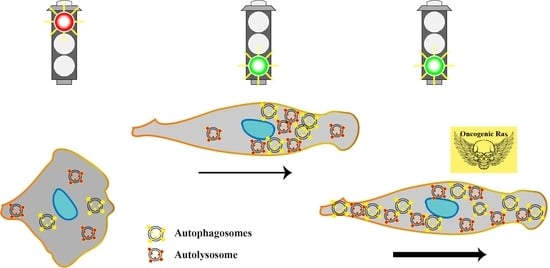
Autophagy Is Polarized toward Cell Front during Migration and Spatially Perturbed by Oncogenic Ras
Abstract
:1. Introduction
2. Materials and Methods
2.1. List of Plasmids
2.2. List of Reagents
2.3. Cell Culture and Transfections
2.4. Surface Patterning
2.5. Sample Preparation
2.6. Image Acquisition
2.7. Image Processing with TopoAutophagy Macro
2.8. Tumor-on-Chip 3D Motility Assay
2.9. Statistical Analysis
3. Results and Discussion
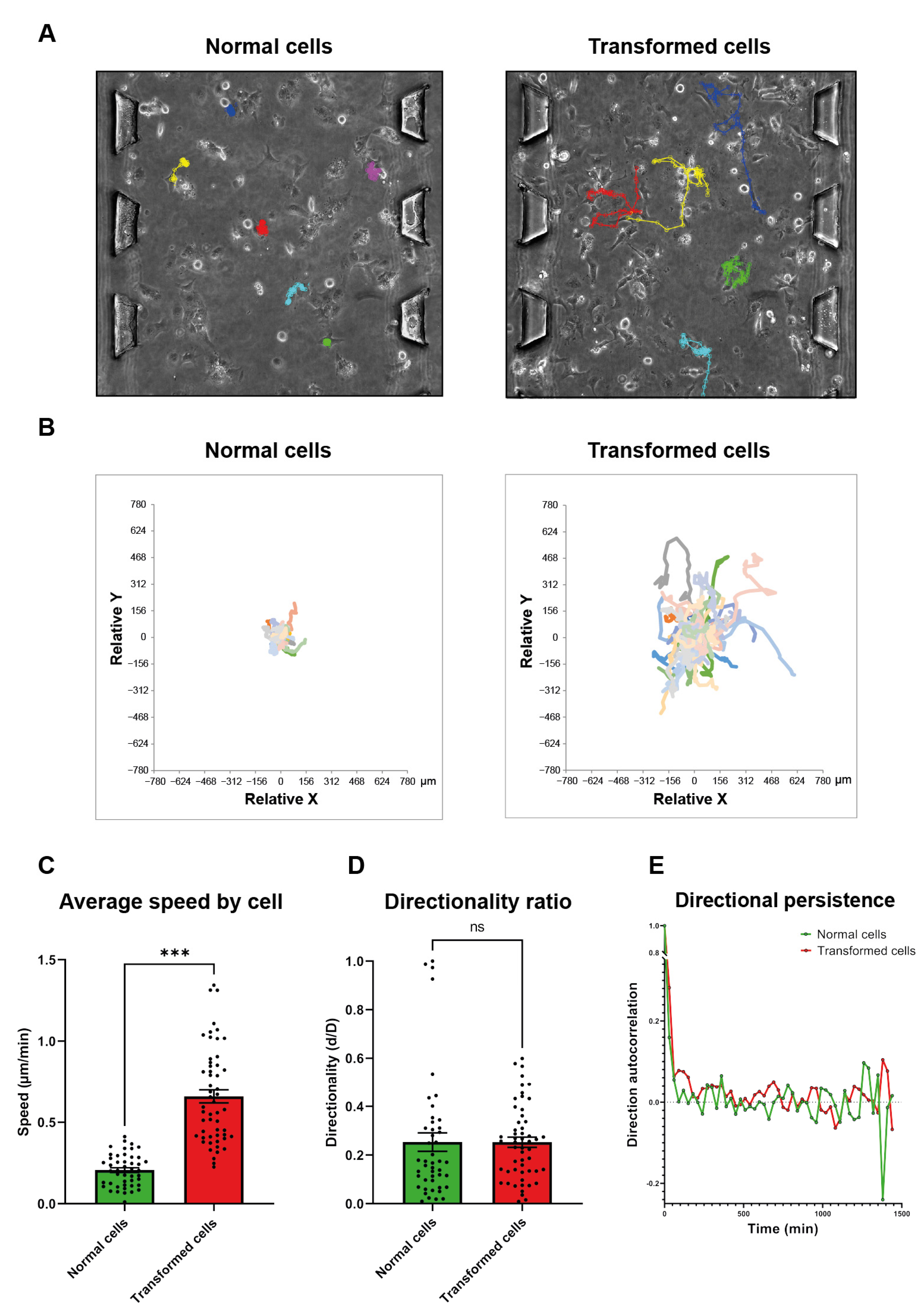
3.1. Ras-Transformation Promotes an Active Random Motility in 3D Collagen Gel
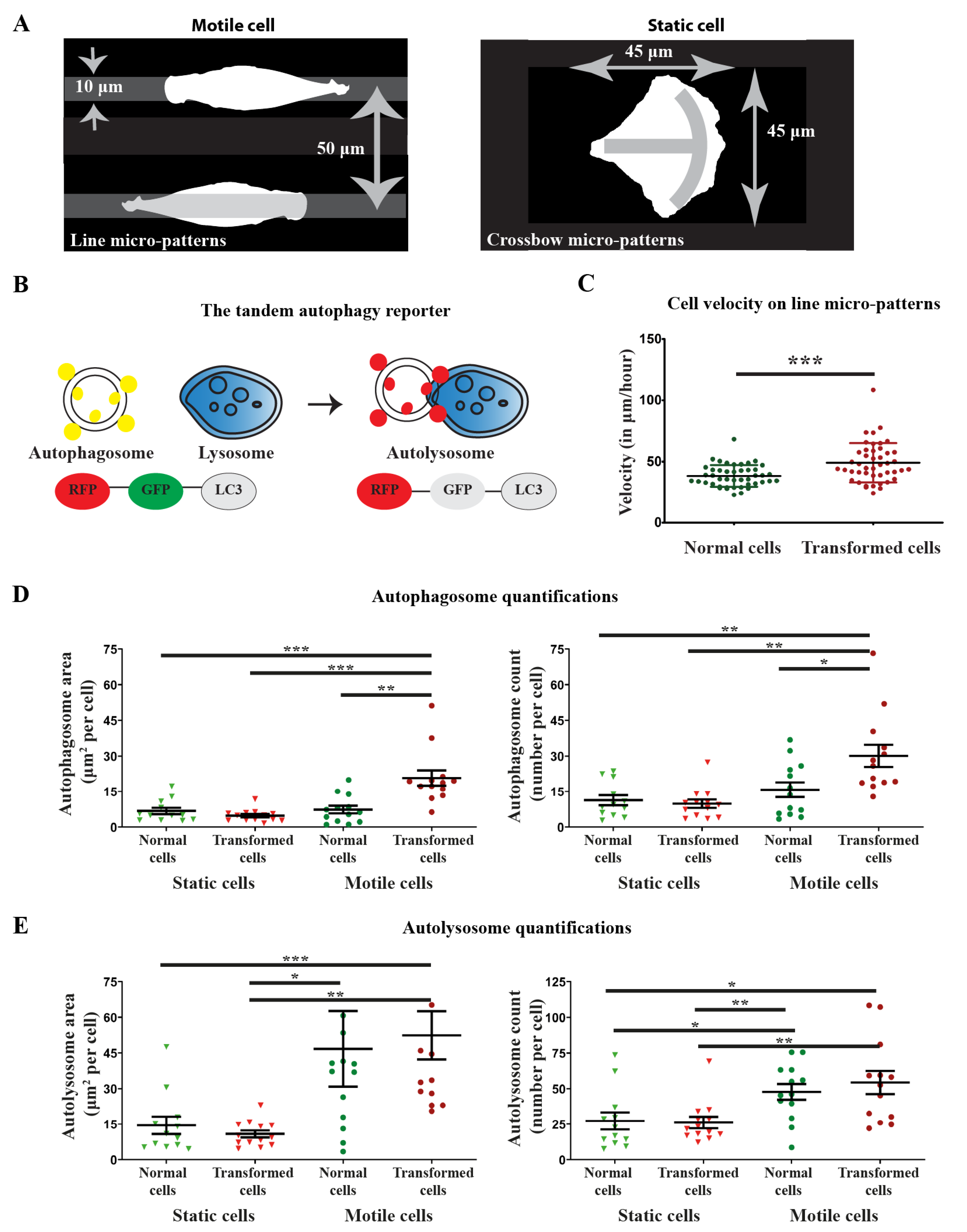
3.2. Development of a Micro-Pattern Strategy to Study Spatial-Temporal Localization of Autophagy Organelles during Directional Migration
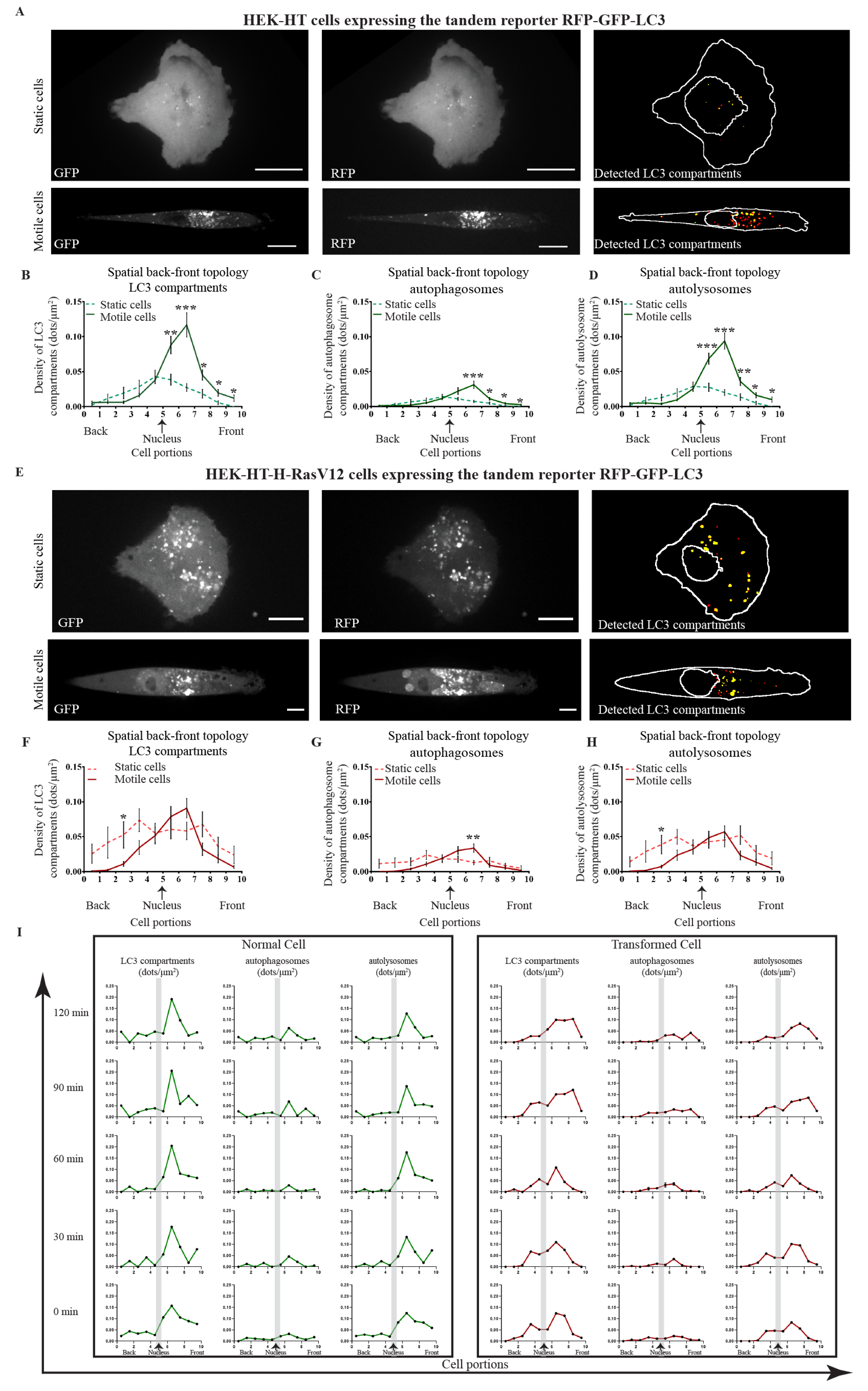
3.3. LC3 Compartments Are Polarized at the Front toward the Direction of Migration and Spatially Perturbed by Oncogenic Ras
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mizushima, N. A Brief History of Autophagy from Cell Biology to Physiology and Disease. Nat. Cell Biol. 2018, 20, 521–527. [Google Scholar] [CrossRef]
- Molino, D.; Zemirli, N.; Codogno, P.; Morel, E. The Journey of the Autophagosome through Mammalian Cell Organelles and Membranes. J. Mol. Biol. 2017, 429, 497–514. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [Green Version]
- Kenific, C.M.; Debnath, J. Cellular and Metabolic Functions for Autophagy in Cancer Cells. Trends Cell Biol. 2015, 25, 37–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mowers, E.E.; Sharifi, M.N.; Macleod, K.F. Autophagy in Cancer Metastasis. Oncogene 2017, 36, 1619–1630. [Google Scholar] [CrossRef]
- Bettoun, A.; Joffre, C.; Zago, G.; Surdez, D.; Vallerand, D.; Gundogdu, R.; Sharif, A.A.D.; Gomez, M.; Cascone, I.; Meunier, B.; et al. Mitochondrial Clearance by the STK38 Kinase Supports Oncogenic Ras-Induced Cell Transformation. Oncotarget 2016, 7, 44142–44160. [Google Scholar] [CrossRef]
- Guo, J.Y.; Chen, H.-Y.; Mathew, R.; Fan, J.; Strohecker, A.M.; Karsli-Uzunbas, G.; Kamphorst, J.J.; Chen, G.; Lemons, J.M.S.; Karantza, V.; et al. Activated Ras Requires Autophagy to Maintain Oxidative Metabolism and Tumorigenesis. Genes Dev. 2011, 25, 460–470. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Wang, X.; Contino, G.; Liesa, M.; Sahin, E.; Ying, H.; Bause, A.; Li, Y.; Stommel, J.M.; Dell’antonio, G.; et al. Pancreatic Cancers Require Autophagy for Tumor Growth. Genes Dev. 2011, 25, 717–729. [Google Scholar] [CrossRef] [Green Version]
- Lefort, S.; Joffre, C.; Kieffer, Y.; Givel, A.-M.; Bourachot, B.; Zago, G.; Bieche, I.; Dubois, T.; Meseure, D.; Vincent-Salomon, A.; et al. Inhibition of Autophagy as a New Means of Improving Chemotherapy Efficiency in High-LC3B Triple-Negative Breast Cancers. Autophagy 2014, 10, 2122–2142. [Google Scholar] [CrossRef]
- Lock, R.; Kenific, C.M.; Leidal, A.M.; Salas, E.; Debnath, J. Autophagy-Dependent Production of Secreted Factors Facilitates Oncogenic RAS-Driven Invasion. Cancer Discov. 2014, 4, 466–479. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Z.; Xie, X.; Cao, H.; Zhou, X.; Zhang, X.D.; Fan, H.; Liu, Z. Autophagy Facilitates TLR4- and TLR3-Triggered Migration and Invasion of Lung Cancer Cells through the Promotion of TRAF6 Ubiquitination. Autophagy 2014, 10, 257–268. [Google Scholar] [CrossRef] [Green Version]
- Kenific, C.M.; Stehbens, S.J.; Goldsmith, J.; Leidal, A.M.; Faure, N.; Ye, J.; Wittmann, T.; Debnath, J. NBR1 Enables Autophagy-Dependent Focal Adhesion Turnover. J. Cell Biol. 2016, 212, 577–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharifi, M.N.; Mowers, E.E.; Drake, L.E.; Collier, C.; Chen, H.; Zamora, M.; Mui, S.; Macleod, K.F. Autophagy Promotes Focal Adhesion Disassembly and Cell Motility of Metastatic Tumor Cells through the Direct Interaction of Paxillin with LC3. Cell Rep. 2016, 15, 1660–1672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuloup-Minguez, V.; Hamaï, A.; Greffard, A.; Nicolas, V.; Codogno, P.; Botti, J. Autophagy Modulates Cell Migration and Β1 Integrin Membrane Recycling. Cell Cycle Georget. Tex. 2013, 12, 3317–3328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiang, L.; Zhao, B.; Ming, M.; Wang, N.; He, T.-C.; Hwang, S.; Thorburn, A.; He, Y.-Y. Regulation of Cell Proliferation and Migration by P62 through Stabilization of Twist1. Proc. Natl. Acad. Sci. USA 2014, 111, 9241–9246. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Tsujioka, M.; Honda, S.; Tanaka, M.; Shimizu, S. Autophagy Suppresses Cell Migration by Degrading GEF-H1, a RhoA GEF. Oncotarget 2016, 7, 34420–34429. [Google Scholar] [CrossRef]
- Azioune, A.; Storch, M.; Bornens, M.; Théry, M.; Piel, M. Simple and Rapid Process for Single Cell Micro-Patterning. Lab Chip 2009, 9, 1640–1642. [Google Scholar] [CrossRef]
- Petrie, R.J.; Doyle, A.D.; Yamada, K.M. Random versus Directionally Persistent Cell Migration. Nat. Rev. Mol. Cell Biol. 2009, 10, 538–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soucy, P.A.; Hoh, M.; Heinz, W.; Hoh, J.; Romer, L. Oriented Matrix Promotes Directional Tubulogenesis. Acta Biomater. 2015, 11, 264–273. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.-H.; Kim, Y.K.; Han, E.D.; Seo, Y.-H.; Kim, B.H.; Mofrad, M.R.K. Passive Control of Cell Locomotion Using Micropatterns: The Effect of Micropattern Geometry on the Migratory Behavior of Adherent Cells. Lab Chip 2012, 12, 2391–2402. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Noda, T.; Yoshimori, T. Dissection of the Autophagosome Maturation Process by a Novel Reporter Protein, Tandem Fluorescent-Tagged LC3. Autophagy 2007, 3, 452–460. [Google Scholar] [CrossRef] [Green Version]
- Hahn, W.C.; Counter, C.M.; Lundberg, A.S.; Beijersbergen, R.L.; Brooks, M.W.; Weinberg, R.A. Creation of Human Tumour Cells with Defined Genetic Elements. Nature 1999, 400, 464–468. [Google Scholar] [CrossRef]
- O’Hayer, K.M.; Counter, C.M. A Genetically Defined Normal Human Somatic Cell System to Study Ras Oncogenesis in Vivo and in Vitro. Methods Enzymol. 2006, 407, 637–647. [Google Scholar] [CrossRef]
- Singh, M.K.; Martin, A.P.J.; Joffre, C.; Zago, G.; Camonis, J.; Coppey, M.; Parrini, M.C. Localization of RalB Signaling at Endomembrane Compartments and Its Modulation by Autophagy. Sci. Rep. 2019, 9, 8910. [Google Scholar] [CrossRef] [Green Version]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next Generation of Scientific Image Data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Nguyen, M.; De Ninno, A.; Mencattini, A.; Mermet-Meillon, F.; Fornabaio, G.; Evans, S.S.; Cossutta, M.; Khira, Y.; Han, W.; Sirven, P.; et al. Dissecting Effects of Anti-Cancer Drugs and Cancer-Associated Fibroblasts by On-Chip Reconstitution of Immunocompetent Tumor Microenvironments. Cell Rep. 2018, 25, 3884–3893.e3. [Google Scholar] [CrossRef] [Green Version]
- Meijering, E.; Dzyubachyk, O.; Smal, I. Methods for Cell and Particle Tracking. Meth. Enzymol. 2012, 504, 183–200. [Google Scholar]
- Gorelik, R.; Gautreau, A. Quantitative and Unbiased Analysis of Directional Persistence in Cell Migration. Nat. Protoc. 2014, 9, 1931–1943. [Google Scholar] [CrossRef]
- Lukinavičius, G.; Blaukopf, C.; Pershagen, E.; Schena, A.; Reymond, L.; Derivery, E.; Gonzalez-Gaitan, M.; D’Este, E.; Hell, S.W.; Wolfram Gerlich, D.; et al. SiR-Hoechst Is a Far-Red DNA Stain for Live-Cell Nanoscopy. Nat. Commun. 2015, 6, 8497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, O.; Saurin, A.T.; Higgins, J.M.G. The Live Cell DNA Stain SiR-Hoechst Induces DNA Damage Responses and Impairs Cell Cycle Progression. Sci. Rep. 2018, 8, 7898. [Google Scholar] [CrossRef]
- Zago, G.; Veith, I.; Singh, M.; Fuhrmann, L.; De Beco, S.; Remorino, A.; Takaoka, S.; Palmeri, M.; Berger, F.; Brandon, N.; et al. RalB Directly Triggers Invasion Downstream Ras by Mobilizing the Wave Complex. eLife 2018, 7, e40474. [Google Scholar] [CrossRef] [PubMed]
- Bodemann, B.O.; Orvedahl, A.; Cheng, T.; Ram, R.R.; Ou, Y.-H.; Formstecher, E.; Maiti, M.; Hazelett, C.C.; Wauson, E.M.; Balakireva, M.; et al. RalB and the Exocyst Mediate the Cellular Starvation Response by Direct Activation of Autophagosome Assembly. Cell 2011, 144, 253–267. [Google Scholar] [CrossRef] [Green Version]
- Joffre, C.; Codogno, P.; Fanto, M.; Hergovich, A.; Camonis, J. STK38 at the Crossroad between Autophagy and Apoptosis. Autophagy 2016, 12, 594–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrini, M.C.; Sadou-Dubourgnoux, A.; Aoki, K.; Kunida, K.; Biondini, M.; Hatzoglou, A.; Poullet, P.; Formstecher, E.; Yeaman, C.; Matsuda, M.; et al. SH3BP1, an Exocyst-Associated RhoGAP, Inactivates Rac1 at the Front to Drive Cell Motility. Mol. Cell 2011, 42, 650–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biondini, M.; Duclos, G.; Meyer-Schaller, N.; Silberzan, P.; Camonis, J.; Parrini, M.C. RalB Regulates Contractility-Driven Cancer Dissemination upon TGFβ Stimulation via the RhoGEF GEF-H1. Sci. Rep. 2015, 5, 11759. [Google Scholar] [CrossRef] [Green Version]
- Latgé, B.; Schauer, K. Determining the Intracellular Organization of Organelles. Meth. Mol. Biol. Clifton NJ 2019, 1862, 263–278. [Google Scholar] [CrossRef]
- Schauer, K.; Duong, T.; Bleakley, K.; Bardin, S.; Bornens, M.; Goud, B. Probabilistic Density Maps to Study Global Endomembrane Organization. Nat. Meth. 2010, 7, 560–566. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, M.K.; Zago, G.; Veith, I.; Camonis, J.; Coppey, M.; Parrini, M.C. Autophagy Is Polarized toward Cell Front during Migration and Spatially Perturbed by Oncogenic Ras. Cells 2021, 10, 2637. https://doi.org/10.3390/cells10102637
Singh MK, Zago G, Veith I, Camonis J, Coppey M, Parrini MC. Autophagy Is Polarized toward Cell Front during Migration and Spatially Perturbed by Oncogenic Ras. Cells. 2021; 10(10):2637. https://doi.org/10.3390/cells10102637
Chicago/Turabian StyleSingh, Manish Kumar, Giulia Zago, Irina Veith, Jacques Camonis, Mathieu Coppey, and Maria Carla Parrini. 2021. "Autophagy Is Polarized toward Cell Front during Migration and Spatially Perturbed by Oncogenic Ras" Cells 10, no. 10: 2637. https://doi.org/10.3390/cells10102637