Cattle In Vitro Induced Pluripotent Stem Cells Generated and Maintained in 5 or 20% Oxygen and Different Supplementation
Abstract
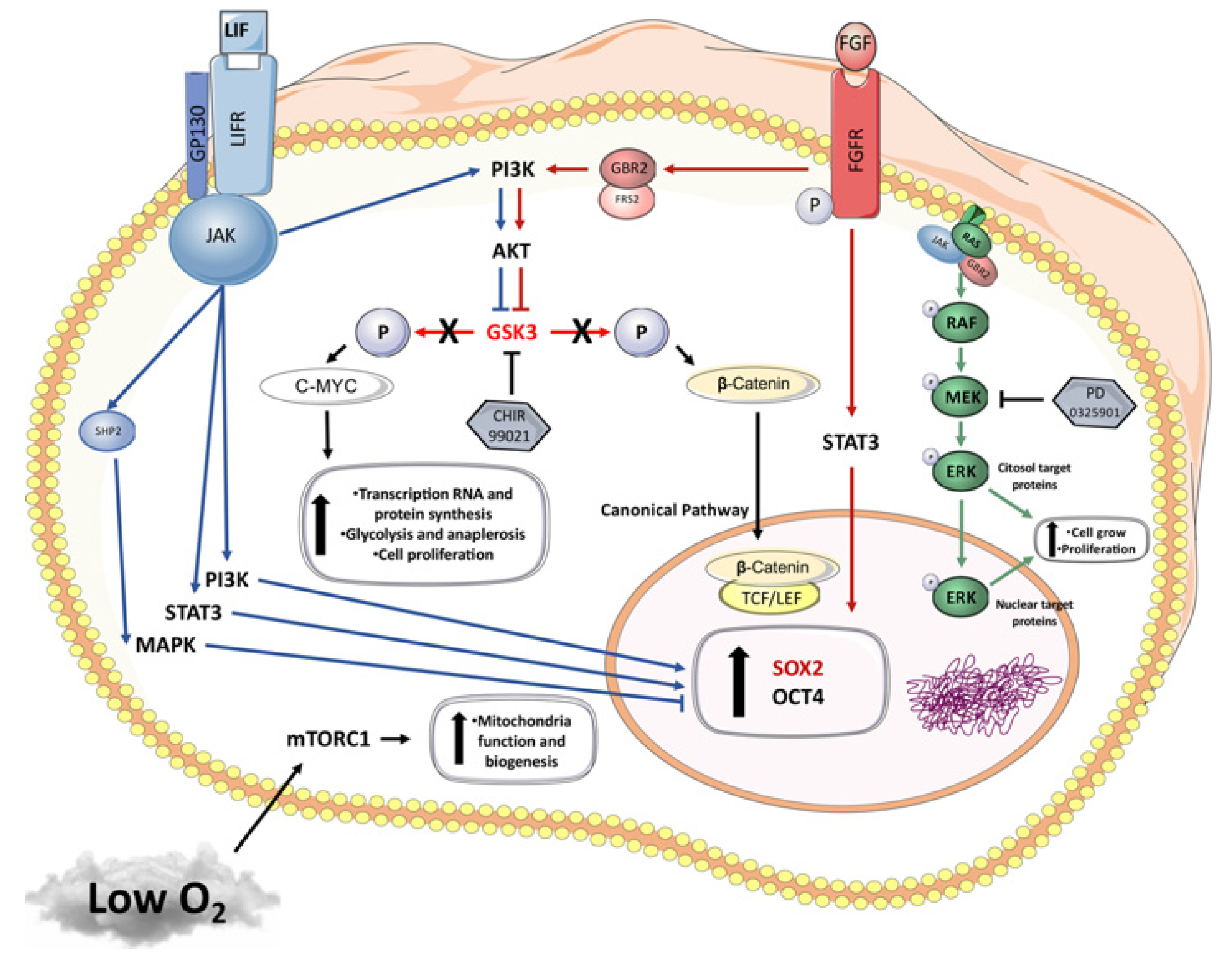
:1. Introduction
2. Materials and Methods
2.1. Cells and Media
2.2. Feeder Cells
2.3. Lentivirus Production
2.4. Bovine Induced Pluripotent Stem Cells Generation
2.5. In Vitro Embryo Production
2.6. Alkaline Phosphatase Activity and Immunofluorescence
2.7. RNA Extraction and Reverse Transcription
2.8. Quantitative PCR
2.9. Undirected Differentiation in Embryoid Bodies
2.10. Statistical Analysis
3. Results
3.1. Influence of Oxygen Tension and Different Pluripotency Inducers during the Reprogramming Process of Bovine Fibroblasts
3.2. Molecular Profile of biPSCs Generated in Different Oxygen Tensions and Different Supplementations
3.3. Undirected Differentiation of biPSCs Generated in Hypoxia and Normoxia Conditions and Different Pluripotent Supplements
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takahashi, K.; Yamanaka, S. Induction of pluripotent st em cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omole, A.E.; Fakoya, A.O.J. Ten years of progress and promise of induced pluripotent stem cells: Historical origins, characteristics, mechanisms, limitations, and potential applications. PeerJ 2018, 6, e4370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pieri, N.C.G.; de Souza, A.F.; Botigelli, R.C.; Machado, L.S.; Ambrosio, C.E.; dos Santos Martins, D.; de Andrade, A.F.C.; Meirelles, F.V.; Hyttel, P.; Bressan, F.F. Stem cells on regenerative and reproductive science in domestic animals. Vet. Res. Commun. 2019, 43, 7–16. [Google Scholar] [CrossRef]
- Nichols, J.; Smith, A. Naive and Primed Pluripotent States. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Ocampo, A.; Belmonte, J.C.I. Cellular Metabolism and Induced Pluripotency. Cell 2016, 166, 1371–1385. [Google Scholar] [CrossRef] [Green Version]
- Navarro, M.; Soto, D.A.; Pinzon, C.A.; Wu, J.; Ross, P.J. Livestock pluripotency is finally captured in vitro. Reprod. Fertil. Dev. 2019, 32, 11–39. [Google Scholar] [CrossRef]
- Soto, D.A.; Ross, P.J. Pluripotent stem cells and livestock genetic engineering. Transgenic Res. 2016, 25, 289–306. [Google Scholar] [CrossRef] [Green Version]
- Kues, W.A.; Niemann, H. The contribution of farm animals to human health. Trends Biotechnol. 2004, 22, 286–294. [Google Scholar] [CrossRef]
- Li, Y.; Cang, M.; Lee, A.S.; Zhang, K.; Liu, D. Reprogramming of sheep fibroblasts into pluripotency under a drug-inducible expression of mouse-derived defined factors. PLoS ONE 2011, 6, e15947. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Balehosur, D.; Murray, B.; Kelly, J.M.; Sumer, H.; Verma, P.J. Generation and characterization of reprogrammed sheep induced pluripotent stem cells. Theriogenology 2012, 77, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Nagy, K.; Sung, H.-K.; Zhang, P.; Laflamme, S.; Vincent, P.; Agha-Mohammadi, S.; Woltjen, K.; Monetti, C.; Michael, I.P.; Smith, L.C.; et al. Induced Pluripotent Stem Cell Lines Derived from Equine Fibroblasts. Stem Cell Rev. Rep. 2011, 7, 693–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitworth, D.J.; Ovchinnikov, D.A.; Sun, J.; Fortuna, P.R.J.; Wolvetang, E.J. Generation and characterization of leukemia inhibitory factor-dependent equine induced pluripotent stem cells from adult dermal fibroblasts. Stem Cells Dev. 2014, 23, 1515–1523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, D.; Li, Z.; Liu, Y.; Gao, Y.; Wang, H. Kinetic Analysis of Porcine Fibroblast Reprogramming toward Pluripotency by Defined Factors. Cell. Reprogramming 2012, 14, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Fujishiro, S.; Nakano, K.; Mizukami, Y.; Azami, T.; Arai, Y.; Matsunari, H.; Ishino, R.; Nishimura, T.; Watanabe, M.; Abe, T.; et al. Generation of Naive-like Porcine Induced Pluripotent Stem Cells Capable of Contributing to Embryonic and Fetal Development. Stem Cells Dev. 2013, 22, 473–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wei, C.; Zhang, P.; Li, X.; Liu, T.; Pu, Y.; Li, Y.; Cao, Z.; Cao, H.; Liu, Y.; et al. Efficient reprogramming of naive-like induced pluripotent stem cells from porcine adipose-derived stem cells with a feeder-independent and serum-free system. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Cao, H.; Yang, P.; Pu, Y.; Sun, X.; Yin, H.; Zhang, Y.; Zhang, Y.; Li, Y.; Liu, Y.; Fang, F.; et al. Characterization of Bovine Induced Pluripotent Stem Cells by Lentiviral Transduction of Reprogramming Factor Fusion Proteins. Int. J. Biol. Sci. 2012. [Google Scholar] [CrossRef]
- Bressan, F.F.; Bassanezze, V.; de Figueiredo Pessôa, L.V.; Sacramento, C.B.; Malta, T.M.; Kashima, S.; Fantinato Neto, P.; Strefezzi, R.D.F.; Pieri, N.C.G.; Krieger, J.E.; et al. Generation of induced pluripotent stem cells from large domestic animals. Stem Cell Res. Ther. 2020, 11, 247. [Google Scholar] [CrossRef]
- Talluri, T.R.; Kumar, D.; Glage, S.; Garrels, W.; Ivics, Z.; Debowski, K.; Behr, R.; Niemann, H.; Kues, W.A. Derivation and characterization of bovine induced pluripotent stem cells by transposon-mediated reprogramming. Cell. Reprogram. 2015, 17, 131–140. [Google Scholar] [CrossRef] [Green Version]
- Pessôa, L.V.F.; Bressan, F.F.; Freude, K.K. Induced pluripotent stem cells throughout the animal kingdom: Availability and applications. World J. Stem Cells 2019, 11. [Google Scholar] [CrossRef]
- Wang, F.; Thirumangalathu, S.; Loeken, M.R. Establishment of new mouse embryonic stem cell lines is improved by physiological glucose and oxygen. Cloning Stem Cells 2006, 8, 108–116. [Google Scholar] [CrossRef]
- Yoshida, Y.; Takahashi, K.; Okita, K.; Ichisaka, T.; Yamanaka, S. Hypoxia Enhances the Generation of Induced Pluripotent Stem Cells. Cell Stem Cell 2009, 5, 237–241. [Google Scholar] [CrossRef] [Green Version]
- Sommer, C.A.; Stadtfeld, M.; Murphy, G.J.; Hochedlinger, K.; Kotton, D.N.; Mostoslavsky, G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells 2009, 27, 543–549. [Google Scholar] [CrossRef] [Green Version]
- Macabelli, C.H.; Ferreira, R.M.; Gimenes, L.U.; De Carvalho, N.A.T.; Soares, J.G.; Ayres, H.; Ferraz, M.L.; Watanabe, Y.F.; Watanabe, O.Y.; Sangalli, J.R.; et al. Reference gene selection for gene expression analysis of oocytes collected from dairy cattle and buffaloes during winter and summer. PLoS ONE 2014, 9, e93287. [Google Scholar] [CrossRef] [Green Version]
- Sumer, H.; Liu, J.; Malaver-Ortega, L.F.; Lim, M.L.; Khodadadi, K.; Verma, P.J. NANOG is a key factor for induction of pluripotency in bovine adult fibroblasts. J. Anim. Sci. 2011, 89, 2708–2716. [Google Scholar] [CrossRef] [Green Version]
- Rossant, J. A mouse is not a cow. Nature 2011, 471, 457–458. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M. Therapeutics Targeting FGF Signaling Network in Human Diseases. Trends Pharmacol. Sci. 2016, 37, 1081–1096. [Google Scholar] [CrossRef]
- Nicola, N.A.; Babon, J.J. Leukemia inhibitory factor (LIF). Cytokine Growth Factor Rev. 2015, 26, 533–544. [Google Scholar] [CrossRef] [Green Version]
- Sim, Y.J.; Kim, M.S.; Nayfeh, A.; Yun, Y.J.; Kim, S.J.; Park, K.T.; Kim, C.H.; Kim, K.S. 2i Maintains a Naive Ground State in ESCs through Two Distinct Epigenetic Mechanisms. Stem Cell Rep. 2017, 8, 1312–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Han, J.; Ding, F.; Cao, S.; Lim, S.S.; Dai, Y.; Zhang, R.; Zhang, Y.; Lim, B.; Li, N. Generation of induced pluripotent stem cells from bovine embryonic fibroblast cells. Cell Res. 2011, 21, 1509–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, B.; Li, T.; Alonso-Gonzalez, L.; Gorre, R.; Keatley, S.; Green, A.; Turner, P.; Kallingappa, P.K.; Verma, V.; Oback, B. A virus-free poly-promoter vector induces pluripotency in quiescent bovine cells under chemically defined conditions of dual kinase inhibition. PLoS ONE 2011, 6, e24501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawaguchi, T.; Tsukiyama, T.; Kimura, K.; Matsuyama, S.; Minami, N.; Yamada, M.; Imai, H. Generation of Naïve Bovine Induced Pluripotent Stem Cells Using PiggyBac Transposition of Doxycycline-Inducible Transcription Factors. PLoS ONE 2015, 10, e0135403. [Google Scholar] [CrossRef] [Green Version]
- Ezashi, T.; Das, P.; Roberts, R.M. Low O2 tensions and the prevention of differentiation of hES cells. Proc. Natl. Acad. Sci. USA 2005, 102, 4783–4788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathieu, J.; Zhou, W.; Xing, Y.; Sperber, H.; Ferreccio, A.; Agoston, Z.; Kuppusamy, K.T.; Moon, R.T.; Ruohola-Baker, H. Hypoxia-Inducible Factors Have Distinct and Stage-Specific Roles during Reprogramming of Human Cells to Pluripotency. Cell Stem Cell 2014, 14, 592–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Castro, R.V.G.; Pieri, N.C.G.; Fantinato Neto, P.; Grizendi, B.M.; Dória, R.G.S.; Meirelles, F.V.; Smith, L.C.; Garcia, J.M.; Bressan, F.F. In Vitro Induction of Pluripotency from Equine Fibroblasts in 20% or 5% Oxygen. Stem Cells Int. 2020, 2020, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lengner, C.J.; Gimelbrant, A.A.; Erwin, J.A.; Cheng, A.W.; Guenther, M.G.; Welstead, G.G.; Alagappan, R.; Frampton, G.M.; Xu, P.; Muffat, J.; et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell 2010, 141, 872–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Liang, J.; Ni, S.; Zhou, T.; Qing, X.; Li, H.; He, W.; Chen, J.; Li, F.; Zhuang, Q.; et al. A mesenchymal-to-Epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010, 7, 51–63. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Lin, L.; Wang, X.; Gao, M.; Cao, S.; Mai, Y.; Wu, F.; Kuang, J.; Liu, H.; Yang, J.; et al. Resolving Cell Fate Decisions during Somatic Cell Reprogramming by Single-Cell RNA-Seq. Mol. Cell 2019, 73, 815–829.e7. [Google Scholar] [CrossRef] [Green Version]
- Burant, C.F.; Bell, G.I. Mammalian Facilitative Glucose Transporters: Evidence for Similar Substrate Recognition Sites in Functionally Monomeric Proteins. Biochemistry 1992, 31, 10414–10420. [Google Scholar] [CrossRef]
- Pantaleon, M.; Harvey, M.B.; Pascoe, W.S.; James, D.E.; Kaye, P.L. Glucose transporter GLUT3: Ontogeny, targeting, and role in the mouse blastocyst. Proc. Natl. Acad. Sci. USA 1997, 94, 3795–3800. [Google Scholar] [CrossRef] [Green Version]
- Harvey, A.J.; Kind, K.L.; Pantaleon, M.; Armstrong, D.T.; Thompson, J.G. Oxygen-regulated gene expression in bovine blastocysts. Biol. Reprod. 2004, 71, 1108–1119. [Google Scholar] [CrossRef]
- Christensen, D.R.; Calder, P.C.; Houghton, F.D. GLUT3 and PKM2 regulate OCT4 expression and support the hypoxic culture of human embryonic stem cells. Sci. Rep. 2015, 5, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Trinh, T.; Aljoufi, A.; Broxmeyer, H.E. Hypoxia Signaling Pathway in Stem Cell Regulation: Good and Evil. Curr. Stem Cell Rep. 2018, 4, 149–157. [Google Scholar] [CrossRef]
- Chen, H.; Aksoy, I.; Gonnot, F.; Osteil, P.; Aubry, M.; Hamela, C.; Rognard, C.; Hochard, A.; Voisin, S.; Fontaine, E.; et al. Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Pimton, P.; Lecht, S.; Stabler, C.T.; Johannes, G.; Schulman, E.S.; Lelkes, P.I. Hypoxia enhances differentiation of mouse embryonic stem cells into definitive endoderm and distal lung cells. Stem Cells Dev. 2015, 24, 663–676. [Google Scholar] [CrossRef] [Green Version]
- Mzoughi, S.; Zhang, J.; Hequet, D.; Teo, S.X.; Fang, H.; Xing, Q.R.; Bezzi, M.; Seah, M.K.Y.; Ong, S.L.M.; Shin, E.M.; et al. PRDM15 safeguards naive pluripotency by transcriptionally regulating WNT and MAPK-ERK signaling. Nat. Genet. 2017, 49, 1354–1363. [Google Scholar] [CrossRef]
- Beach, K.M.; Wang, J.; Otteson, D.C. Regulation of Stem Cell Properties of Müller Glia by JAK/STAT and MAPK Signaling in the Mammalian Retina. Stem Cells Int. 2017, 2017. [Google Scholar] [CrossRef] [Green Version]



| Primer | Transcript ID | Foward (5′ → 3′) | Reverse (5′ → 3′) | Product Size (bp) |
|---|---|---|---|---|
| ACTB | NM_173979.3 | CAGCAGATGTGGATCAGCAAGC | AACGCAGCTAACAGTCCGCC | 89 |
| PPIA | NM_178320.2 | CATACAGGTCCTGGCATC | CACGTGCTTGCCATCCAA | 107 |
| OCT4 | NM_174580.2 | GCAAACGATCAAGCAGTGACTAC | GGCGCCAGAGGAGAGGATACG | 93 |
| SOX2 | NM_001105463.2 | ATGGGCTCGGTGGTGAAGT | TGGTAGTGCTGGGACATGTGA | 178 |
| STELLA | NM_001111109.2 | AGTGAGCGGAGGTACAGGAT | TCGCACTCTTGATCGAATCTCA | 132 |
| GLUT1 | NM_174602.2 | ATCCTCATTGCCGTGGTGCT | ACGATGCCAGAGCCGATGGT | 133 |
| GLUT3 | NM_174603.3 | CGCCTTTGGCACTCTCAAC | GCACTGGATGATGGCTGGTAA | 88 |
| TUBB3 | NM_001077127.1 | GGATAGACCCCAGTGGCAAT | TTGTGTGAAGAAGCCTCGTTG | 88 |
| VIM | NM_173969.3 | CTCCTACCGCAGGATGTTCG | TGGATGTGGTCACGTAGCTC | 140 |
| PECAM1 | NM_174571.3 | AATCAGAGCGTGGGCTCAAA | ATCCACTGGGGCTATCACCT | 147 |
| BMP4 | NM_001045877.1 | AGCTTCCACCACGAAGAACAT | CACCTCGTTCTCTGGGATGC | 102 |
| AFP | NM_001034262.2 | CGGACCTTCCGAGCCATAAC | CTCTTTCCCCATCCTGCAGAC | 154 |
| FOXA2 | XM_025001047.1 | CGAGCCCGAGGGCTACTC | GTACGTGTTCATGCCGTTCA | 92 |
| mOSKM | ACGAGCCACAAGCTCACCTCT | GGCATTAAAGCAGCGTATCC | 221 |
| 5% O2 | 20% O2 | Total | |
|---|---|---|---|
| bFGF | 0.02% | 0.005% | 0.0125% |
| bFL2i | 0.075% | 0.07% | 0.0725% |
| Total | 0.0476% | 0.0375% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bessi, B.W.; Botigelli, R.C.; Pieri, N.C.G.; Machado, L.S.; Cruz, J.B.; de Moraes, P.; de Souza, A.F.; Recchia, K.; Barbosa, G.; de Castro, R.V.G.; et al. Cattle In Vitro Induced Pluripotent Stem Cells Generated and Maintained in 5 or 20% Oxygen and Different Supplementation. Cells 2021, 10, 1531. https://doi.org/10.3390/cells10061531
Bessi BW, Botigelli RC, Pieri NCG, Machado LS, Cruz JB, de Moraes P, de Souza AF, Recchia K, Barbosa G, de Castro RVG, et al. Cattle In Vitro Induced Pluripotent Stem Cells Generated and Maintained in 5 or 20% Oxygen and Different Supplementation. Cells. 2021; 10(6):1531. https://doi.org/10.3390/cells10061531
Chicago/Turabian StyleBessi, Brendon Willian, Ramon Cesar Botigelli, Naira Caroline Godoy Pieri, Lucas Simões Machado, Jessica Brunhara Cruz, Pamela de Moraes, Aline Fernanda de Souza, Kaiana Recchia, Gabriela Barbosa, Raquel Vasconcelos Guimarães de Castro, and et al. 2021. "Cattle In Vitro Induced Pluripotent Stem Cells Generated and Maintained in 5 or 20% Oxygen and Different Supplementation" Cells 10, no. 6: 1531. https://doi.org/10.3390/cells10061531








