Secreted Frizzled-Related Protein 1 Promotes Odontoblastic Differentiation and Reparative Dentin Formation in Dental Pulp Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Immunohistochemical Staining
2.2. Cell Culture
2.3. Semi-Quantitative Reverse Transcription Polymerase Chain Reaction
2.4. Odontoblastic Differentiation of hDPCs
2.5. Alizarin Red S Staining
2.6. Small Interfering RNA Transfection
2.7. Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction
2.8. Direct Pulp Capping Model
2.9. Immunofluorescence Staining
2.10. Western Blotting Analysis
2.11. Statistical Analysis
3. Results
3.1. Localization of Sfrp1 in Dental Tissues and Expression of SFRP1 mRNA in Dental Cells
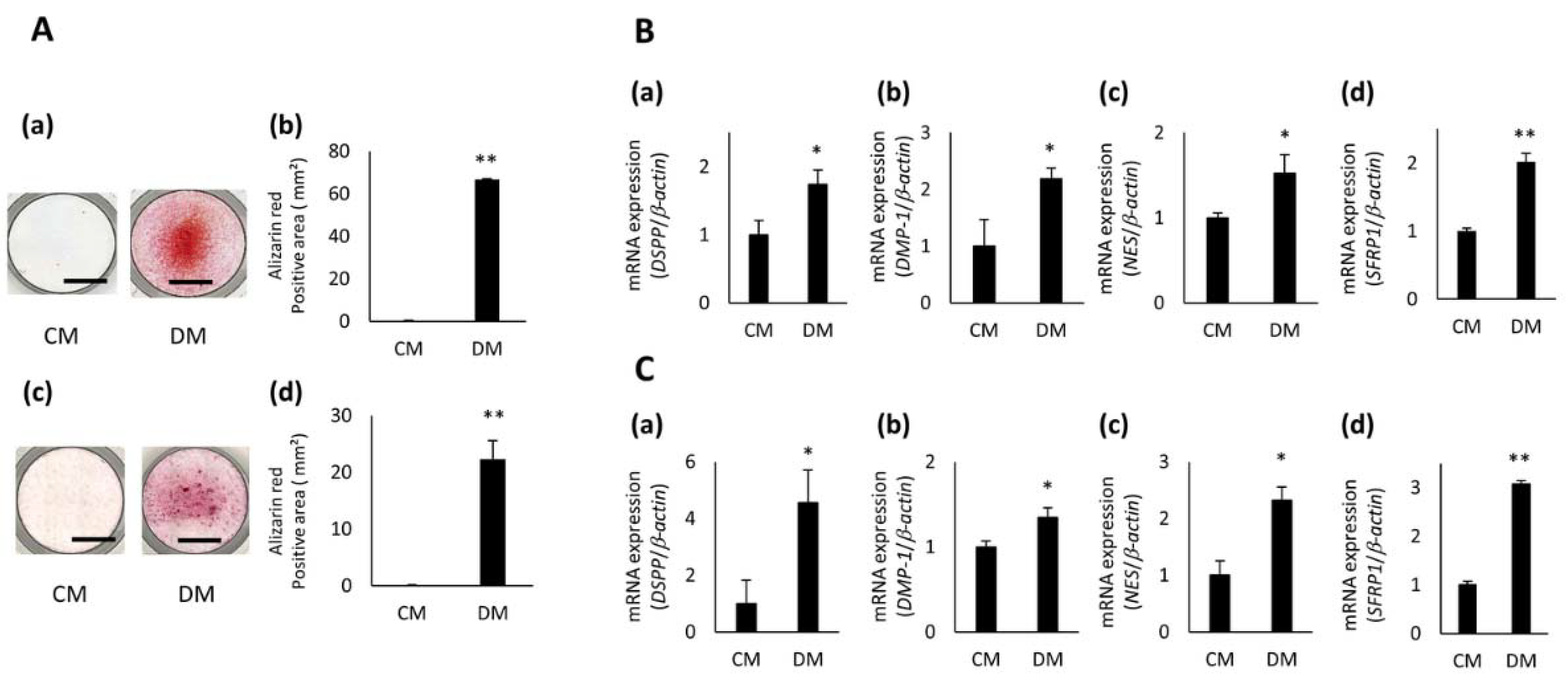
3.2. Expression of SFRP1 during Odontoblastic Differentiation of hDPCs
3.3. Effects of SFRP1 Down-Regulation on Odontoblastic Differentiation of hDPCs
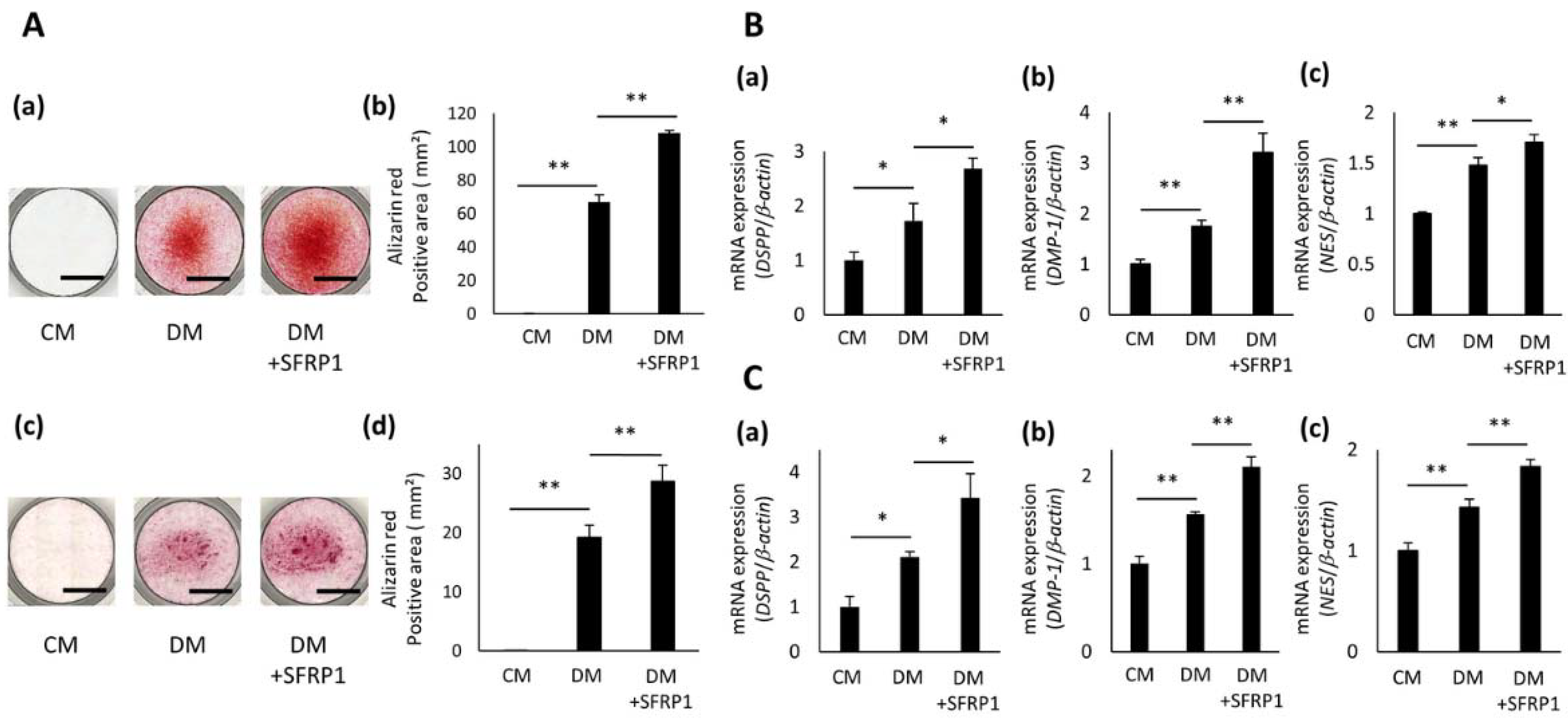
3.4. Effects of SFRP1 Stimulation on Odontoblastic Differentiation of hDPCs
3.5. Effects of SFRP1 Regulation on BMP-2 Gene Expression in hDPCs
3.6. Effects of SFRP1 on Reparative Dentin Formation after Direct Pulp Capping Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Yildirim, S. Dental Pulp Is a Connective Tissue. In Dental Pulp Stem Cells; Kalkstein, A., Hutter, R., Eds.; Springer: New York, NY, USA, 2013; pp. 17–24. [Google Scholar]
- Zhang, W.; Zhang, J.; Zhao, L.B.; Jiang, Y.J.; Hu, X.R.; Li, Z.G.; Liu, B. Comparison of neurovascular relationships between human pulp and rat pulp. Int. J. Clin. Exp. Med. 2017, 10, 6017–6028. [Google Scholar]
- Nakashima, M.; Akamine, A. The application of tissue engineering to regeneration of pulp and dentin in endodontics. J. Endod. 2005, 31, 711–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khasnis, S.A.; Kidiyoor, K.H.; Patil, A.B.; Kenganal, S.B. Vertical root fractures and their management. J. Conserv. Dent. 2014, 17, 103–110. [Google Scholar] [CrossRef]
- Malhotra, N.; Kundabala, M.; Acharaya, S. A review of root fractures: Diagnosis, treatment and prognosis. Dent. Update 2011, 38, 615–616. [Google Scholar] [CrossRef] [Green Version]
- Accorinte, M.L.; Loguercio, A.D.; Reis, A.; Carneiro, E.; Grande, R.H.; Murata, S.S.; Holland, R. Response of human dental pulp capped with MTA and calcium hydroxide powder. Oper. Dent. 2008, 33, 488–495. [Google Scholar] [CrossRef]
- Lewis, B.A.; Burgess, J.O.; Gray, S.E. Mechanical properties of dental base materials. Am. J. Dent. 1992, 5, 69–72. [Google Scholar]
- Mooney, G.C.; North, S. The current opinions and use of MTA for apical barrier formation of non-vital immature permanent incisors by consultants in paediatric dentistry in the UK. Dent. Traumatol. 2008, 24, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Al-Hezaimi, K.; Salameh, Z.; Al-Fouzan, K.; Al Rejaie, M.; Tay, F.R. Histomorphometric and micro-computed tomography analysis of pulpal response to three different pulp capping materials. J. Endod. 2011, 37, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Magloire, H.; Joffre, A.; Bleicher, F. An in vitro model of human dental pulp repair. J. Dent. Res. 1996, 75, 1971–1978. [Google Scholar] [CrossRef]
- Marrelli, M.; Falisi, G.; Apicella, A.; Apicella, D.; Amantea, M.; Cielo, A.; Bonanome, L.; Palmieri, F.; Santacroce, L.; Giannini, S.; et al. Behaviour of dental pulp stem cells on different types of innovative mesoporous and nanoporous silicon scaffolds with different functionalizations of the surfaces. J. Biol. Regul. Homeost. Agents 2015, 29, 991–997. [Google Scholar]
- Ballini, A.; Boccaccio, A.; Saini, R.; Van Pham, P.; Tatullo, M. Dental-Derived Stem Cells and Their Secretome and Interactions with Bioscaffolds/Biomaterials in Regenerative Medicine: From the In Vitro Research to Translational Applications. Stem Cells Int. 2017, 2017, 6975251. [Google Scholar] [CrossRef]
- Yoshida, S.; Wada, N.; Hasegawa, D.; Miyaji, H.; Mitarai, H.; Tomokiyo, A.; Hamano, S.; Maeda, H. Semaphorin 3A Induces Odontoblastic Phenotype in Dental Pulp Stem Cells. J. Dent. Res. 2016, 95, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Roberts-Clark, D.J.; Smith, A.J. Angiogenic growth factors in human dentine matrix. Arch. Oral Biol. 2000, 45, 1013–1016. [Google Scholar] [CrossRef]
- Bovolenta, P.; Esteve, P.; Ruiz, J.M.; Cisneros, E.; Lopez-Rios, J. Beyond Wnt inhibition: New functions of secreted Frizzled-related proteins in development and disease. J. Cell. Sci. 2008, 121, 737–746. [Google Scholar] [CrossRef] [Green Version]
- Bodine, P.V.; Billiard, J.; Moran, R.A.; Ponce-de-Leon, H.; McLarney, S.; Mangine, A.; Scrimo, M.J.; Bhat, R.A.; Stauffer, B.; Green, J.; et al. The Wnt antagonist secreted frizzled-related protein-1 controls osteoblast and osteocyte apoptosis. J. Cell. Biochem. 2005, 96, 1212–1230. [Google Scholar] [CrossRef]
- Li, J.; Huang, X.; Xu, X.; Mayo, J.; Bringas, P., Jr.; Jiang, R.; Wang, S.; Chai, Y. SMAD4-mediated WNT signaling controls the fate of cranial neural crest cells during tooth morphogenesis. Development 2011, 138, 1977–1989. [Google Scholar] [CrossRef] [Green Version]
- Wada, N.; Maeda, H.; Tanabe, K.; Tsuda, E.; Yano, K.; Nakamuta, H.; Akamine, A. Periodontal ligament cells secrete the factor that inhibits osteoclastic differentiation and function: The factor is osteoprotegerin/osteoclastogenesis inhibitory factor. J. Periodontal Res. 2001, 36, 56–63. [Google Scholar] [CrossRef]
- Wada, N.; Maeda, H.; Yoshimine, Y.; Akamine, A. Lipopolysaccharide stimulates expression of osteoprotegerin and receptor activator of NF-kappa B ligand in periodontal ligament fibroblasts through the induction of interleukin-1 beta and tumor necrosis factor-alpha. Bone 2004, 35, 629–635. [Google Scholar] [CrossRef]
- Mizumachi, H.; Yoshida, S.; Tomokiyo, A.; Hasegawa, D.; Hamano, S.; Yuda, A.; Sugii, H.; Serita, S.; Mitarai, H.; Koori, K.; et al. Calcium-sensing receptor-ERK signaling promotes odontoblastic differentiation of human dental pulp cells. Bone 2017, 101, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Satoh, W.; Gotoh, T.; Tsunematsu, Y.; Aizawa, S.; Shimono, A. Sfrp1 and Sfrp2 regulate anteroposterior axis elongation and somite segmentation during mouse embryogenesis. Development 2006, 133, 989–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteve, P.; Crespo, I.; Kaimakis, P.; Sandonís, A.; Bovolenta, P. Sfrp1 Modulates Cell-signaling Events Underlying Telencephalic Patterning, Growth and Differentiation. Cereb. Cortex. 2019, 29, 1059–1074. [Google Scholar] [CrossRef]
- Foronjy, R.; Imai, K.; Shiomi, T.; Mercer, B.; Sklepkiewicz, P.; Thankachen, J.; Bodine, P.; D’Armiento, J. The divergent roles of secreted frizzled related protein-1 (SFRP1) in lung morphogenesis and emphysema. Am. J. Pathol. 2010, 177, 598–607. [Google Scholar] [CrossRef]
- Kele, J.; Andersson, E.R.; Villaescusa, J.C.; Cajanek, L.; Parish, C.L.; Bonilla, S.; Toledo, E.M.; Bryja, V.; Rubin, J.S.; Shimono, A.; et al. SFRP1 and SFRP2 dose-dependently regulate midbrain dopamine neuron development in vivo and in embryonic stem cells. Stem Cells 2012, 30, 865–875. [Google Scholar] [CrossRef]
- Ghuman, M.S.; Al-Masri, M.; Xavier, G.; Cobourne, M.T.; McKay, I.J.; Hughes, F.J. Gingival fibroblasts prevent BMP-mediated osteoblastic differentiation. J. Periodontal Res. 2019, 54, 300–309. [Google Scholar] [CrossRef]
- Yokota, T.; Oritani, K.; Garrett, K.P.; Kouro, T.; Nishida, M.; Takahashi, I.; Ichii, M.; Satoh, Y.; Kincade, P.W.; Kanakura, Y. Soluble frizzled-related protein 1 is estrogen inducible in bone marrow stromal cells and suppresses the earliest events in lymphopoiesis. J. Immunol. 2008, 181, 6061–6072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopinathan, G.; Foyle, D.; Luan, X.; Diekwisch, T.G.H. The Wnt Antagonist SFRP1: A Key Regulator of Periodontal Mineral Homeostasis. Stem Cells Dev. 2019, 28, 1004–1014. [Google Scholar] [CrossRef] [PubMed]
- Rombouts, C.; Jeanneau, C.; Bakopoulou, A.; About, I. Dental Pulp Stem Cell Recruitment Signals within Injured Dental Pulp Tissue. Dent. J. 2016, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Couve, E.; Schmachtenberg, O. Schwann Cell Responses and Plasticity in Different Dental Pulp Scenarios. Front. Cell. Neurosci. 2018, 12, 299. [Google Scholar] [CrossRef] [PubMed]
- Eramo, S.; Natali, A.; Pinna, R.; Milia, E. Dental pulp regeneration via cell homing. Int. Endod. J. 2018, 51, 405–419. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, M.; Farges, J.C.; Lacerda-Pinheiro, S.; Six, N.; Jegat, N.; Decup, F.; Septier, D.; Carrouel, F.; Durand, S.; Chaussain-Miller, C.; et al. Inflammatory and immunological aspects of dental pulp repair. Pharmacol. Res. 2008, 58, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2000, 97, 13625–13630. [Google Scholar] [CrossRef] [Green Version]
- Neves, V.C.; Babb, R.; Chandrasekaran, D.; Sharpe, P.T. Promotion of natural tooth repair by small molecule GSK3 antagonists. Sci. Rep. 2017, 7, 39654. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, Y.; Shelley, E.J.; Wen, L.; Stump, R.J.; Shimono, A.; Lovicu, F.J.; McAvoy, J.W. Sfrp1 and Sfrp2 are not involved in Wnt/β-catenin signal silencing during lens induction but are required for maintenance of Wnt/β-catenin signaling in lens epithelial cells. Dev. Biol. 2013, 384, 181–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, J.; Esteve, P.; Weinl, C.; Ruiz, J.M.; Fermin, Y.; Trousse, F.; Dwivedy, A.; Holt, C.; Bovolenta, P. SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. Nat. Neurosci. 2005, 8, 1301–1309. [Google Scholar] [CrossRef]
- Kawano, Y.; Diez, S.; Uysal-Onganer, P.; Darrington, R.S.; Waxman, J.; Kypta, R.M. Secreted Frizzled-related protein-1 is a negative regulator of androgen receptor activity in prostate cancer. Br. J. Cancer 2009, 100, 1165–1174. [Google Scholar] [CrossRef] [Green Version]
- Gauger, K.J.; Chenausky, K.L.; Murray, M.E.; Schneider, S.S. SFRP1 reduction results in an increased sensitivity to TGF-β signaling. BMC Cancer 2011, 11, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandyopadhyay, A.; Tsuji, K.; Cox, K.; Harfe, B.D.; Rosen, V.; Tabin, C.J. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006, 2, e216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aberg, T.; Wozney, J.; Thesleff, I. Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Dev. Dyn. 1997, 210, 383–396. [Google Scholar] [CrossRef]
- Yang, X.; van der Kraan, P.M.; Bian, Z.; Fan, M.; Walboomers, X.F.; Jansen, J.A. Mineralized tissue formation by BMP2-transfected pulp stem cells. J. Dent. Res. 2009, 88, 1020–1025. [Google Scholar] [CrossRef]
- Cho, Y.D.; Yoon, W.J.; Woo, K.M.; Baek, J.H.; Park, J.C.; Ryoo, H.M. The canonical BMP signaling pathway plays a crucial part in stimulation of dentin sialophosphoprotein expression by BMP-2. J. Biol. Chem. 2010, 285, 36369–36376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Harris, M.A.; Cui, Y.; Mishina, Y.; Harris, S.E.; Gluhak-Heinrich, J. Bmp2 Is Required for Odontoblast Differentiation and Pulp Vasculogenesis. J. Dent. Res. 2012, 91, 58–64. [Google Scholar] [CrossRef]
- Guo, F.; Feng, J.; Wang, F.; Li, W.; Gao, Q.; Chen, Z.; Shoff, L.; Donly, K.J.; Gluhak-Heinrich, J.; Chun, Y.H.P.; et al. Bmp2 Deletion Causes an Amelogenesis Imperfecta Phenotype Via Regulating Enamel Gene Expression. J. Cell. Physiol. 2015, 230, 1871–1882. [Google Scholar] [CrossRef] [Green Version]
- Shigetani, Y.; Yoshiba, K.; Kuratate, M.; Takei, E.; Yoshiba, N.; Yamanaka, Y.; Ohshima, H.; Okiji, T. Temporospatial localization of dentine matrix protein 1 following direct pulp capping with calcium hydroxide in rat molars. Int. Endod. J. 2015, 48, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Bodine, P.V.; Zhao, W.; Kharode, Y.P.; Bex, F.J.; Lambert, A.J.; Goad, M.B.; Gaur, T.; Stein, G.S.; Lian, J.B.; Komm, B.S. The Wnt antagonist secreted frizzled-related protein-1 is a negative regulator of trabecular bone formation in adult mice. Mol. Endocrinol. 2004, 18, 1222–1237. [Google Scholar] [CrossRef] [Green Version]
- Debiais, F.; Hott, M.; Graulet, A.M.; Marie, P.J. The effects of fibroblast growth factor-2 on human neonatal calvaria osteoblastic cells are differentiation stage specific. J. Bone. Miner. Res. 1998, 13, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Kawashima, N.; Suzuk, N.; Yamamoto, M.; Ohnishi, K.; Katsube, K.; Tanabe, H.; Kudo, A.; Saito, M.; Suda, H. Periostin is a negative regulator of mineralization in the dental pulp tissue. Odontology 2015, 103, 152–159. [Google Scholar] [CrossRef]
- Tatullo, M.; Spagnuolo, G.; Codispoti, B.; Zamparini, F.; Zhang, A.; Esposti, M.D.; Aparicio, C.; Rengo, C.; Nuzzolese, M.; Manzoli, L.; et al. PLA-Based Mineral-Doped Scaffolds Seeded with Human Periapical Cyst-Derived MSCs: A Promising Tool for Regenerative Healing in Dentistry. Materials 2019, 12, 597. [Google Scholar] [CrossRef] [Green Version]
- Aulino, P.; Costa, A.; Chiaravalloti, E.; Perniconi, B.; Adamo, S.; Coletti, D.; Marrelli, M.; Tatullo, M.; Teodori, L. Muscle extracellular matrix scaffold is a multipotent environment. Int. J. Med. Sci. 2015, 12, 336–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammarström, L.; Heijl, L.; Gestrelius, S. Periodontal regeneration in a buccal dehiscence model in monkeys after application of enamel matrix proteins. J. Clin. Periodontol. 1997, 24, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Takayama, S.; Kitamura, M.; Shimabukuro, Y.; Yanagi, K.; Ikezawa, K.; Saho, T.; Nozaki, T.; Okada, H. Recombinant human basic fibroblast growth factor (bFGF) stimulates periodontal regeneration in class II furcation defects created in beagle dogs. J. Periodontal Res. 2003, 38, 97–103. [Google Scholar] [CrossRef]
- Cox, C.F.; Sübay, R.K.; Ostro, E.; Suzuki, S.; Suzuki, S.H. Tunnel defects in dentin bridges: Their formation following direct pulp capping. Oper. Dent. 1996, 21, 4–11. [Google Scholar]
- Six, N.; Lasfargues, J.J.; Goldberg, M. Differential repair responses in the coronal and radicular areas of the exposed rat molar pulp induced by recombinant human bone morphogenetic protein 7 (osteogenic protein 1). Arch. Oral. Biol. 2002, 47, 177–187. [Google Scholar] [CrossRef]




| Target Gene (Abbreviation) | Forward (Top) and Reverse (Bottom) Primer Sequences | Size of Amplified Products (bp) | Annealing Temperature (°C) | Cycles | Sequence ID |
|---|---|---|---|---|---|
| SFRP1 | AAAGCAAGGGCCATTTAGATTAG TTCTGGGCTTGACCTTAATTGTA | 328 | 55 | 27 | NM_003012.5 |
| GAPDH | ACCACAGTCCATGCCATCCAC TCCACCACCCTGTTGCTGTA | 452 | 60 | 18 | NM_001256799.2 |
| Target Gene (Abbreviation) | Forward (Top) and Reverse (Bottom) Primer Sequences | Size of Amplified Products (bp) | Annealing Temperature (°C) | Cycles | Sequence ID |
|---|---|---|---|---|---|
| SFRP1 | CAAGAAGAAGAAGCCCCTGA AAGTGGTGGCTGAGGTTGTC | 123 | 60 | 40 | NM_003012.5 |
| DSPP | ATATTGAGGGCTGGAATGGGGA TTTGTGGCTCCAGCATTGTCA | 136 | 60 | 40 | NM_014208.3 |
| DMP-1 | CCCTTGGAGAGCAGTGAGTC CTCCTTTTCCTGTGCTCCTG | 166 | 60 | 40 | NM_004407.4 |
| NES | TGGCCACGTACAGGACCCTCC AGATCCAAGACGCCGGCCCT | 143 | 60 | 40 | NM_006617.1 |
| BMP-2 | TCCACTAATCATGCCATTGTTCAGA GGGACACAGCATGCCTTAGGA | 74 | 60 | 40 | NM_001200.4 |
| β-actin | ATTGCCGACAGGATGCAGA GAGTACTTGCGCTCAGGAGGA | 89 | 60 | 40 | NM_001101.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ipposhi, K.; Tomokiyo, A.; Ono, T.; Yamashita, K.; Alhasan, M.A.; Hasegawa, D.; Hamano, S.; Yoshida, S.; Sugii, H.; Itoyama, T.; et al. Secreted Frizzled-Related Protein 1 Promotes Odontoblastic Differentiation and Reparative Dentin Formation in Dental Pulp Cells. Cells 2021, 10, 2491. https://doi.org/10.3390/cells10092491
Ipposhi K, Tomokiyo A, Ono T, Yamashita K, Alhasan MA, Hasegawa D, Hamano S, Yoshida S, Sugii H, Itoyama T, et al. Secreted Frizzled-Related Protein 1 Promotes Odontoblastic Differentiation and Reparative Dentin Formation in Dental Pulp Cells. Cells. 2021; 10(9):2491. https://doi.org/10.3390/cells10092491
Chicago/Turabian StyleIpposhi, Keita, Atsushi Tomokiyo, Taiga Ono, Kozue Yamashita, Muhammad Anas Alhasan, Daigaku Hasegawa, Sayuri Hamano, Shinichiro Yoshida, Hideki Sugii, Tomohiro Itoyama, and et al. 2021. "Secreted Frizzled-Related Protein 1 Promotes Odontoblastic Differentiation and Reparative Dentin Formation in Dental Pulp Cells" Cells 10, no. 9: 2491. https://doi.org/10.3390/cells10092491






