The Retinoblastoma Tumor Suppressor Is Required for the NUP98-HOXA9-Induced Aberrant Nuclear Envelope Phenotype
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfections
2.2. Plasmids
2.3. Antibodies
2.4. Immunofluorescence Microscopy
2.5. Calculation of Pearson’s Coefficients
2.6. Foci Counting
2.7. GFP-Trap Magnetic Agarose Assays
2.8. Western Blotting
2.9. Proximity Ligation Assays
2.10. Statistical Analyses
2.11. Image Design
3. Results
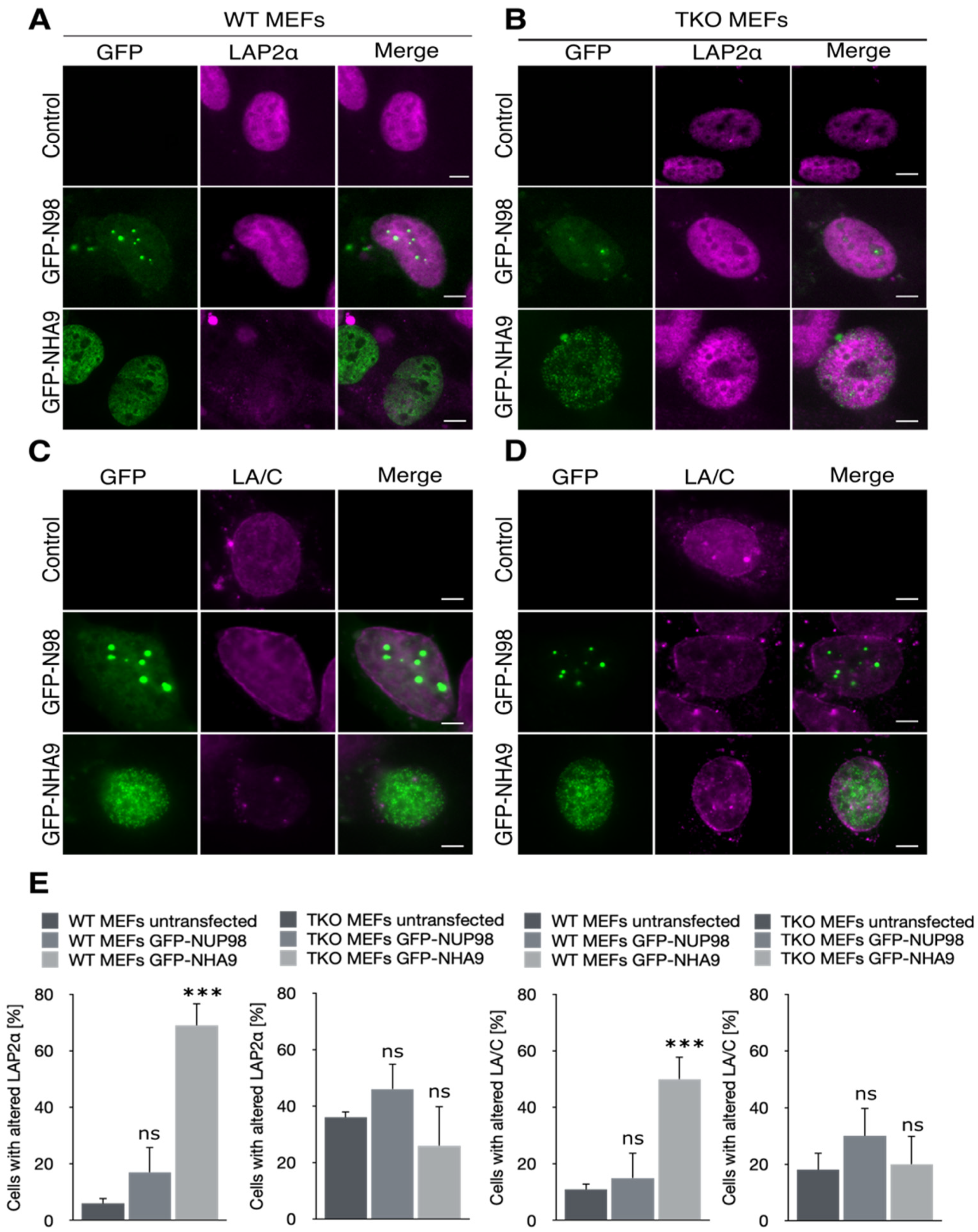
3.1. NHA9 Alters the Nuclear Architecture in Mouse Embryonic Fibroblasts
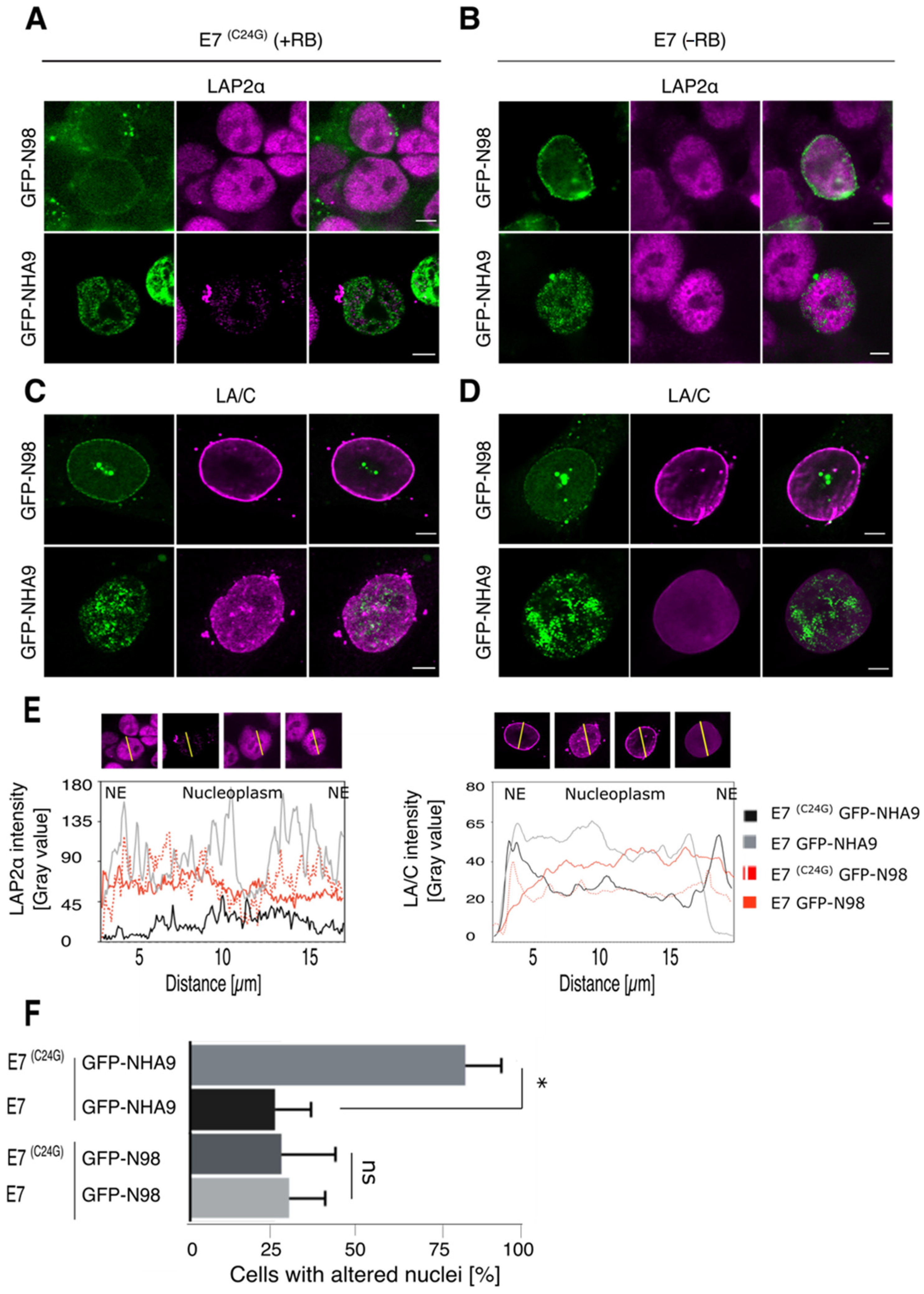
3.2. NHA9 Provokes Changes in Nuclear Architecture Only in the Presence of the Pocket Proteins
3.3. Epigenetic Dysregulation Correlating with NHA9 Expression
3.4. Association of Polycomb-Group Proteins with NHA9 Appears to Depend on RB
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Gough, S.M.; Slape, C.I.; Aplan, P.D. NUP98 Gene Fusions and Hematopoietic Malignancies: Common Themes and New Biologic Insights. Blood 2011, 118, 6247–6257. [Google Scholar] [CrossRef] [Green Version]
- Fahrenkrog, B. Nucleoporin Gene Fusions and Hematopoietic Malignancies. N. J. Sci. 2014, 2014, 468306. [Google Scholar] [CrossRef]
- Martins, N. On the Effects of Leukemogenic Nucleoporin Fusion Proteins on Nucleocytoplasmic Transport and Gene Expression. In Nuclear-Cytoplasmic Transport; Yang, W., Ed.; Nucleic Acids and Molecular Biology; Springer International Publishing: Cham, Switzerland, 2018; pp. 223–248. ISBN 978-3-319-77309-4. [Google Scholar]
- Xu, H.; Valerio, D.G.; Eisold, M.E.; Sinha, A.; Koche, R.P.; Hu, W.; Chen, C.-W.; Chu, S.H.; Brien, G.L.; Park, C.Y.; et al. NUP98-Fusion Proteins Interact With the NSL and MLL1 Complexes To Drive Leukemogenesis. Cancer Cell 2016, 30, 863–878. [Google Scholar] [CrossRef] [Green Version]
- Rosenblum, J.S.; Blobel, G. Autoproteolysis in Nucleoporin Biogenesis. Proc. Natl. Acad. Sci. USA 1999, 96, 11370–11375. [Google Scholar] [CrossRef] [Green Version]
- Fontoura, B.M.; Blobel, G.; Yaseen, N.R. The Nucleoporin Nup98 Is a Site for GDP/GTP Exchange on Ran and Termination of Karyopherin Beta 2-Mediated Nuclear Import. J. Biol. Chem. 2000, 275, 31289–31296. [Google Scholar] [CrossRef] [Green Version]
- Griffis, E.R.; Craige, B.; Dimaano, C.; Ullman, K.S.; Powers, M.A. Distinct Functional Domains within Nucleoporins Nup153 and Nup98 Mediate Transcription-Dependent Mobility. Mol. Biol. Cell 2004, 15, 1991–2002. [Google Scholar] [CrossRef] [Green Version]
- Powers, M.A.; Forbes, D.J.; Dahlberg, J.E.; Lund, E. The Vertebrate GLFG Nucleoporin, Nup98, Is an Essential Component of Multiple RNA Export Pathways. J. Cell Biol. 1997, 136, 241–250. [Google Scholar] [CrossRef]
- Celetti, G.; Paci, G.; Caria, J.; VanDelinder, V.; Bachand, G.; Lemke, E.A. The Liquid State of FG-Nucleoporins Mimics Permeability Barrier Properties of Nuclear Pore Complexes. J. Cell Biol. 2019, 219, e201907157. [Google Scholar] [CrossRef]
- Capelson, M.; Liang, Y.; Schulte, R.; Mair, W.; Wagner, U.; Hetzer, M.W. Chromatin-Bound Nuclear Pore Components Regulate Gene Expression in Higher Eukaryotes. Cell 2010, 140, 372–383. [Google Scholar] [CrossRef] [Green Version]
- Kalverda, B.; Pickersgill, H.; Shloma, V.V.; Fornerod, M. Nucleoporins Directly Stimulate Expression of Developmental and Cell-Cycle Genes inside the Nucleoplasm. Cell 2010, 140, 360–371. [Google Scholar] [CrossRef] [Green Version]
- Franks, T.M.; Benner, C.; Narvaiza, I.; Marchetto, M.C.N.; Young, J.M.; Malik, H.S.; Gage, F.H.; Hetzer, M.W. Evolution of a Transcriptional Regulator from a Transmembrane Nucleoporin. Genes Dev. 2016, 10, 15–30. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Franks, T.M.; Marchetto, M.C.; Gage, F.H.; Hetzer, M.W. Dynamic Association of NUP98 with the Human Genome. PLoS Genet. 2013, 9, e1003308. [Google Scholar] [CrossRef]
- Light, W.H.; Freaney, J.; Sood, V.; Thompson, A.; D’Urso, A.; Horvath, C.M.; Brickner, J.H. A Conserved Role for Human Nup98 in Altering Chromatin Structure and Promoting Epigenetic Transcriptional Memory. PLoS Biol. 2013, 11, e1001524. [Google Scholar] [CrossRef] [Green Version]
- Panda, D.; Pascual-Garcia, P.; Dunagin, M.; Tudor, M.; Hopkins, K.C.; Xu, J.; Gold, B.; Raj, A.; Capelson, M.; Cherry, S. Nup98 Promotes Antiviral Gene Expression to Restrict RNA Viral Infection in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, E3890–E3899. [Google Scholar] [CrossRef] [Green Version]
- Cross, M.K.; Powers, M.A. Nup98 Regulates Bipolar Spindle Assembly through Association with Microtubules and Opposition of MCAK. Mol. Biol. Cell 2011, 22, 661–672. [Google Scholar] [CrossRef]
- Jeganathan, K.B.; Malureanu, L.; van Deursen, J.M. The Rae1-Nup98 Complex Prevents Aneuploidy by Inhibiting Securin Degradation. Nature 2005, 438, 1036–1039. [Google Scholar] [CrossRef]
- Jeganathan, K.B.; Baker, D.J.; van Deursen, J.M. Securin Associates with APCCdh1 in Prometaphase but Its Destruction Is Delayed by Rae1 and Nup98 until the Metaphase/Anaphase Transition. Cell Cycle 2006, 5, 366–370. [Google Scholar] [CrossRef]
- Xu, S.; Powers, M.A. Nup98-Homeodomain Fusions Interact with Endogenous Nup98 during Interphase and Localize to Kinetochores and Chromosome Arms during Mitosis. MBoC 2010, 21, 1585–1596. [Google Scholar] [CrossRef] [Green Version]
- Yassin, E.R.; Abdul-Nabi, A.M.; Takeda, A.; Yaseen, N.R. Effects of the NUP98–DDX10 Oncogene on Primary Human CD34+ Cells: Role of a Conserved Helicase Motif. Leukemia 2010, 24, 1001–1011. [Google Scholar] [CrossRef] [Green Version]
- Oka, M.; Asally, M.; Yasuda, Y.; Ogawa, Y.; Tachibana, T.; Yoneda, Y. The Mobile FG Nucleoporin Nup98 Is a Cofactor for Crm1-Dependent Protein Export. MBoC 2010, 21, 1885–1896. [Google Scholar] [CrossRef] [Green Version]
- Jankovic, D.; Gorello, P.; Liu, T.; Ehret, S.; La Starza, R.; Desjobert, C.; Baty, F.; Brutsche, M.; Jayaraman, P.-S.; Santoro, A.; et al. Leukemogenic Mechanisms and Targets of a NUP98/HHEX Fusion in Acute Myeloid Leukemia. Blood 2008, 111, 5672–5682. [Google Scholar] [CrossRef]
- Fahrenkrog, B.; Martinelli, V.; Nilles, N.; Fruhmann, G.; Chatel, G.; Juge, S.; Sauder, U.; Di Giacomo, D.; Mecucci, C.; Schwaller, J. Expression of Leukemia-Associated Nup98 Fusion Proteins Generates an Aberrant Nuclear Envelope Phenotype. PLoS ONE 2016, 11, e0152321. [Google Scholar] [CrossRef] [Green Version]
- Calvo, K.R.; Sykes, D.B.; Pasillas, M.P.; Kamps, M.P. Nup98-HoxA9 Immortalizes Myeloid Progenitors, Enforces Expression of Hoxa9, Hoxa7 and Meis1, and Alters Cytokine-Specific Responses in a Manner Similar to That Induced by Retroviral Co-Expression of Hoxa9 and Meis1. Oncogene 2002, 21, 4247–4256. [Google Scholar] [CrossRef] [Green Version]
- Chung, K.Y.; Morrone, G.; Schuringa, J.J.; Plasilova, M.; Shieh, J.-H.; Zhang, Y.; Zhou, P.; Moore, M.A.S. Enforced Expression of NUP98-HOXA9 in Human CD34+ Cells Enhances Stem Cell Proliferation. Cancer Res. 2006, 66, 11781–11791. [Google Scholar] [CrossRef] [Green Version]
- Ghannam, G.; Takeda, A.; Camarata, T.; Moore, M.A.; Viale, A.; Yaseen, N.R. The Oncogene Nup98-HOXA9 Induces Gene Transcription in Myeloid Cells. J. Biol. Chem. 2004, 279, 866–875. [Google Scholar] [CrossRef] [Green Version]
- Takeda, A.; Goolsby, C.; Yaseen, N.R. NUP98-HOXA9 Induces Long-Term Proliferation and Blocks Differentiation of Primary Human CD34+ Hematopoietic Cells. Cancer Res. 2006, 66, 6628–6637. [Google Scholar] [CrossRef] [Green Version]
- Markiewicz, E.; Dechat, T.; Foisner, R.; Quinlan, R.A.; Hutchison, C.J. Lamin A/C Binding Protein LAP2α Is Required for Nuclear Anchorage of Retinoblastoma Protein. Mol. Biol. Cell 2002, 13, 4401–4413. [Google Scholar] [CrossRef] [Green Version]
- Johnson, B.R.; Nitta, R.T.; Frock, R.L.; Mounkes, L.; Barbie, D.A.; Stewart, C.L.; Harlow, E.; Kennedy, B.K. A-Type Lamins Regulate Retinoblastoma Protein Function by Promoting Subnuclear Localization and Preventing Proteasomal Degradation. Proc. Natl. Acad. Sci. USA 2004, 101, 9677–9682. [Google Scholar] [CrossRef] [Green Version]
- Dorner, D.; Vlcek, S.; Foeger, N.; Gajewski, A.; Makolm, C.; Gotzmann, J.; Hutchison, C.J.; Foisner, R. Lamina-Associated Polypeptide 2α Regulates Cell Cycle Progression and Differentiation via the Retinoblastoma–E2F Pathway. J. Cell Biol. 2006, 173, 83–93. [Google Scholar] [CrossRef] [Green Version]
- Dyer, M.A.; Schweers, B.; Gray, J.; Zhang, J. Compensation by P107 Following Rb Gene Inactivation Prevents Retinoblastoma in Mice But Not Humans. Investig. Ophthalmol. Vis. Sci. 2005, 46, 3552. [Google Scholar]
- Donovan, S.L.; Schweers, B.; Martins, R.; Johnson, D.; Dyer, M.A. Compensation by Tumor Suppressor Genes during Retinal Development in Mice and Humans. BMC Biol. 2006, 4, 14. [Google Scholar] [CrossRef] [Green Version]
- Norrie, J.L.; Nityanandam, A.; Lai, K.; Chen, X.; Wilson, M.; Stewart, E.; Griffiths, L.; Jin, H.; Wu, G.; Orr, B.; et al. Retinoblastoma from Human Stem Cell-Derived Retinal Organoids. Nat. Commun. 2021, 12, 4535. [Google Scholar] [CrossRef]
- Naetar, N.; Korbei, B.; Kozlov, S.; Kerenyi, M.A.; Dorner, D.; Kral, R.; Gotic, I.; Fuchs, P.; Cohen, T.V.; Bittner, R.; et al. Loss of Nucleoplasmic LAP2α–Lamin A Complexes Causes Erythroid and Epidermal Progenitor Hyperproliferation. Nat. Cell Biol. 2008, 10, 1341–1348. [Google Scholar] [CrossRef]
- Narita, M.; Nũnez, S.; Heard, E.; Narita, M.; Lin, A.W.; Hearn, S.A.; Spector, D.L.; Hannon, G.J.; Lowe, S.W. Rb-Mediated Heterochromatin Formation and Silencing of E2F Target Genes during Cellular Senescence. Cell 2003, 113, 703–716. [Google Scholar] [CrossRef] [Green Version]
- Breiling, A.; Turner, B.M.; Bianchi, M.E.; Orlando, V. General Transcription Factors Bind Promoters Repressed by Polycomb Group Proteins. Nature 2001, 412, 651–655. [Google Scholar] [CrossRef]
- Zhang, S.; Schones, D.E.; Malicet, C.; Rochman, M.; Zhou, M.; Foisner, R.; Bustin, M. High Mobility Group Protein N5 (HMGN5) and Lamina-Associated Polypeptide 2α (LAP2α) Interact and Reciprocally Affect Their Genome-Wide Chromatin Organization. J. Biol. Chem. 2013, 288, 18104–18109. [Google Scholar] [CrossRef] [Green Version]
- Gesson, K.; Rescheneder, P.; Skoruppa, M.P.; von Haeseler, A.; Dechat, T.; Foisner, R. A-Type Lamins Bind Both Hetero- and Euchromatin, the Latter Being Regulated by Lamina-Associated Polypeptide 2 Alpha. Genome Res. 2016, 26, 462–473. [Google Scholar] [CrossRef] [Green Version]
- Dechat, T.; Pfleghaar, K.; Sengupta, K.; Shimi, T.; Shumaker, D.K.; Solimando, L.; Goldman, R.D. Nuclear Lamins: Major Factors in the Structural Organization and Function of the Nucleus and Chromatin. Genes Dev. 2008, 22, 832–853. [Google Scholar] [CrossRef] [Green Version]
- Gesson, K.; Vidak, S.; Foisner, R. Lamina-Associated Polypeptide (LAP)2α and Nucleoplasmic Lamins in Adult Stem Cell Regulation and Disease. Semin. Cell Dev. Biol. 2014, 29, 116–124. [Google Scholar] [CrossRef] [Green Version]
- Pascual-Garcia, P.; Jeong, J.; Capelson, M. Nucleoporin Nup98 Associates with Trx/MLL and NSL Histone-Modifying Complexes and Regulates Hox Gene Expression. Cell Rep. 2014, 9, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.G.; Song, J.; Wang, Z.; Dormann, H.L.; Casadio, F.; Li, H.; Luo, J.-L.; Patel, D.J.; Allis, C.D. Haematopoietic Malignancies Caused by Dysregulation of a Chromatin-Binding PHD Finger. Nature 2009, 459, 847–851. [Google Scholar] [CrossRef] [Green Version]
- Gough, S.M.; Lee, F.; Yang, F.; Walker, R.L.; Zhu, Y.J.; Pineda, M.; Onozawa, M.; Chung, Y.J.; Bilke, S.; Wagner, E.K.; et al. NUP98–PHF23 Is a Chromatin-Modifying Oncoprotein That Causes a Wide Array of Leukemias Sensitive to Inhibition of PHD Histone Reader Function. Cancer Discov. 2014, 4, 564–577. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Guo, Y.; Gough, S.M.; Zhang, J.; Vann, K.R.; Li, K.; Cai, L.; Shi, X.; Aplan, P.D.; Wang, G.G.; et al. Mechanistic Insights into Chromatin Targeting by Leukemic NUP98-PHF23 Fusion. Nat. Commun. 2020, 11, 3339. [Google Scholar] [CrossRef]
- Dick, F.A.; Goodrich, D.W.; Sage, J.; Dyson, N.J. Non-Canonical Functions of the RB Protein in Cancer. Nat. Rev. Cancer 2018, 18, 442–451. [Google Scholar] [CrossRef]
- Ishak, C.A.; Marshall, A.E.; Passos, D.T.; White, C.R.; Kim, S.J.; Cecchini, M.J.; Ferwati, S.; MacDonald, W.A.; Howlett, C.J.; Welch, I.D.; et al. An RB-EZH2 Complex Mediates Silencing of Repetitive DNA Sequences. Mol. Cell 2016, 64, 1074–1087. [Google Scholar] [CrossRef] [Green Version]
- Montoya-Durango, D.E.; Ramos, K.A.; Bojang, P.; Ruiz, L.; Ramos, I.N.; Ramos, K.S. LINE-1 Silencing by Retinoblastoma Proteins Is Effected through the Nucleosomal and Remodeling Deacetylase Multiprotein Complex. BMC Cancer 2016, 16, 38. [Google Scholar] [CrossRef] [Green Version]
- Loi, M.; Cenni, V.; Duchi, S.; Squarzoni, S.; Otin, C.L.-; Foisner, R.; Lattanzi, G.; Capanni, C. Barrier-to-Autointegration Factor (BAF) Involvement in Prelamin A-Related Chromatin Organization Changes. Oncotarget 2015, 7, 15662–15677. [Google Scholar] [CrossRef] [PubMed]
- Grace, M.; Munger, K. Proteomic Analysis of the Gamma Human Papillomavirus Type 197 E6 and E7 Associated Cellular Proteins. Virology 2017, 500, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Mcdonald, J.H.; Dunn, K.W. Statistical Tests for Measures of Colocalization in Biological Microscopy. J. Microsc. 2013, 252, 295–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikawa, T.; Masuda, K.; Endo, T.A.; Endo, M.; Isono, K.; Koseki, Y.; Nakagawa, R.; Kometani, K.; Takano, J.; Agata, Y.; et al. Conversion of T Cells to B Cells by Inactivation of Polycomb-Mediated Epigenetic Suppression of the B-Lineage Program. Genes Dev. 2016, 30, 2475–2485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Münger, K.; Werness, B.A.; Dyson, N.; Phelps, W.C.; Harlow, E.; Howley, P.M. Complex Formation of Human Papillomavirus E7 Proteins with the Retinoblastoma Tumor Suppressor Gene Product. EMBO J. 1989, 8, 4099–4105. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, S.L.; Stremlau, M.; He, X.; Basile, J.R.; Munger, K. Degradation of the Retinoblastoma Tumor Suppressor by the Human Papillomavirus Type 16 E7 Oncoprotein Is Important for Functional Inactivation and Is Separable from Proteasomal Degradation of E7. J. Virol. 2001, 75, 7583–7591. [Google Scholar] [CrossRef] [Green Version]
- Vélez-Cruz, R.; Johnson, D.G. The Retinoblastoma (RB) Tumor Suppressor: Pushing Back against Genome Instability on Multiple Fronts. Int. J. Mol. Sci. 2017, 18, 1776. [Google Scholar] [CrossRef] [PubMed]
- Aaron, J.S.; Taylor, A.B.; Chew, T.-L. Image Co-Localization—Co-Occurrence versus Correlation. J. Cell Sci. 2018, 131, jcs211847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, J.H.; Davis, E.S.; Daugird, T.A.; Zhao, S.; Quiroga, I.Y.; Uryu, H.; Li, J.; Storey, A.J.; Tsai, Y.-H.; Keeley, D.P.; et al. Phase Separation Drives Aberrant Chromatin Looping and Cancer Development. Nature 2021, 595, 591–595. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Fursova, N.A.; Kelley, J.R.; Huseyin, M.K.; Feldmann, A.; Klose, R.J. PRC1 Catalytic Activity Is Central to Polycomb System Function. Mol. Cell 2020, 77, 857–874.e9. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of Histone H2A Ubiquitination in Polycomb Silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef]
- Min, J.; Zhang, Y.; Xu, R.-M. Structural Basis for Specific Binding of Polycomb Chromodomain to Histone H3 Methylated at Lys. Genes Dev. 1823, 17, 1823–1828. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.S. Proximity Ligation Assay (PLA). Curr. Protoc. Immunol. 2018, 123, e58. [Google Scholar] [CrossRef] [PubMed]
- Franks, T.M.; McCloskey, A.; Shokhirev, M.N.; Benner, C.; Rathore, A.; Hetzer, M.W. Nup98 Recruits the Wdr82–Set1A/COMPASS Complex to Promoters to Regulate H3K4 Trimethylation in Hematopoietic Progenitor Cells. Genes Dev. 2017, 31, 2222–2234. [Google Scholar] [CrossRef] [Green Version]
- Terlecki-Zaniewicz, S.; Humer, T.; Eder, T.; Schmoellerl, J.; Heyes, E.; Manhart, G.; Kuchynka, N.; Parapatics, K.; Liberante, F.G.; Müller, A.C.; et al. Biomolecular Condensation of NUP98 Fusion Proteins Drives Leukemogenic Gene Expression. Nat. Struct. Mol. Biol. 2021, 28, 190–201. [Google Scholar] [CrossRef]
- Kasper, L.H.; Brindle, P.K.; Schnabel, C.A.; Pritchard, C.E.J.; Cleary, M.L.; van Deursen, J.M.A. CREB Binding Protein Interacts with Nucleoporin-Specific FG Repeats That Activate Transcription and Mediate NUP98-HOXA9 Oncogenicity. Mol. Cell. Biol. 1999, 19, 764–776. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.G.; Cai, L.; Pasillas, M.P.; Kamps, M.P. NUP98-NSD1 Links H3K36 Methylation to Hox-A Gene Activation and Leukaemogenesis. Nat. Cell Biol. 2007, 9, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Oka, M.; Mura, S.; Otani, M.; Miyamoto, Y.; Nogami, J.; Maehara, K.; Harada, A.; Tachibana, T.; Yoneda, Y.; Ohkawa, Y. Chromatin-Bound CRM1 Recruits SET-Nup214 and NPM1c onto HOX Clusters Causing Aberrant HOX Expression in Leukemia Cells. eLife 2019, 8, e46667. [Google Scholar] [CrossRef]
- Shima, Y.; Yumoto, M.; Katsumoto, T.; Kitabayashi, I. MLL Is Essential for NUP98-HOXA9-Induced Leukemia. Leukemia 2017, 31, 2200–2210. [Google Scholar] [CrossRef]
- Coschi, C.H.; Ishak, C.A.; Gallo, D.; Marshall, A.; Talluri, S.; Wang, J.; Cecchini, M.J.; Martens, A.L.; Percy, V.; Welch, I.; et al. Haploinsufficiency of an RB–E2F1–Condensin II Complex Leads to Aberrant Replication and Aneuploidy. Cancer Discov. 2014, 4, 840–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, C.T.; Hess, J.L. Role of HOXA9 in Leukemia: Dysregulation, Cofactors and Essential Targets. Oncogene 2016, 35, 1090–1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, C.T.; Hess, J.L. Deregulation of the HOXA9/MEIS1 Axis in Acute Leukemia. Curr. Opin. Hematol. 2016, 23, 354–361. [Google Scholar] [CrossRef]
- Riz, I.; Hawley, R.G. G1/S Transcriptional Networks Modulated by the HOX11/TLX1 Oncogene of T-Cell Acute Lymphoblastic Leukemia. Oncogene 2005, 24, 5561–5575. [Google Scholar] [CrossRef] [Green Version]
- Trojer, P.; Reinberg, D. Facultative Heterochromatin: Is There a Distinctive Molecular Signature? Mol. Cell 2007, 28, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Plass, C.; Oakes, C.; Blum, W.; Marcucci, G. Epigenetics in Acute Myeloid Leukemia. Semin. Oncol. 2008, 35, 378–387. [Google Scholar] [CrossRef] [Green Version]
- Dahiya, A.; Wong, S.; Gonzalo, S.; Gavin, M.; Dean, D.C. Linking the Rb and Polycomb Pathways. Mol. Cell 2001, 8, 557–569. [Google Scholar] [CrossRef]
- Bracken, A.P.; Pasini, D.; Capra, M.; Prosperini, E.; Colli, E.; Helin, K. EZH2 Is Downstream of the PRB-E2F Pathway, Essential for Proliferation and Amplified in Cancer. EMBO J. 2003, 22, 5323–5335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lees, J.A.; Saito, M.; Vidal, M.; Valentine, M.; Look, T.; Harlow, E.; Dyson, N.; Helin, K. The Retinoblastoma Protein Binds to a Family of E2F Transcription Factors. Mol. Cell Biol. 1993, 13, 7813–7825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNeer, N.A.; Philip, J.; Geiger, H.; Ries, R.E.; Lavallée, V.-P.; Walsh, M.; Shah, M.; Arora, K.; Emde, A.-K.; Robine, N.; et al. Genetic Mechanisms of Primary Chemotherapy Resistance in Pediatric Acute Myeloid Leukemia. Leukemia 2019, 33, 1934–1943. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yam, J.C.; Tham, C.C.; Pang, C.P.; Chu, W.K. RB Regulates DNA Double Strand Break Repair Pathway Choice by Mediating CtIP Dependent End Resection. Int. J. Mol. Sci. 2020, 21, 9176. [Google Scholar] [CrossRef] [PubMed]
- Vélez-Cruz, R.; Manickavinayaham, S.; Biswas, A.K.; Clary, R.W.; Premkumar, T.; Cole, F.; Johnson, D.G. RB Localizes to DNA Double-Strand Breaks and Promotes DNA End Resection and Homologous Recombination through the Recruitment of BRG1. Genes Dev. 2016, 30, 2500–2512. [Google Scholar] [CrossRef] [Green Version]
- Garsed, D.W.; Alsop, K.; Fereday, S.; Emmanuel, C.; Kennedy, C.J.; Etemadmoghadam, D.; Gao, B.; Gebski, V.; Garès, V.; Christie, E.L.; et al. Homologous Recombination DNA Repair Pathway Disruption and Retinoblastoma Protein Loss Are Associated with Exceptional Survival in High-Grade Serous Ovarian Cancer. Clin. Cancer Res. 2018, 24, 569–580. [Google Scholar] [CrossRef] [Green Version]
- Thangavel, C.; Boopathi, E.; Ciment, S.; Liu, Y.; O’Neill, R.; Sharma, A.; McMahon, S.B.; Mellert, H.; Addya, S.; Ertel, A.; et al. The Retinoblastoma Tumor Suppressor Modulates DNA Repair and Radioresponsiveness. Clin. Cancer Res. 2014, 20, 5468–5482. [Google Scholar] [CrossRef] [Green Version]
- Puthiyaveetil, A.G.; Reilly, C.M.; Pardee, T.S.; Caudell, D.L. Non-Homologous End Joining Mediated DNA Repair Is Impaired in the NUP98-HOXD13 Mouse Model for Myelodysplastic Syndrome. Leuk. Res. 2013, 37, 112–116. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vaz, M.; Fahrenkrog, B. The Retinoblastoma Tumor Suppressor Is Required for the NUP98-HOXA9-Induced Aberrant Nuclear Envelope Phenotype. Cells 2021, 10, 2851. https://doi.org/10.3390/cells10112851
Vaz M, Fahrenkrog B. The Retinoblastoma Tumor Suppressor Is Required for the NUP98-HOXA9-Induced Aberrant Nuclear Envelope Phenotype. Cells. 2021; 10(11):2851. https://doi.org/10.3390/cells10112851
Chicago/Turabian StyleVaz, Marcela, and Birthe Fahrenkrog. 2021. "The Retinoblastoma Tumor Suppressor Is Required for the NUP98-HOXA9-Induced Aberrant Nuclear Envelope Phenotype" Cells 10, no. 11: 2851. https://doi.org/10.3390/cells10112851






