential for Normal Inner Ear Development
7.2. N-Myc Is Essential to Proper Functionality of Organ of Corti Hair Cells
8. Neither N-Myc nor L-Myc Are Essential for the Long-Term Survival of Organ of Corti Hair Cells
N-Myc and L-Myc Are Necessary for Proper Neurosensory Developmental Timing
9. Conclusions
10. Translational Significance and Future Directions
10.1. Hearing Loss Prevention Is Key, but N-Myc and L-Myc Are Unlikely to Play a Role
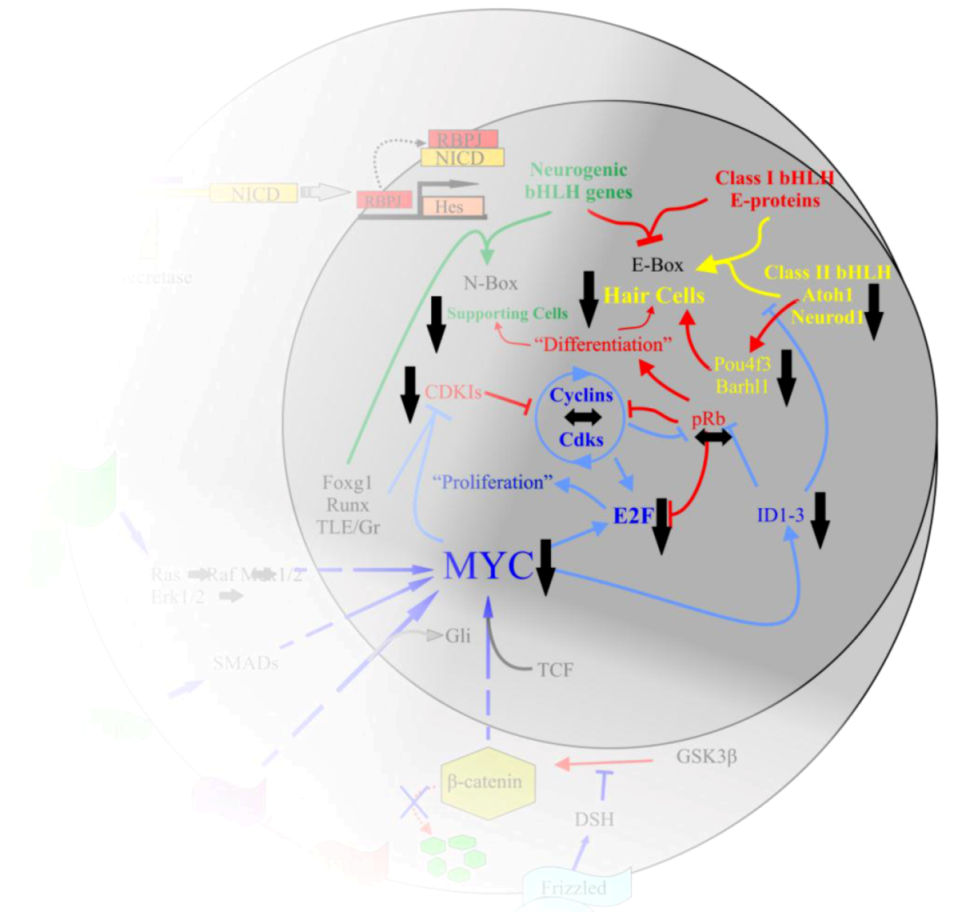
10.2. Proper Manipulation of N-Myc and L-Myc Is Likely Required for Regenerating Hair Cells
10.3. Regeneration of Hair Cells through Forced Cell Cycle Re-Entry of Supporting Cells
- (1)
- The Mycs are essential, non-redundant, untested proto-oncogenes necessary for proliferation. The inner ear appears to be resistant to cancer formation [144] and therefore provides an ample ground to test post-mitotic cell cycle re-entry either through inhibition of tumor suppressor proteins (i.e., pRb, Ink, Kip/Cip) or an increase in proto-oncogenes proteins (i.e., E7). Despite this, forced cell cycle re‑entry has focused on the inhibition of tumor suppressors with very few studies manipulating proto‑oncogenes and none assessing the Myc node, despite its known importance elsewhere in the body. Furthermore, N-Myc and L-Myc are expressed strongly in the inner ear until at least P0.
- (2)
- Myc manipulation may ameliorate cell death mechanisms. Pathways currently being manipulated are essential to the direct progression of the cell cycle and forced cell cycle re-entry is being performed through bypassing normal cell cycle checkpoints, such that damaged DNA that would normally trigger cell cycle discontinuation (through p53), now is propagated until cell death. Cells that re-enter the cell cycle in the absence of p53 have a longer survival [43,44,158]. However, maintaining cell cycle checkpoints (through not removing the Kips/Cips or pRb) may be necessary to ensure the accuracy of newly synthesized DNA. In addition to indirect cell cycle control, the Mycs are essential for cellular metabolism and DNA synthesis. Myc upregulates both ribonucleotide reductase, which is needed for the synthesis of deoxyribonucleotide triphosphates [159] and serine hydroxymethyltransferase, which is required in nucleotide metabolism [160]. Together, manipulation of the Mycs would allow some functioning of the cell cycle checkpoints and the resulting DNA replication may be more robust.
- (3)
- In non-mammalian systems, shortly after hair cell damage, both Atoh1 and Notch1 pathways are activated [161,162] indicating a necessity for properly timed onset of terminal differentiation. We have shown that the timing of proliferation and differentiation is in part driven by the Myc node and that this timing is necessary for proper neurosensory differentiation as viability of hair cells may depend on a proper delay between cell cycle exit and onset of Atoh1-mediated differentiation that varies systematically along the organ of Corti [75]. Forcing proliferation and then inducing differentiation without ensuring that the newly formed progenitor cell populations have had enough time after exiting the cell cycle and are receptive to differentiation is a possible cause of the initial formation of hair cell-like cells followed by hair cell death seen in forced cell cycle re-entry and similar to the pattern that was seen in the Pax2-Cre N-Myc CKO. Thus, manipulation of Myc to force a post-mitotic cell into cell cycle entry, if possible, may make the transition back into the post-mitotic and subsequent differentiated state more long-lasting, achieving in effect the opposite of what was found in the N-Myc CKOs.
10.4. Regeneration of Hair Cells from Stem Cells
10.5. Further Assessment of Functional Redundancy in the Myc/Max/Mad Network
Acknowledgements
Conflict of Interest
References
- Stevens, G.; Flaxman, S.; Brunskill, E.; Mascarenhas, M.; Mathers, C.D.; Finucane, M. Global and regional hearing impairment prevalence: An analysis of 42 studies in 29 countries. Eur. J. Public Health 2011. [Google Scholar]
- Pauley, S.; Kopecky, B.; Beisel, K.; Soukup, G.; Fritzsch, B. Stem cells and molecular strategies to restore hearing. Panminerva Med. 2008, 50, 41–53. [Google Scholar]
- Corwin, J.T.; Cotanche, D.A. Regeneration of sensory hair cells after acoustic trauma. Science 1988, 240, 1772–1774. [Google Scholar]
- Ryals, B.M.; Rubel, E.W. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science 1988, 240, 1774–1776. [Google Scholar]
- OTOscope. Available online: http://www.healthcare.uiowa.edu/labs/morl/otoscope/home.html (accessed on 19 June 2012).
- Shearer, A.E.; Hildebrand, M.S.; Sloan, C.M.; Smith, R.J. Deafness in the genomics era. Hear. Res. 2011, 282, 1–9. [Google Scholar]
- Hone, S.W.; Smith, R.J. Genetics of hearing impairment. Semin. Neonatol. 2001, 6, 531–541. [Google Scholar] [CrossRef]
- Sharma, A.; Campbell, J. A sensitive period for cochlear implantation in deaf children. J. Matern. Fetal Neonatal Med. 2011, 24, 151–153. [Google Scholar]
- Weichbold, V.; Tsiakpini, L.; Coninx, F.; DHaese, P. Development of a parent questionnaire for assessment of auditory behaviour of infants up to two years of age. Laryngorhinootologie 2005, 84, 328–334. [Google Scholar]
- Flynn, M.C. Challenges and recent developments in sound processing for Baha(R). Adv. Otorhinolaryngol. 2011, 71, 112–123. [Google Scholar]
- Colquitt, J.L.; Loveman, E.; Baguley, D.M.; Mitchell, T.E.; Sheehan, P.Z.; Harris, P.; Proops, D.W.; Jones, J.; Clegg, A.J.; et al. Bone-anchored hearing aids for people with bilateral hearing impairment: A systematic review. Clin. Otolaryngol. 2011, 36, 419–441. [Google Scholar] [CrossRef]
- Hausler, R.; Stieger, C.; Bernhard, H.; Kompis, M. A novel implantable hearing system with direct acoustic cochlear stimulation. Audiol. Neurootol. 2008, 13, 247–256. [Google Scholar]
- Muller, M.; von Hunerbein, K.; Hoidis, S.; Smolders, J.W. A physiological place-frequency map of the cochlea in the CBA/J mouse. Hear. Res. 2005, 202, 63–73. [Google Scholar]
- Peterson, N.R.; Pisoni, D.B.; Miyamoto, R.T. Cochlear implants and spoken language processing abilities: Review and assessment of the literature. Restor. Neurol. Neurosci. 2010, 28, 237–250. [Google Scholar]
- Chang, Y.P.; Fu, Q.J. Effects of talker variability on vowel recognition in cochlear implants. J. Speech Lang. Hear. Res. 2006, 49, 1331–1341. [Google Scholar] [CrossRef]
- Bent, T.; Buchwald, A.; Pisoni, D.B. Perceptual adaptation and intelligibility of multiple talkers for two types of degraded speech. J. Acoust. Soc. Am. 2009, 126, 2660–2669. [Google Scholar]
- Gifford, R.H.; Revit, L.J. Speech perception for adult cochlear implant recipients in a realistic background noise: Effectiveness of preprocessing strategies and external options for improving speech recognition in noise. J. Am. Acad. Audiol. 2010, 21, 441–451, quiz 487–448. [Google Scholar]
- Colletti, L.; Mandala, M.; Zoccante, L.; Shannon, R.V.; Colletti, V. Infants versus older children fitted with cochlear implants: Performance over 10 years. Int. J. Pediatr. Otorhinolaryngol. 2011, 75, 504–509. [Google Scholar] [CrossRef]
- Geers, A.E.; Moog, J.S.; Biedenstein, J.; Brenner, C.; Hayes, H. Spoken language scores of children using cochlear implants compared to hearing age-mates at school entry. J. Deaf Stud. Deaf Educ. 2009, 14, 371–385. [Google Scholar]
- Govaerts, P.J.; De Beukelaer, C.; Daemers, K.; De Ceulaer, G.; Yperman, M.; Somers, T.; Schatteman, I.; Offeciers, F.E. Outcome of cochlear implantation at different ages from 0 to 6 years. Otol Neurotol. 2002, 23, 885–890. [Google Scholar]
- Alam, S.A.; Robinson, B.K.; Huang, J.; Green, S.H. Prosurvival and proapoptotic intracellular signaling in rat spiral ganglion neurons in vivo after the loss of hair cells. J. Comp. Neurol. 2007, 503, 832–852. [Google Scholar] [CrossRef]
- Shibata, S.B.; Budenz, C.L.; Bowling, S.A.; Pfingst, B.E.; Raphael, Y. Nerve maintenance and regeneration in the damaged cochlea. Hear. Res. 2011, 281, 56–64. [Google Scholar]
- Nadol, J.B., Jr. Patterns of neural degeneration in the human cochlea and auditory nerve: Implications for cochlear implantation. Otolaryngol. Head Neck Surg. 1997, 117, 220–228. [Google Scholar]
- Wise, A.K.; Fallon, J.B.; Neil, A.J.; Pettingill, L.N.; Geaney, M.S.; Skinner, S.J.; Shepherd, R.K. Combining cell-based therapies and neural prostheses to promote neural survival. Neurotherapeutics 2011, 8, 774–787. [Google Scholar]
- Puligilla, C.; Kelley, M.W. Building the world’s best hearing aid; regulation of cell fate in the cochlea. Curr. Opin. Genet. Dev. 2009, 19, 368–373. [Google Scholar] [CrossRef]
- Bermingham, N.A.; Hassan, B.A.; Price, S.D.; Vollrath, M.A.; Ben-Arie, N.; Eatock, R.A.; Bellen, H.J.; Lysakowski, A.; Zoghbi, H.Y. Math1: An essential gene for the generation of inner ear hair cells. Science 1999, 284, 1837–1841. [Google Scholar]
- Pan, N.; Jahan, I.; Kersigo, J.; Kopecky, B.; Santi, P.; Johnson, S.; Schmitz, H.; Fritzsch, B. Conditional deletion of Atoh1 using Pax2-Cre results in viable mice without differentiated cochlear hair cells that have lost most of the organ of Corti. Hear. Res. 2011, 275, 66–80. [Google Scholar]
- Pan, N.; Jahan, I.; Kersigo, J.; Duncan, J.S.; Kopecky, B.; Fritzsch, B. A novel atoh1 "self-terminating" mouse model reveals the necessity of proper atoh1 level and duration for hair cell differentiation and viability. PLoS One 2012, 7, e30358. [Google Scholar]
- Clough, R.L.; Sud, R.; Davis-Silberman, N.; Hertzano, R.; Avraham, K.B.; Holley, M.; Dawson, S.J. Brn-3c (POU4F3) regulates BDNF and NT-3 promoter activity. Biochem. Biophys. Res. Commun. 2004, 324, 372–381. [Google Scholar]
- Hertzano, R.; Montcouquiol, M.; Rashi-Elkeles, S.; Elkon, R.; Yucel, R.; Frankel, W.N.; Rechavi, G.; Moroy, T.; Friedman, T.B.; Kelley, M.W.; et al. Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum. Mol. Genet. 2004, 13, 2143–2153. [Google Scholar] [CrossRef]
- Chellappa, R.; Li, S.; Pauley, S.; Jahan, I.; Jin, K.; Xiang, M. Barhl1 regulatory sequences required for cell-specific gene expression and autoregulation in the inner ear and central nervous system. Mol. Cell Biol. 2008, 28, 1905–1914. [Google Scholar]
- Henderson, D.; Bielefeld, E.C.; Harris, K.C.; Hu, B.H. The role of oxidative stress in noise-induced hearing loss. Ear Hear. 2006, 27, 1–19. [Google Scholar]
- Kopecky, B.; Fritzsch, B. Regeneration of hair cells: Making sense of all the noise. Pharmaceuticals (Basel) 2011, 4, 848–879. [Google Scholar]
- Pan, N.; Kopecky, B.; Jahan, I.; Fritzsch, B. Understanding the evolution and development of neurosensory transcription factors of the ear to enhance therapeutic translation. Cell Tissue Res. 2012, 349, 415–432. [Google Scholar] [CrossRef]
- Warchol, M.E.; Corwin, J.T. Regenerative proliferation in organ cultures of the avian cochlea: Identification of the initial progenitors and determination of the latency of the proliferative response. J. Neurosci. 1996, 16, 5466–5477. [Google Scholar]
- Warchol, M.E.; Corwin, J.T. Supporting cells in avian vestibular organs proliferate in serum-free culture. Hear. Res. 1993, 71, 28–36. [Google Scholar]
- Collado, M.S.; Burns, J.C.; Hu, Z.; Corwin, J.T. Recent advances in hair cell regeneration research. Curr. Opin. Otolaryngol. Head Neck Surg. 2008, 16, 465–471. [Google Scholar]
- Slattery, E.L.; Warchol, M.E. Cisplatin ototoxicity blocks sensory regeneration in the avian inner ear. J. Neurosci. 2010, 30, 3473–3481. [Google Scholar]
- Warchol, M.E. Characterization of supporting cell phenotype in the avian inner ear: Implications for sensory regeneration. Hear. Res. 2007, 227, 11–18. [Google Scholar] [CrossRef]
- Lin, V.; Golub, J.S.; Nguyen, T.B.; Hume, C.R.; Oesterle, E.C.; Stone, J.S. Inhibition of Notch activity promotes nonmitotic regeneration of hair cells in the adult mouse utricles. J. Neurosci. 2011, 31, 15329–15339. [Google Scholar]
- de Felipe, M.M.; Feijoo Redondo, A.F.; Garcia-Sancho, J.; Schimmang, T.; Alonso, M.B. Cell- and gene-therapy approaches to inner ear repair. Histol. Histopathol. 2011, 26, 923–940. [Google Scholar]
- Jacques, B.E.; Dabdoub, A.; Kelley, M.W. Fgf signaling regulates development and transdifferentiation of hair cells and supporting cells in the basilar papilla. Hear. Res. 2012, 289, 27–39. [Google Scholar]
- Loponen, H.; Ylikoski, J.; Albrecht, J.H.; Pirvola, U. Restrictions in cell cycle progression of adult vestibular supporting cells in response to ectopic cyclin D1 expression. PLoS One 2011, 6, e27360. [Google Scholar]
- Laine, H.; Doetzlhofer, A.; Mantela, J.; Ylikoski, J.; Laiho, M.; Roussel, M.F.; Segil, N.; Pirvola, U. p19(Ink4d) and p21(Cip1) collaborate to maintain the postmitotic state of auditory hair cells, their codeletion leading to DNA damage and p53-mediated apoptosis. J. Neurosci. 2007, 27, 1434–1444. [Google Scholar]
- Rocha-Sanchez, S.M.; Scheetz, L.R.; Contreras, M.; Weston, M.D.; Korte, M.; McGee, J.; Walsh, E.J. Mature mice lacking Rbl2/p130 gene have supernumerary inner ear hair cells and supporting cells. J. Neurosci. 2011, 31, 8883–8893. [Google Scholar] [CrossRef]
- Mantela, J.; Jiang, Z.; Ylikoski, J.; Fritzsch, B.; Zacksenhaus, E.; Pirvola, U. The retinoblastoma gene pathway regulates the postmitotic state of hair cells of the mouse inner ear. Development 2005, 132, 2377–2388. [Google Scholar]
- Oshima, K.; Shin, K.; Diensthuber, M.; Peng, A.W.; Ricci, A.J.; Heller, S. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell 2010, 141, 704–716. [Google Scholar] [CrossRef]
- Duncan, J.S.; Lim, K.C.; Engel, J.D.; Fritzsch, B. Limited inner ear morphogenesis and neurosensory development are possible in the absence of GATA3. Int. J. Dev. Biol. 2011, 55, 297–303. [Google Scholar]
- Kelly, M.C.; Chang, Q.; Pan, A.; Lin, X.; Chen, P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J. Neurosci. 2012, 32, 6699–6710. [Google Scholar] [CrossRef]
- Liu, Z.; Dearman, J.A.; Cox, B.C.; Walters, B.J.; Zhang, L.; Ayrault, O.; Zindy, F.; Gan, L.; Roussel, M.F.; Zuo, J. Age-dependent in vivo conversion of mouse cochlear pillar and deiters’ cells to immature hair cells by Atoh1 ectopic expression. J. Neurosci. 2012, 32, 6600–6610. [Google Scholar] [CrossRef]
- Bouchard, M.; de Caprona, D.; Busslinger, M.; Xu, P.; Fritzsch, B. Pax2 and Pax8 cooperate in mouse inner ear morphogenesis and innervation. BMC Dev. Biol. 2010, 10, 89. [Google Scholar] [CrossRef]
- Ahmed, M.; Xu, J.; Xu, P.X. EYA1 and SIX1 drive the neuronal developmental program in cooperation with the SWI/SNF chromatin-remodeling complex and SOX2 in the mammalian inner ear. Development 2012, 139, 1965–1977. [Google Scholar]
- Ahmed, M.; Wong, E.Y.; Sun, J.; Xu, J.; Wang, F.; Xu, P.X. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev. Cell 2012, 22, 377–390. [Google Scholar]
- Ohyama, T. Unraveling inner ear induction by gene manipulation using Pax2-Cre BAC transgenic mice. Brain Res. 2009, 1277, 84–89. [Google Scholar] [CrossRef]
- Fritzsch, B.; Beisel, K.W.; Hansen, L.A. The molecular basis of neurosensory cell formation in ear development: A blueprint for hair cell and sensory neuron regeneration? Bioessays 2006, 28, 1181–1193. [Google Scholar] [CrossRef]
- Bok, J.; Chang, W.; Wu, D.K. Patterning and morphogenesis of the vertebrate inner ear. Int. J. Dev. Biol. 2007, 51, 521–533. [Google Scholar] [CrossRef]
- Kopecky, B.; Fritzsch, B. Embryology of the ear. In Hereditary Hearing Loss and Its Syndromes; Smith, S., Ed.; Oxford Press: Oxford, UK, 2013. [Google Scholar]
- Fritzsch, B.; Jahan, I.; Pan, N.; Kersigo, J.; Duncan, J.; Kopecky, B. Dissecting the molecular basis of organ of Corti development: Where are we now? Hear. Res. 2011, 276, 16–26. [Google Scholar] [CrossRef]
- Groves, A.K.; Fekete, D.M. Shaping sound in space: The regulation of inner ear patterning. Development 2012, 139, 245–257. [Google Scholar] [CrossRef]
- Fritzsch, B.; Kopecky, B. Neurosensory specification. In Encyclopedia of Genetics; Academic Press: New York, NY, USA, 2012. [Google Scholar]
- Pauley, S.; Lai, E.; Fritzsch, B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev. Dyn. 2006, 235, 2470–2482. [Google Scholar] [CrossRef]
- Jahan, I.; Pan, N.; Kersigo, J.; Calisto, L.E.; Morris, K.A.; Kopecky, B.; Duncan, J.S.; Beisel, K.W.; Fritzsch, B. Expression of Neurog1 instead of Atoh1 can partially rescue organ of Corti cell survival. PLoS One 2012, 7, e30853. [Google Scholar]
- Doetzlhofer, A.; Basch, M.L.; Ohyama, T.; Gessler, M.; Groves, A.K.; Segil, N. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev. Cell 2009, 16, 58–69. [Google Scholar] [CrossRef]
- Kiernan, A.E.; Xu, J.; Gridley, T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006, 2, e4. [Google Scholar] [CrossRef]
- Brooker, R.; Hozumi, K.; Lewis, J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development 2006, 133, 1277–1286. [Google Scholar] [CrossRef]
- Giraldez, F.; Fritzsch, B. The molecular biology of ear development—“Twenty years are nothing”. Int. J. Dev. Biol. 2007, 51, 429–438. [Google Scholar] [CrossRef]
- Coffman, J.A. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol. Int. 2003, 27, 315–324. [Google Scholar] [CrossRef]
- Xia, K.; Xue, H.; Dong, D.; Zhu, S.; Wang, J.; Zhang, Q.; Hou, L.; Chen, H.; Tao, R.; Huang, Z.; et al. Identification of the proliferation/differentiation switch in the cellular network of multicellular organisms. PLoS Comput. Biol. 2006, 2, e145. [Google Scholar] [CrossRef]
- Parker, S.B.; Eichele, G.; Zhang, P.; Rawls, A.; Sands, A.T.; Bradley, A.; Olson, E.N.; Harper, J.W.; Elledge, S.J. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 1995, 267, 1024–1027. [Google Scholar]
- Han, J.D.; Bertin, N.; Hao, T.; Goldberg, D.S.; Berriz, G.F.; Zhang, L.V.; Dupuy, D.; Walhout, A.J.; Cusick, M.E.; Roth, F.P.; et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 2004, 430, 88–93. [Google Scholar]
- Rocha-Sanchez, S.M.; Beisel, K.W. Pocket proteins and cell cycle regulation in inner ear development. Int. J. Dev. Biol. 2007, 51, 585–595. [Google Scholar] [CrossRef]
- Zindy, F.; Cunningham, J.J.; Sherr, C.J.; Jogal, S.; Smeyne, R.J.; Roussel, M.F. Postnatal neuronal proliferation in mice lacking Ink4d and Kip1 inhibitors of cyclin-dependent kinases. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 13462–13467. [Google Scholar]
- Cunningham, J.J.; Levine, E.M.; Zindy, F.; Goloubeva, O.; Roussel, M.F.; Smeyne, R.J. The cyclin-dependent kinase inhibitors p19(Ink4d) and p27(Kip1) are coexpressed in select retinal cells and act cooperatively to control cell cycle exit. Mol. Cell. Neurosci. 2002, 19, 359–374. [Google Scholar] [CrossRef]
- Lee, Y.S.; Liu, F.; Segil, N. A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development 2006, 133, 2817–2826. [Google Scholar] [CrossRef]
- Matei, V.; Pauley, S.; Kaing, S.; Rowitch, D.; Beisel, K.W.; Morris, K.; Feng, F.; Jones, K.; Lee, J.; Fritzsch, B. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev. Dyn. 2005, 234, 633–650. [Google Scholar] [CrossRef]
- Kruman, II.; Wersto, R.P.; Cardozo-Pelaez, F.; Smilenov, L.; Chan, S.L.; Chrest, F.J.; Emokpae, R., Jr.; Gorospe, M.; Mattson, M.P. Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron 2004, 41, 549–561. [Google Scholar] [CrossRef]
- Chen, P.; Segil, N. p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development 1999, 126, 1581–1590. [Google Scholar]
- Lowenheim, H.; Furness, D.N.; Kil, J.; Zinn, C.; Gultig, K.; Fero, M.L.; Frost, D.; Gummer, A.W.; Roberts, J.M.; Rubel, E.W.; et al. Gene disruption of p27(Kip1) allows cell proliferation in the postnatal and adult organ of Corti. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 4084–4088. [Google Scholar]
- Nelsen, C.J.; Rickheim, D.G.; Timchenko, N.A.; Stanley, M.W.; Albrecht, J.H. Transient expression of cyclin D1 is sufficient to promote hepatocyte replication and liver growth in vivo. Cancer Res. 2001, 61, 8564–8568. [Google Scholar]
- Zheng, J.L.; Helbig, C.; Gao, W.Q. Induction of cell proliferation by fibroblast and insulin-like growth factors in pure rat inner ear epithelial cell cultures. J. Neurosci. 1997, 17, 216–226. [Google Scholar]
- Hume, C.R.; Kirkegaard, M.; Oesterle, E.C. ErbB expression: The mouse inner ear and maturation of the mitogenic response to heregulin. J. Assoc. Res. Otolaryngol. 2003, 4, 422–443. [Google Scholar] [CrossRef]
- Montcouquiol, M.; Corwin, J.T. Brief treatments with forskolin enhance s-phase entry in balance epithelia from the ears of rats. J. Neurosci. 2001, 21, 974–982. [Google Scholar]
- Lu, Z.; Corwin, J.T. The influence of glycogen synthase kinase 3 in limiting cell addition in the mammalian ear. Dev. Neurobiol. 2008, 68, 1059–1075. [Google Scholar] [CrossRef]
- Gu, R.; Montcouquiol, M.; Marchionni, M.; Corwin, J.T. Proliferative responses to growth factors decline rapidly during postnatal maturation of mammalian hair cell epithelia. Eur. J. Neurosci. 2007, 25, 1363–1372. [Google Scholar]
- Farah, M.H.; Olson, J.M.; Sucic, H.B.; Hume, R.I.; Tapscott, S.J.; Turner, D.L. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development 2000, 127, 693–702. [Google Scholar]
- Le, T.T.; Wroblewski, E.; Patel, S.; Riesenberg, A.N.; Brown, N.L. Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev. Biol. 2006, 295, 764–778. [Google Scholar]
- Battiste, J.; Helms, A.W.; Kim, E.J.; Savage, T.K.; Lagace, D.C.; Mandyam, C.D.; Eisch, A.J.; Miyoshi, G.; Johnson, J.E. Ascl1 defines sequentially generated lineage-restricted neuronal and oligodendrocyte precursor cells in the spinal cord. Development 2007, 134, 285–293. [Google Scholar]
- Jones, J.M.; Montcouquiol, M.; Dabdoub, A.; Woods, C.; Kelley, M.W. Inhibitors of differentiation and DNA binding (Ids) regulate Math1 and hair cell formation during the development of the organ of Corti. J. Neurosci. 2006, 26, 550–558. [Google Scholar]
- Kopecky, B.; Santi, P.; Johnson, S.; Schmitz, H.; Fritzsch, B. Conditional deletion of N-Myc disrupts neurosensory and non-sensory development of the ear. Dev. Dyn. 2011, 240, 1373–1390. [Google Scholar]
- Grandori, C.; Cowley, S.M.; James, L.P.; Eisenman, R.N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000, 16, 653–699. [Google Scholar]
- Levens, D. Disentangling the MYC web. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 5757–5759. [Google Scholar]
- Liu, J.; Levens, D. Making myc. Curr. Top. Microbiol. Immunol. 2006, 302, 1–32. [Google Scholar]
- Gallant, P.; Shiio, Y.; Cheng, P.F.; Parkhurst, S.M.; Eisenman, R.N. Myc and Max homologs in Drosophila. Science 1996, 274, 1523–1527. [Google Scholar]
- Schreiber-Agus, N.; Horner, J.; Torres, R.; Chiu, F.C.; DePinho, R.A. Zebra fish myc family and max genes: Differential expression and oncogenic activity throughout vertebrate evolution. Mol. Cell Biol. 1993, 13, 2765–2775. [Google Scholar]
- Schreiber-Agus, N.; Torres, R.; Horner, J.; Lau, A.; Jamrich, M.; DePinho, R.A. Comparative analysis of the expression and oncogenic activities of Xenopus c-, N-, and L-myc homologs. Mol. Cell Biol. 1993, 13, 2456–2468. [Google Scholar]
- Malynn, B.A.; de Alboran, I.M.; O'Hagan, R.C.; Bronson, R.; Davidson, L.; DePinho, R.A.; Alt, F.W. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000, 14, 1390–1399. [Google Scholar]
- Pirity, M.; Blanck, J.K.; Schreiber-Agus, N. Lessons learned from Myc/Max/Mad knockout mice. Curr. Top. Microbiol. Immunol. 2006, 302, 205–234. [Google Scholar]
- Facchini, L.M.; Penn, L.Z. The molecular role of Myc in growth and transformation: Recent discoveries lead to new insights. FASEB J. 1998, 12, 633–651. [Google Scholar]
- Cole, M.D.; Mango, S.E. cis-acting determinants of c-myc mRNA stability. Enzyme 1990, 44, 167–180. [Google Scholar]
- Wisdom, R.; Lee, W. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 1991, 5, 232–243. [Google Scholar]
- Lemm, I.; Ross, J. Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol. Cell Biol. 2002, 22, 3959–3969. [Google Scholar]
- Luscher, B.; Kuenzel, E.A.; Krebs, E.G.; Eisenman, R.N. Myc oncoproteins are phosphorylated by casein kinase II. EMBO J. 1989, 8, 1111–1119. [Google Scholar]
- Lutterbach, B.; Hann, S.R. Overexpression of c-Myc and cell immortalization alters c-Myc phosphorylation. Oncogene 1997, 14, 967–975. [Google Scholar]
- Dang, C.V.; Barrett, J.; Villa-Garcia, M.; Resar, L.M.; Kato, G.J.; Fearon, E.R. Intracellular leucine zipper interactions suggest c-Myc hetero-oligomerization. Mol. Cell Biol. 1991, 11, 954–962. [Google Scholar]
- Lee, L.A.; Dang, C.V. Myc target transcriptomes. Curr. Top. Microbiol. Immunol. 2006, 302, 145–167. [Google Scholar]
- Ayer, D.E.; Eisenman, R.N. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 1993, 7, 2110–2119. [Google Scholar]
- Zervos, A.S.; Gyuris, J.; Brent, R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell 1993, 72, 223–232. [Google Scholar]
- Wechsler, D.S.; Shelly, C.A.; Petroff, C.A.; Dang, C.V. MXI1, a putative tumor suppressor gene, suppresses growth of human glioblastoma cells. Cancer Res. 1997, 57, 4905–4912. [Google Scholar]
- Iritani, B.M.; Delrow, J.; Grandori, C.; Gomez, I.; Klacking, M.; Carlos, L.S.; Eisenman, R.N. Modulation of T-lymphocyte development, growth and cell size by the Myc antagonist and transcriptional repressor Mad1. EMBO J. 2002, 21, 4820–4830. [Google Scholar]
- Schreiber-Agus, N.; Chin, L.; Chen, K.; Torres, R.; Rao, G.; Guida, P.; Skoultchi, A.I.; DePinho, R.A. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell 1995, 80, 777–786. [Google Scholar]
- Hassig, C.A.; Fleischer, T.C.; Billin, A.N.; Schreiber, S.L.; Ayer, D.E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 1997, 89, 341–347. [Google Scholar]
- Laherty, C.D.; Yang, W.M.; Sun, J.M.; Davie, J.R.; Seto, E.; Eisenman, R.N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 1997, 89, 349–356. [Google Scholar] [CrossRef]
- McMahon, S.B.; Wood, M.A.; Cole, M.D. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell Biol. 2000, 20, 556–562. [Google Scholar]
- Saleh, A.; Schieltz, D.; Ting, N.; McMahon, S.B.; Litchfield, D.W.; Yates, J.R., 3rd; Lees-Miller, S.P.; Cole, M.D.; Brandl, C.J. Tra1p is a component of the yeast Ada.Spt transcriptional regulatory complexes. J. Biol. Chem. 1998, 273, 26559–26565. [Google Scholar]
- Blackwell, T.K.; Huang, J.; Ma, A.; Kretzner, L.; Alt, F.W.; Eisenman, R.N.; Weintraub, H. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol. Cell Biol. 1993, 13, 5216–5224. [Google Scholar]
- Haggerty, T.J.; Zeller, K.I.; Osthus, R.C.; Wonsey, D.R.; Dang, C.V. A strategy for identifying transcription factor binding sites reveals two classes of genomic c-Myc target sites. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 5313–5318. [Google Scholar]
- Nair, S.K.; Burley, S.K. Structural aspects of interactions within the Myc/Max/Mad network. Curr. Top. Microbiol. Immunol. 2006, 302, 123–143. [Google Scholar]
- Hedges, S.B. The origin and evolution of model organisms. Nat. Rev. Genet. 2002, 3, 838–849. [Google Scholar]
- Gallant, P. Myc/Max/Mad in invertebrates: The evolution of the Max network. Curr. Top. Microbiol. Immunol. 2006, 302, 235–253. [Google Scholar]
- Amati, B.; Frank, S.R.; Donjerkovic, D.; Taubert, S. Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim. Biophys. Acta 1471, 1471, M135–M145. [Google Scholar]
- Elend, M.; Eilers, M. Cell growth: Downstream of Myc—To grow or to cycle? Curr. Biol. 1999, 9, R936–R938. [Google Scholar] [CrossRef]
- Benassayag, C.; Montero, L.; Colombie, N.; Gallant, P.; Cribbs, D.; Morello, D. Human c-Myc isoforms differentially regulate cell growth and apoptosis in Drosophila melanogaster. Mol. Cell Biol. 2005, 25, 9897–9909. [Google Scholar] [CrossRef]
- DePinho, R.A.; Legouy, E.; Feldman, L.B.; Kohl, N.E.; Yancopoulos, G.D.; Alt, F.W. Structure and expression of the murine N-myc gene. Proc. Natl. Acad. Sci. U. S. A. 1986, 83, 1827–1831. [Google Scholar]
- Peyrefitte, S.; Kahn, D.; Haenlin, M. New members of the Drosophila Myc transcription factor subfamily revealed by a genome-wide examination for basic helix-loop-helix genes. Mech. Dev. 2001, 104, 99–104. [Google Scholar]
- Yuan, J.; Tirabassi, R.S.; Bush, A.B.; Cole, M.D. The C. elegans MDL-1 and MXL-1 proteins can functionally substitute for vertebrate MAD and MAX. Oncogene 1998, 17, 1109–1118. [Google Scholar]
- Shen-Li, H.; O'Hagan, R.C.; Hou, H., Jr.; Horner, J.W., 2nd; Lee, H.W.; DePinho, R.A. Essential role for Max in early embryonic growth and development. Genes Dev. 2000, 14, 17–22. [Google Scholar]
- Charron, J.; Malynn, B.A.; Robertson, E.J.; Goff, S.P.; Alt, F.W. High-frequency disruption of the N-myc gene in embryonic stem and pre-B cell lines by homologous recombination. Mol. Cell Biol. 1990, 10, 1799–1804. [Google Scholar]
- Sawai, S.; Shimono, A.; Hanaoka, K.; Kondoh, H. Embryonic lethality resulting from disruption of both N-myc alleles in mouse zygotes. New Biol. 1991, 3, 861–869. [Google Scholar]
- Stanton, B.R.; Reid, S.W.; Parada, L.F. Germ line transmission of an inactive N-myc allele generated by homologous recombination in mouse embryonic stem cells. Mol. Cell Biol. 1990, 10, 6755–6758. [Google Scholar]
- Knoepfler, P.S.; Cheng, P.F.; Eisenman, R.N. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002, 16, 2699–2712. [Google Scholar]
- Davis, A.C.; Wims, M.; Spotts, G.D.; Hann, S.R.; Bradley, A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993, 7, 671–682. [Google Scholar] [CrossRef]
- Romand, R.; Hirning-Folz, U.; Ehret, G. N-myc expression in the embryonic cochlea of the mouse. Hear. Res. 1994, 72, 53–58. [Google Scholar]
- Dominguez-Frutos, E.; Lopez-Hernandez, I.; Vendrell, V.; Neves, J.; Gallozzi, M.; Gutsche, K.; Quintana, L.; Sharpe, J.; Knoepfler, P.S.; Eisenman, R.N.; et al. N-myc controls proliferation, morphogenesis, and patterning of the inner ear. J. Neurosci. 2011, 31, 7178–7189. [Google Scholar]
- Hatton, K.S.; Mahon, K.; Chin, L.; Chiu, F.C.; Lee, H.W.; Peng, D.; Morgenbesser, S.D.; Horner, J.; DePinho, R.A. Expression and activity of L-Myc in normal mouse development. Mol. Cell Biol. 1996, 16, 1794–1804. [Google Scholar]
- Ohyama, T.; Groves, A.K. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis 2004, 38, 195–199. [Google Scholar]
- Pfeffer, P.L.; Gerster, T.; Lun, K.; Brand, M.; Busslinger, M. Characterization of three novel members of the zebrafish Pax2/5/8 family: Dependency of Pax5 and Pax8 expression on the Pax2.1 (noi) function. Development 1998, 125, 3063–3074. [Google Scholar]
- Kopecky, B.; Decook, R.; Fritzsch, B. Mutational ataxia resulting from abnormal vestibular acquisition and processing is partially compensated for. Behav. Neurosci. 2012, 126, 301–313. [Google Scholar] [CrossRef]
- Rabbitts, P.H.; Watson, J.V.; Lamond, A.; Forster, A.; Stinson, M.A.; Evan, G.; Fischer, W.; Atherton, E.; Sheppard, R.; Rabbitts, T.H. Metabolism of c-myc gene products: c-myc mRNA and protein expression in the cell cycle. EMBO J. 1985, 4, 2009–2015. [Google Scholar]
- Waters, C.M.; Littlewood, T.D.; Hancock, D.C.; Moore, J.P.; Evan, G.I. c-myc protein expression in untransformed fibroblasts. Oncogene 1991, 6, 797–805. [Google Scholar]
- Kopecky, B.; DeCook, R.; Fritzsch, B. N-Myc and L-Myc are Essential for Hair Cell Formation but not Maintenance. Brain Res. 2012. submitted for publication.. [Google Scholar]
- Kopecky, B.; Jahan, I.; Fritzsch, B. Correct timing of proliferation and differentiation is necessary for normal inner ear development and auditory hair cell viability. Dev. Dyn. 2012. submitted for publication.. [Google Scholar]
- Campbell, K.; Claussen, A.; Meech, R.; Verhulst, S.; Fox, D.; Hughes, L. D-methionine (D-met) significantly rescues noise-induced hearing loss: Timing studies. Hear. Res. 2011, 282, 138–144. [Google Scholar]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar]
- Sulg, M.; Kirjavainen, A.; Pajusola, K.; Bueler, H.; Ylikoski, J.; Laiho, M.; Pirvola, U. Differential sensitivity of the inner ear sensory cell populations to forced cell cycle re-entry and p53 induction. J. Neurochem. 2010, 112, 1513–1526. [Google Scholar] [CrossRef]
- Brigande, J.V.; Heller, S. Quo vadis, hair cell regeneration? Nat. Neurosci. 2009, 12, 679–685. [Google Scholar] [CrossRef]
- Izumikawa, M.; Minoda, R.; Kawamoto, K.; Abrashkin, K.A.; Swiderski, D.L.; Dolan, D.F.; Brough, D.E.; Raphael, Y. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005, 11, 271–276. [Google Scholar] [CrossRef]
- Izumikawa, M.; Batts, S.A.; Miyazawa, T.; Swiderski, D.L.; Raphael, Y. Response of the flat cochlear epithelium to forced expression of Atoh1. Hear. Res. 2008, 240, 52–56. [Google Scholar]
- Batts, S.A.; Shoemaker, C.R.; Raphael, Y. Notch signaling and Hes labeling in the normal and drug-damaged organ of Corti. Hear. Res. 2009, 249, 15–22. [Google Scholar]
- Ronaghi, M.; Nasr, M.; Heller, S. Concise review: Inner ear stem cells—An oxymoron, but why? Stem. Cells 2012, 30, 69–74. [Google Scholar] [CrossRef]
- Dallos, P. Cochlear amplification, outer hair cells and prestin. Curr. Opin. Neurobiol. 2008, 18, 370–376. [Google Scholar]
- Sage, C.; Huang, M.; Karimi, K.; Gutierrez, G.; Vollrath, M.A.; Zhang, D.S.; Garcia-Anoveros, J.; Hinds, P.W.; Corwin, J.T.; Corey, D.P.; et al. Proliferation of functional hair cells in vivo in the absence of the retinoblastoma protein. Science 2005, 307, 1114–1118. [Google Scholar]
- Weber, T.; Corbett, M.K.; Chow, L.M.; Valentine, M.B.; Baker, S.J.; Zuo, J. Rapid cell-cycle reentry and cell death after acute inactivation of the retinoblastoma gene product in postnatal cochlear hair cells. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 781–785. [Google Scholar]
- Cardinaal, R.M.; de Groot, J.C.; Huizing, E.H.; Veldman, J.E.; Smoorenburg, G.F. Cisplatin-induced ototoxicity: Morphological evidence of spontaneous outer hair cell recovery in albino guinea pigs? Hear. Res. 2000, 144, 147–156. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, W.; Ding, D.; Salvi, R. Pifithrin-alpha suppresses p53 and protects cochlear and vestibular hair cells from cisplatin-induced apoptosis. Neuroscience 2003, 120, 191–205. [Google Scholar]
- Ceballos, E.; Munoz-Alonso, M.J.; Berwanger, B.; Acosta, J.C.; Hernandez, R.; Krause, M.; Hartmann, O.; Eilers, M.; Leon, J. Inhibitory effect of c-Myc on p53-induced apoptosis in leukemia cells. Microarray analysis reveals defective induction of p53 target genes and upregulation of chaperone genes. Oncogene 2005, 24, 4559–4571. [Google Scholar] [CrossRef]
- Ceballos, E.; Delgado, M.D.; Gutierrez, P.; Richard, C.; Muller, D.; Eilers, M.; Ehinger, M.; Gullberg, U.; Leon, J. c-Myc antagonizes the effect of p53 on apoptosis and p21WAF1 transactivation in K562 leukemia cells. Oncogene 2000, 19, 2194–2204. [Google Scholar]
- Fulda, S.; Lutz, W.; Schwab, M.; Debatin, K.M. MycN sensitizes neuroblastoma cells for drug-triggered apoptosis. Med. Pediatr. Oncol. 2000, 35, 582–584. [Google Scholar]
- Laine, H.; Sulg, M.; Kirjavainen, A.; Pirvola, U. Cell cycle regulation in the inner ear sensory epithelia: Role of cyclin D1 and cyclin-dependent kinase inhibitors. Dev. Biol. 2010, 337, 134–146. [Google Scholar]
- Bello-Fernandez, C.; Packham, G.; Cleveland, J.L. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 7804–7808. [Google Scholar] [CrossRef]
- Nikiforov, M.A.; Chandriani, S.; O'Connell, B.; Petrenko, O.; Kotenko, I.; Beavis, A.; Sedivy, J.M.; Cole, M.D. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol. Cell Biol. 2002, 22, 5793–5800. [Google Scholar]
- Cafaro, J.; Lee, G.S.; Stone, J.S. Atoh1 expression defines activated progenitors and differentiating hair cells during avian hair cell regeneration. Dev. Dyn. 2007, 236, 156–170. [Google Scholar]
- Daudet, N.; Gibson, R.; Shang, J.; Bernard, A.; Lewis, J.; Stone, J. Notch regulation of progenitor cell behavior in quiescent and regenerating auditory epithelium of mature birds. Dev. Biol. 2009, 326, 86–100. [Google Scholar] [CrossRef]
- Oshima, K.; Grimm, C.M.; Corrales, C.E.; Senn, P.; Martinez Monedero, R.; Geleoc, G.S.; Edge, A.; Holt, J.R.; Heller, S. Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. J. Assoc. Res. Otolaryngol. 2007, 8, 18–31. [Google Scholar]
- Li, H.; Corrales, C.E.; Edge, A.; Heller, S. Stem cells as therapy for hearing loss. Trends Mol. Med. 2004, 10, 309–315. [Google Scholar]
- Nesbit, C.E.; Tersak, J.M.; Prochownik, E.V. MYC oncogenes and human neoplastic disease. Oncogene 1999, 18, 3004–3016. [Google Scholar]
- Cole, M.D.; McMahon, S.B. The Myc oncoprotein: A critical evaluation of transactivation and target gene regulation. Oncogene 1999, 18, 2916–2924. [Google Scholar]
- Berberich, S.; Hyde-DeRuyscher, N.; Espenshade, P.; Cole, M. max encodes a sequence-specific DNA-binding protein and is not regulated by serum growth factors. Oncogene 1992, 7, 775–779. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kopecky, B.; Fritzsch, B. The Myc Road to Hearing Restoration. Cells 2012, 1, 667-698. https://doi.org/10.3390/cells1040667
Kopecky B, Fritzsch B. The Myc Road to Hearing Restoration. Cells. 2012; 1(4):667-698. https://doi.org/10.3390/cells1040667
Chicago/Turabian StyleKopecky, Benjamin, and Bernd Fritzsch. 2012. "The Myc Road to Hearing Restoration" Cells 1, no. 4: 667-698. https://doi.org/10.3390/cells1040667



