Genetic Characterization of Rat Hepatic Stellate Cell Line HSC-T6 for In Vitro Cell Line Authentication
Abstract
:1. Introduction
2. Materials and Methods
2.1. Literature Search
2.2. Cell Culture
2.3. Electron Microscopic Analysis
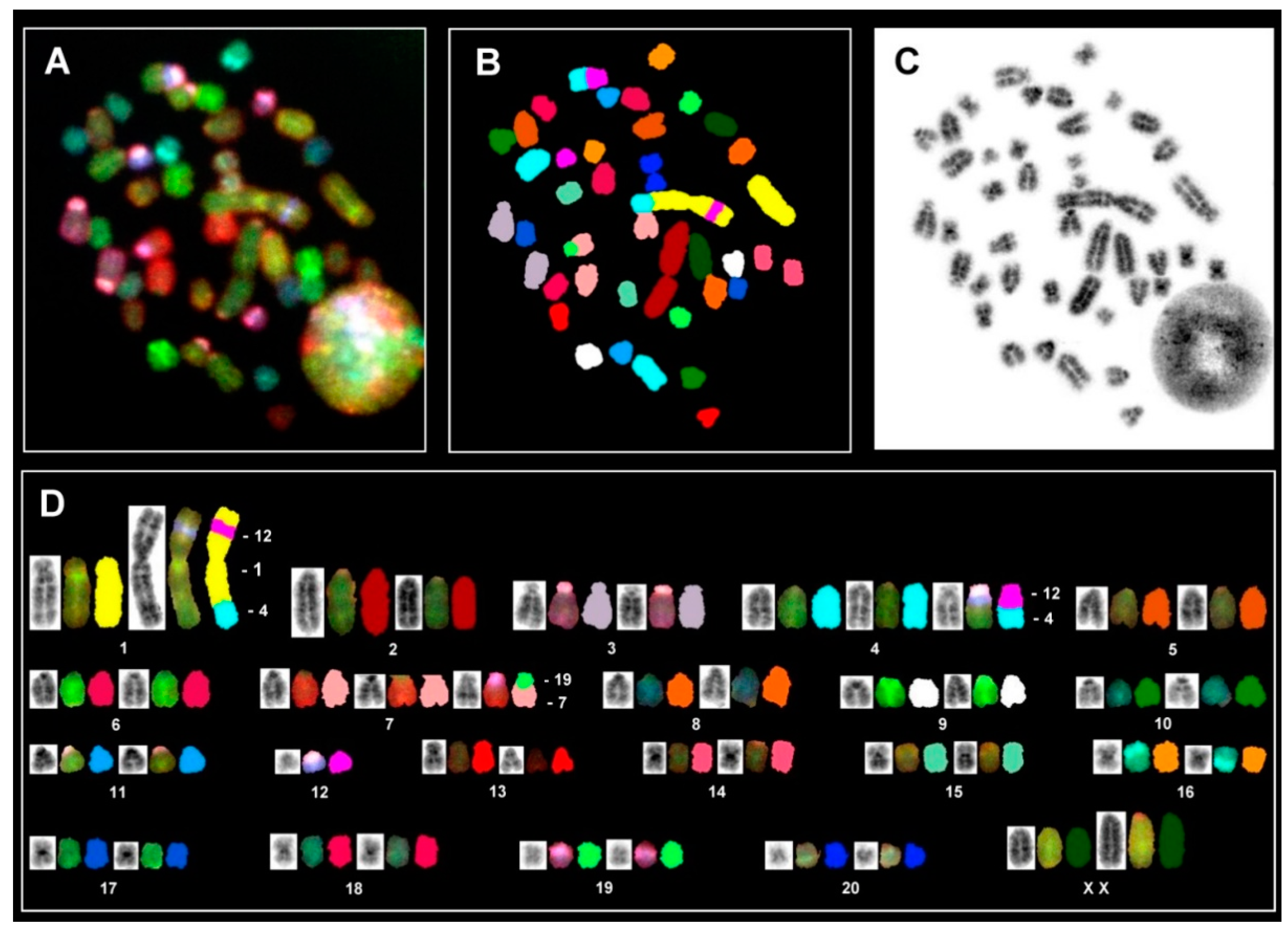
2.4. Preparation of HSC-T6 Metaphase Chromosomes and Karyotyping
2.5. In Situ Hybrdization and Spectral Imaging
2.6. Short Tandem Repeat (STR) Profiling
2.7. Next Generation Sequencing and Data Analysis
2.8. Western Blot Analysis
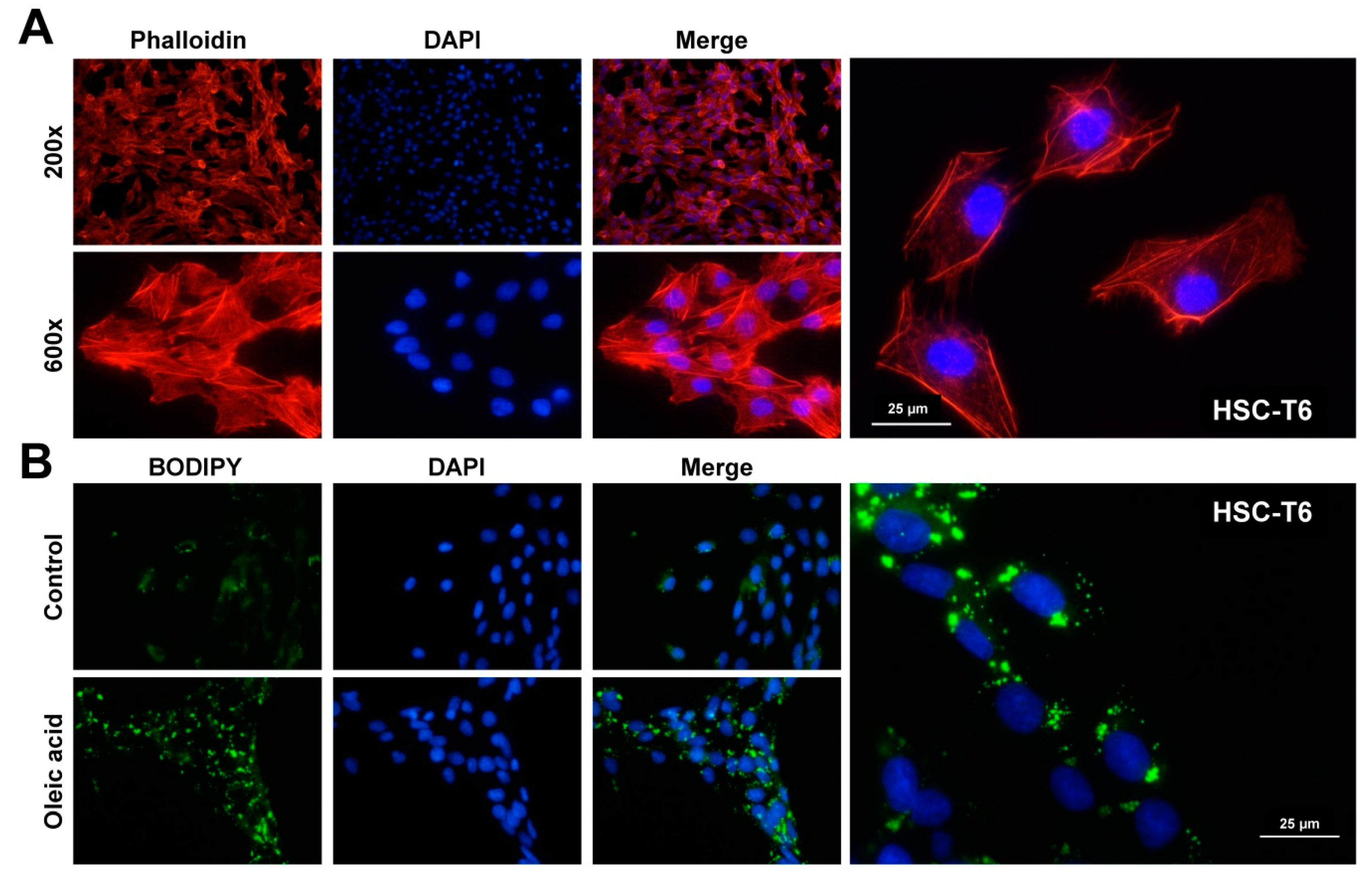
2.9. Rhodamine-Phalloidin Stain
2.10. Stimulation with Fatty Acids and Lipid Droplet Stain
2.11. Stimulation of HSC-T6 Cells
2.12. Reverse Transcription Polymerase Chain Reaction
3. Results
3.1. Usage of HSC-T6 Cells in Biomedical Research
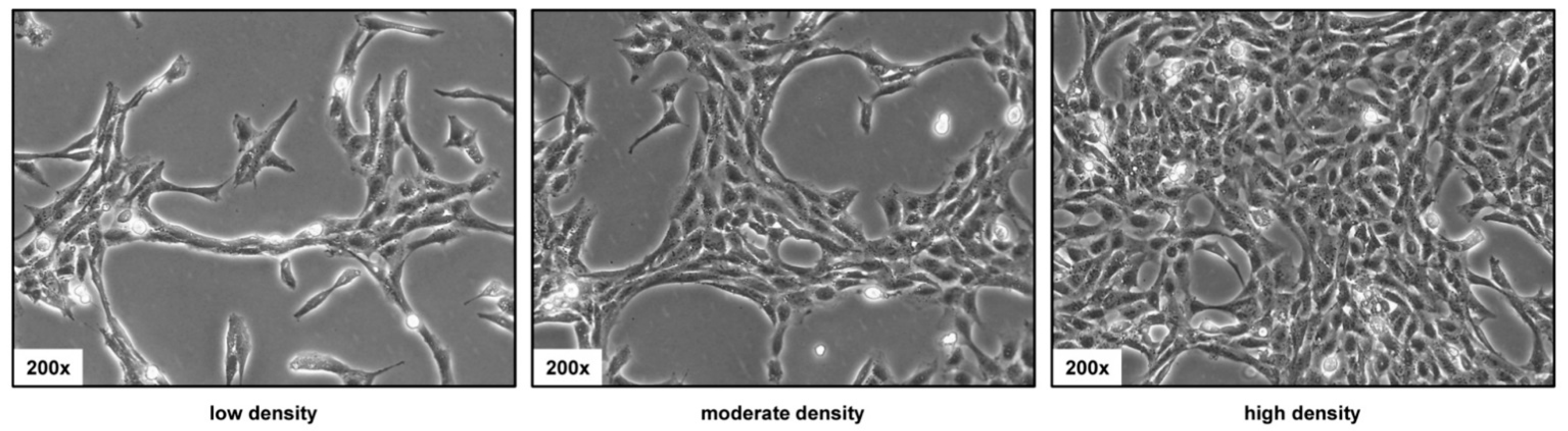
3.2. Phenotypic Appearance of HSC-T6 Cells
3.3. Electronmicroscopic Analysis of HSC-T6 Cells
3.4. Expression of Typical Hepatic Stellate Cell Markers in HSC-T6 Cells
3.5. Convential Genetic Analysis of HSC-T6 Cells
3.6. In Situ Hybridization and Spectral Karyotype Analysis
3.7. Short Tandem Repeat Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herrmann, J.; Gressner, A.M.; Weiskirchen, R. Immortal hepatic stellate cell lines: Useful tools to study hepatic stellate cell biology and function? J. Cell. Mol. Med. 2007, 11, 704–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Ratziu, V.; Choi, S.-G.; Lalazar, A.; Theiss, G.; Dang, Q.; Kim, S.-J.; Friedman, S.L. Transcriptional activation of transforming growth factor beta1 and its receptors by the Kruppel-like factor Zf9/core promoter-binding protein and Sp1. Potential mechanisms for autocrine fibrogenesis in response to injury. J. Biol. Chem. 1998, 273, 33750–33758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, S.; Piantedosi, R.; Frank, J.; Lalazar, A.; Rockey, D.C.; Friedman, S.L.; Blaner, W.S. An immortalized rat liver stellate cell line (HSC-T6): A new cell model for the study of retinoid metabolism in vitro. J. Lipid Res. 2000, 41, 882–893. [Google Scholar] [CrossRef]
- Almeida, J.L.; Hill, C.R.; Cole, K.D. Mouse cell line authentication. Cytotechnology 2013, 66, 133–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, J.L.; Cole, K.D.; Plant, A.L. Standards for cell line authentication and beyond. PLoS Biol. 2016, 14, e1002476. [Google Scholar] [CrossRef]
- Capes-Davis, A.; Neve, R.M. Authentication: A standard problem or a problem of standards? PLoS Biol. 2016, 14, e1002477. [Google Scholar] [CrossRef] [Green Version]
- Weiskirchen, R.; Weimer, J.; Meurer, S.K.; Kron, A.; Seipel, B.; Vater, I.; Arnold, N.; Siebert, R.; Xu, L.; Friedman, S.L.; et al. Genetic characteristics of the human hepatic stellate cell line LX-2. PLoS ONE 2013, 8, e75692. [Google Scholar] [CrossRef] [Green Version]
- Schröder, S.K.; Schüler, H.M.; Petersen, K.V.; Tesauro, C.; Knudsen, B.R.; Pedersen, F.S.; Krus, F.; Buhl, E.M.; Roeb, E.; Roderfeld, M.; et al. Genetic and molecular characterization of the immortalized murine hepatic stellate cell line GRX. Cells 2022, 11, 1504. [Google Scholar] [CrossRef]
- National Library of Medicine. Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 1 April 2022).
- Meurer, S.K.; Alsamman, M.; Sahin, H.; Wasmuth, H.E.; Kisseleva, T.; Brenner, D.A.; Trautwein, C.; Weiskirchen, R.; Scholten, D. Overexpression of endoglin modulates TGF-β1-signalling pathways in a novel immortalized mouse hepatic stellate cell line. PLoS One 2013, 8, e56116. [Google Scholar] [CrossRef] [Green Version]
- Hamta, A.; Adamovic, T.; Samuelson, E.; Helou, K.; Behboudi, A.; Levan, G. Chromosome ideograms of the laboratory rat (Rattus norvegicus) based on high-resolution banding, and anchoring of the cytogenetic map to the DNA sequence by FISH in sample chromosomes. Cytogenet. Genome Res. 2006, 115, 158–168. [Google Scholar] [CrossRef]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Buwe, A.; Steinlein, C.; Koehler, M.; Bar-Am, I.; Katzin, N.; Schmid, M. Multicolor spectral karyotyping of rat chromosomes. Cytogenet. Genome Res. 2003, 103, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Babraham Bioinformatics-FastQC a Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 April 2022).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527, Erratum in Nat. Biotechnol. 2016, 34, 888. [Google Scholar] [CrossRef] [PubMed]
- Enbsembl. Rattus Norvegicus Rnor_6.0 Reference Transcriptome. Available online: http://ftp.ensembl.org/pub/release-104/fasta/rattus_norvegicus/cdna/Rattus_norvegicus.Rnor_6.0.cdna.all.fa.gz (accessed on 24 April 2022).
- Enbsembl. Rattus Norvegicus Rnor_6.0 Transcriptome Information. Available online: http://ftp.ensembl.org/pub/release-104/gtf/rattus_norvegicus/Rattus_norvegicus.Rnor_6.0.104.gtf.gz (accessed on 24 April 2022).
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef] [PubMed]
- Schröder, S.K.; Asimakopoulou, A.; Tillmann, S.; Koschmieder, S.; Weiskirchen, R. TNF-α controls Lipocalin-2 expression in PC-3 prostate cancer cells. Cytokine 2020, 135, 155214. [Google Scholar] [CrossRef]
- Qiu, B.; Simon, M.C. BODIPY 493/503 staining of neutral lipid droplets for microscopy and quantification by flow cytometry. Bio-Protocol 2016, 6, e1912. [Google Scholar] [CrossRef] [Green Version]
- Koreivienė, J. Microalgae lipid staining with fluorescent BODIPY Dye. Methods Mol. Biol. 2017, 1980, 47–53. [Google Scholar] [CrossRef]
- Greenwel, P.; Schwartz, M.; Rosas, M.; Peyrol, S.; Grimaud, J.A.; Rojkind, M. Characterization of fat-storing cell lines derived from normal and CCl4-cirrhotic livers. Differences in the production of interleukin-6. Lab. Investig. 1991, 65, 644–653. [Google Scholar]
- Greenwel, P.; Rubin, J.; Schwartz, M.; Hertzberg, E.L.; Rojkind, M. Liver fat-storing cell clones obtained from a CCl4-cirrhotic rat are heterogeneous with regard to proliferation, expression of extracellular matrix components, interleukin-6, and connexin 43. Lab. Investig. 1993, 69, 210–216. [Google Scholar]
- Sauvant, P.; Sapin, V.; Abergel, A.; Schmidt, C.K.; Blanchon, L.; Alexandre-Gouabau, M.-C.; Rosenbaum, J.; Bommelaer, G.; Rock, E.; Dastugue, B.; et al. PAV-1, a new rat hepatic stellate cell line converts retinol into retinoic acid, a process altered by ethanol. Int. J. Biochem. Cell Biol. 2002, 34, 1017–1029. [Google Scholar] [CrossRef]
- Pan, Q.; Li, D.-G.; Lu, H.-M.; Wang, Y.-Q.; Zhang, W.-Z.; Xu, Q.-F. A new immortalized rat cell line, hepatic stellate cell-PQ, exhibiting characteristics of hepatic stellate cell. Hepatobiliary Pancreat. Dis. Int. 2005, 4, 281–284. [Google Scholar] [PubMed]
- Xiong, S.; Yavrom, S.; Hazra, S.; Wu, D.; She, H. Spontaneously immortalized cell line from rat liver fibrosis with phenotypes of both hepatic stellate cells and myofibroblasts. Hepatology 2001, 34, 520A. [Google Scholar]
- Sung, C.K.; She, H.; Xiong, S.; Tsukamoto, H. Tumor necrosis factor-α inhibits peroxisome proliferator-activated receptor γ activity at a posttranslational level in hepatic stellate cells. Am. J. Physiol. Liver Physiol. 2004, 286, G722–G729. [Google Scholar] [CrossRef]
- Isono, M.; Soda, M.; Inoue, A.; Akiyoshi, H.; Sato, K. Reverse transformation of hepatic myofibroblast-like cells by TGFβ1/LAP. Biochem. Biophys. Res. Commun. 2003, 311, 959–965. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.Y.; Kim, K.M.; Nan, J.-X.; Zhao, Y.Z.; Park, P.-H.; Lee, S.J.; Sohn, D.H. Induction of apoptosis by Tanshinone I via cytochrome c release in activated hepatic stellate cells. Pharmacol. Toxicol. 2003, 92, 195–200. [Google Scholar] [CrossRef]
- Li, T.; Leng, X.-S.; Zhu, J.-Y.; Wang, F.-S. Establishment and characterization of an immortalized rat hepatic stellate cell line. Int. J. Clin. Exp. Pathol. 2015, 8, 12064–12074. [Google Scholar]
- Fausther, M.; Goree, J.R.; Lavoie, G.; Graham, A.L.; Sévigny, J.; Dranoff, J.A. Establishment and characterization of rat portal myofibroblast cell lines. PLoS ONE 2015, 10, e0121161. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-W.; Chiu, Y.-T.; Fu, S.-L.; Huang, Y.-T. Osthole ameliorates hepatic fibrosis and inhibits hepatic stellate cell activation. J. Biomed. Sci. 2015, 22, 63. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.; Zhang, X.; Yin, Q.; Meng, A.; Su, S.; Jing, X.; Li, H.; Guan, X.; Li, X.; Liu, S.; et al. F-actin cytoskeleton reorganization is associated with hepatic stellate cell activation. Mol. Med. Rep. 2014, 9, 1641–1647. [Google Scholar] [CrossRef] [Green Version]
- Routray, I.; Mahmood, A.; Ngwa, N.E.; Tasleem, M.; Sahin, K.; Kucuk, O.; Ali, S. Cell line cross-contamination and accidental co-culture. J. Stem Cell Res. Ther. 2016, 1, 31. [Google Scholar]
- da Silva, J.A.T. Incorrect cell line validation and verification. Ann. Transl. Med. 2018, 6, 136. [Google Scholar] [CrossRef] [PubMed]
- American Type Culture Collection Standards Development Organization Workgroup ASN-0002. Cell line misidentification: The beginning of the end. Nat. Cancer 2010, 10, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Horbach, S.P.J.M.; Halffman, W. The ghosts of HeLa: How cell line misidentification contaminates the scientific literature. PLoS ONE 2017, 12, e0186281. [Google Scholar] [CrossRef] [PubMed]
- Hughes, P.; Marshall, D.; Reid, Y.; Parkes, H.; Gelber, C. The costs of using unauthenticated, over-passaged cell lines: How much more data do we need? Biotechniques 2007, 43, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Ávila, J.A.; Catalina, P.; Aguilar-Quesada, R. Quality control of cell lines using DNA as target. DNA 2022, 2, 4. [Google Scholar] [CrossRef]
- Nims, R.W.; Sykes, G.; Cottrill, K.; Ikonomi, P.; Elmore, E. Short tandem repeat profiling: Part of an overall strategy for reducing the frequency of cell misidentification. Vitr. Cell. Dev. Biol.-Anim. 2010, 46, 811–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, J.L.; Dakic, A.; Kindig, K.; Kone, M.; Letham, D.L.D.; Langdon, S.; Peat, R.; Holding-Pillai, J.; Hall, E.M.; Ladd, M.; et al. Interlaboratory study to validate a STR profiling method for intraspecies identification of mouse cell lines. PLoS ONE 2019, 14, e0218412. [Google Scholar] [CrossRef] [Green Version]
- Szpirer, J.; Defeo-Jones, D.; Ellis, R.W.; Szpirer, C. Assignment of three rat cellularRAS oncogenes to chromosomes 1, 4, and X. Somat. Cell Mol. Genet. 1985, 11, 93–97. [Google Scholar] [CrossRef]
- Falck, E.; Hedberg, C.; Klinga-Levan, K.; Behboudi, A. SKY analysis revealed recurrent numerical and structural chromosome changes in BDII rat endometrial carcinomas. Cancer Cell Int. 2011, 11, 20. [Google Scholar] [CrossRef] [Green Version]
- Acharya, P.; Chouhan, K.; Weiskirchen, S.; Weiskirchen, R. Cellular mechanisms of liver fibrosis. Front. Pharmacol. 2021, 12, 671640. [Google Scholar] [CrossRef] [PubMed]
- Mancardi, D.; Mezzanotte, M.; Arrigo, E.; Barinotti, A.; Roetto, A. Iron overload, oxidative stress, and ferroptosis in the failing heart and liver. Antioxidants 2021, 10, 1864. [Google Scholar] [CrossRef] [PubMed]
- Liau, G.; Chan, L.M.; Feng, P. Increased ferritin gene expression is both promoted by cAMP and a marker of growth arrest in rabbit vascular smooth muscle cells. J. Biol. Chem. 1991, 266, 18819–18826. [Google Scholar] [CrossRef]
- Hein, J.; Boichuk, S.; Wu, J.; Cheng, Y.; Freire, R.; Jat, P.S.; Roberts, T.M.; Gjoerup, O.V. Simian virus 40 large T antigen disrupts genome integrity and activates a DNA damage response via Bub1 binding. J. Virol. 2009, 83, 117–127. [Google Scholar] [CrossRef] [Green Version]




| Transcript Id | Gene Id | Gene | Gene Description | TPM | Remarks |
|---|---|---|---|---|---|
| ENSRNOT00000083468.1 | ENSRNOG00000058039.1 | Acta2 | Actin alpha 2, smooth muscle | 2.377 | HSC marker |
| ENSRNOT00000005311.6 | ENSRNOG00000003897.6 | Col1a1 | Collagen type I alpha 1 chain | 2033.74 | HSC marker |
| ENSRNOT00000004956.4 | ENSRNOG00000003357.4 | Col3a1 | Collagen type III alpha 1 chain | 2128.9 | HSC marker |
| ENSRNOT00000024430.5 | ENSRNOG00000018087.5 | Vim | Vimentin | 2054.81 | HSC marker |
| ENSRNOT00000057585.4 | ENSRNOG00000014288.8 | Fn1 | Fibronectin 1 | 143.953 | HSC marker |
| ENSRNOT00000019772.6 | 352.585 | ||||
| ENSRNOT00000013538.6 | ENSRNOG00000009884.6 | Lgals1 | Galectin 1 | 4801.86 | HSC marker |
| ENSRNOT00000017486.7 | ENSRNOG00000012840.7 | Sparc | Secreted protein acidic and cysteine rich | 2188.34 | HSC marker |
| ENSRNOT00000067011.2 | ENSRNOG00000003772.7 | Csrp2 | Cysteine and glycine-rich protein 2 | 195.244 | HSC marker |
| ENSRNOT00000013745.7 | ENSRNOG00000010208.7 | Timp1 | TIMP metallopeptidase inhibitor | 5518.57 | Inhibitor of MMPs |
| ENSRNOT00000004290.4 | ENSRNOG00000003148.5 | Timp2 | TIMP metallopeptidase inhibitor 2 | 369.667 | Inhibitor of MMPs |
| ENSRNOT00000010180.5 | ENSRNOG00000007650.5 | Cd63 | Cd63 molecule | 1639.48 | TIMP1 receptor |
| ENSRNOT00000011208.7 | ENSRNOG00000008301.7 | Tagln2 | Transgelin 2 | 754.93 | Smooth muscle marker |
| ENSRNOT00000015962.6 | ENSRNOG00000020660.7 | Cfl1 | Cofilin 1 | 1670.37 | Cytoskeleton |
| ENSRNOT00000042459.4 | ENSRNOG00000034254.4 | Actb | Actin, beta | 4379.5 | Cytoskeleton |
| ENSRNOT00000080216.1 | 1820.7 | ||||
| ENSRNOT00000078250.1 | ENSRNOG00000056836.1 | Cav1 | Caveolin 1 | 167.988 | Scaffolding protein |
| ENSRNOT00000008659.4 | ENSRNOG00000009972.7 | Rara | Retinoic acid receptor, alpha | 24.8697 | Nuclear receptors |
| ENSRNOT00000084644.1 | 23.8544 | ||||
| ENSRNOT00000033048.6 | ENSRNOG00000024061.7 | Rarb | Retinoic acid receptor, beta | 0.541197 | Nuclear receptors |
| ENSRNOT00000016801.5 | ENSRNOG00000012499.7 | Rarg | Retinoic acid receptor, gamma | 64.5919 | Nuclear receptors |
| ENSRNOT00000017096.7 | 39.1824 | ||||
| ENSRNOT00000012892.4 | ENSRNOG00000009446.4 | Rxra | Retinoid X receptor alpha | 18.9052 | Nuclear receptors |
| ENSRNOT00000041613.5 | ENSRNOG00000000464.7 | Rxrb | Retinoid X receptor beta | 20.0384 | Nuclear receptors |
| ENSRNOT00000087670.1 | 1.79029 | ||||
| ENSRNOT00000086978.1 | 1.08069 | ||||
| ENSRNOT00000081588.1 | 0.804912 | ||||
| ENSRNOT00000091182.1 | 0.723898 | ||||
| ENSRNOT00000087895.1 | 0.688452 | ||||
| ENSRNOT00000084638.1 | 0.499241 | ||||
| ENSRNOT00000019571.3 | ENSRNOG00000014090.3 | Retsat | Retinol saturase | 52.476 | Retinoid metabolism |
| ENSRNOT00000081756.1 | ENSRNOG00000053850.1 | Rdh5 | Retinol dehydrogenase 5 | 0.0383589 | Retinoid metabolism |
| ENSRNOT00000032076.4 | ENSRNOG00000025767.4 | Rdh8 | Retinol dehydrogenase 8 | 0.112568 | Retinoid metabolism |
| ENSRNOT00000009096.4 | ENSRNOG00000006681.4 | Rdh10 | Retinol dehydrogenase 10 | 3.87175 | Retinoid metabolism |
| ENSRNOT00000085927.1 | ENSRNOG00000054770.1 | Rdh11 | Retinol dehydrogenase 11 | 26.8725 | Retinoid metabolism |
| ENSRNOT00000078436.1 | 0.15904 | ||||
| ENSRNOT00000089162.1 | ENSRNOG00000056553.1 | Rdh12 | Retinol dehydrogenase 12 | 1.06243 | Retinoid metabolism |
| ENSRNOT00000031462.5 | ENSRNOG00000027919.5 | Rdh13 | Retinol dehydrogenase 13 | 1.96789 | Retinoid metabolism |
| ENSRNOT00000006020.6 | ENSRNOG00000039551.3 | Rdh14 | Retinol dehydrogenase 14 | 8.16116 | Retinoid metabolism |
| ENSRNOT00000093003.1 | 2.97058 | ||||
| ENSRNOT00000092948.1 | 0.857514 | ||||
| ENSRNOT00000018622.4 | ENSRNOG00000013794.4 | Rbp1 | Retinol binding protein 1 | 25.3842 | Retinoid metabolism |
| ENSRNOT00000018755.6 | ENSRNOG00000013932.6 | Rbp2 | Retinol binding protein 2 | 0.309974 | Retinoid metabolism |
| ENSRNOT00000021055.7 | ENSRNOG00000015518.7 | Rbp4 | Retinol binding protein 4 | 0.0363529 | Retinoid metabolism |
| ENSRNOT00000021348.5 | ENSRNOG00000015850.5 | Rbp7 | Retinol binding protein 7 | 0.244971 | Retinoid metabolism |
| ENSRNOT00000082156.1 | ENSRNOG00000025608.4 | Lrat | lecithin retinol acyltransferase | 0.0125274 | Retinoid metabolism |
| ENSRNOT00000022113.4 | ENSRNOG00000016275.4 | Ttr | Transthyretin | 0.161195 | RBP transporter |
| ENSRNOT00000030919.5 | ENSRNOG00000022619.6 | Fth1 | Ferritin heavy chain 1 | 9890.55 | Iron storage |
| ENSRNOT00000087162.1 | ENSRNOG00000059900.1 | Bst2 | Bone marrow stromal cell antigen 2 | 1681.66 | Antiviral response |
| ENSRNOT00000091906.1 | 589.283 | ||||
| ENSRNOT00000080988.1 | ENSRNOG00000052802.1 | Aldoa | Aldolase, fructose-bisphophate | 1471.21 | Glycolysis |
| ENSRNOT00000087928.1 | 492.926 | ||||
| ENSRNOT00000088473.1 | 183.94 | ||||
| ENSRNOT00000015332.7 | ENSRNOG00000011329.7 | Pkm | Pyruvate kinase M1/2 | 1437.52 | Glycolysis |
| ENSRNOT00000083666.1 | 455,466 | ||||
| ENSRNOT00000077604.1 | ENSRNOG00000058249.1 | Pgk1 | Phosphoglycerate kinase 1 | 1404.31 | Glycolysis |
| ENSRNOT00000085653.1 | 0.204793 | ||||
| ENSRNOT00000050443.4 | ENSRNOG00000018630.7 | LOC108351137 | Glyceraldehyde-3-phosphate dehydrogenase | 4598.86 | Glycolysis |
| ENSRNOT00000041328.3 | ENSRNOG00000030963.3 | 4243.2 |
| Cell | Chromosome Numbers | der(1)t(1;4;12) | der(1)t(1;4) | der(1)t(1;15) | der(3)t(3;18) | der(4)t(4;12) | der(7)t(7;18) | der(7)t(7;19) | der(7) t(7;20) | der(10)t(10;12;13) | der(12)t(12:18) | der(13)t(13;17) | der(15)t(15;17) | der(15)t(15;18) | der(X)t(X;20) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 41,XX | ● | ● | ● | ● | ||||||||||
| 2 | 40,XX | ● | ● | ● | ● | ● | ● | ● | |||||||
| 3 | 44,XX | ● | ● | ● | |||||||||||
| 4 | 41,XX | ● | ● | ● | |||||||||||
| 5 | 41,XX | ● | ● | ● | |||||||||||
| 6 | 40,XX | ● | ● | ● | |||||||||||
| 7 | 43,XX | ● | ● | ● | |||||||||||
| 8 | 43,XX | ● | ● | ● | ● | ||||||||||
| 9 | 42,XX | ● | ● | ● | ● | ● | |||||||||
| 10 | 43,XX | ● | ● | ● | ● | ||||||||||
| 11 | 43,XX | ● | ● | ● | ● | ||||||||||
| 12 | 43,XX | ● | ● | ● | |||||||||||
| 13 | 42,XX | ● | ● | ● | |||||||||||
| 14 | 42,XX | ● | ● | ● | ● | ||||||||||
| 15 | 43,XX | ● | ● | ● | ● | ||||||||||
| 16 | 43,XX | ● | ● | ● | |||||||||||
| 17 | 43,XX | ● | ● | ● | |||||||||||
| 18 | 43,XX | ● | ● | ● | ● | ||||||||||
| 19 | 41,XX | ● | ● | ● | ● | ||||||||||
| 20 | 43,XX | ● | ● | ● |
| SN | Marker Name 1 | Chromosomal Location | Allele Sizes (bp) |
|---|---|---|---|
| 1 | 73 | 1 | 194 |
| 2 | 8 | 2 | 234 |
| 3 | 2 | 2 | 127 |
| 4 | 4 | 3 | 238 |
| 5 | 3 | 3 | 160, 162 |
| 6 | 26 | 4 | 166 |
| 7 | 19 | 4 | 175 |
| 8 | 81 | 5 | 130, 132 |
| 9 | 34 | 6 | 188 |
| 10 | 30 | 7 | 186, 192 |
| 11 | 24 | 8 | 249, 253 |
| 12 | 59 | 9 | 143, 146, 180 2 |
| 13 | 62 | 9 | 177 |
| 14 | 1 | 10 | 96 |
| 15 | 55 | 10 | 210, 218 |
| 16 | 36 | 11 | 234 |
| 17 | 67 | 11 | 165 |
| 18 | 13 | 12 | 121, 135 |
| 19 | 35 | 13 | 197, 203 |
| 20 | 42 | 13 | 127 |
| 21 | 70 | 14 | 175, 179 |
| 22 | 61 | 15 | 128 |
| 23 | 79 | 15 | 172 |
| 24 | 90 | 16 | 174 |
| 25 | 69 | 16 | 139 |
| 26 | 78 | 17 | 147, 151 |
| 27 | 15 | 18 | 232 |
| 28 | 16 | 18 | 247, 251 |
| 29 | 75 | 19 | 144, 184 |
| 30 | 96 | 20 | 210, 212 |
| 31 | 91 | 20 | 205, 211 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nanda, I.; Steinlein, C.; Haaf, T.; Buhl, E.M.; Grimm, D.G.; Friedman, S.L.; Meurer, S.K.; Schröder, S.K.; Weiskirchen, R. Genetic Characterization of Rat Hepatic Stellate Cell Line HSC-T6 for In Vitro Cell Line Authentication. Cells 2022, 11, 1783. https://doi.org/10.3390/cells11111783
Nanda I, Steinlein C, Haaf T, Buhl EM, Grimm DG, Friedman SL, Meurer SK, Schröder SK, Weiskirchen R. Genetic Characterization of Rat Hepatic Stellate Cell Line HSC-T6 for In Vitro Cell Line Authentication. Cells. 2022; 11(11):1783. https://doi.org/10.3390/cells11111783
Chicago/Turabian StyleNanda, Indrajit, Claus Steinlein, Thomas Haaf, Eva M. Buhl, Domink G. Grimm, Scott L. Friedman, Steffen K. Meurer, Sarah K. Schröder, and Ralf Weiskirchen. 2022. "Genetic Characterization of Rat Hepatic Stellate Cell Line HSC-T6 for In Vitro Cell Line Authentication" Cells 11, no. 11: 1783. https://doi.org/10.3390/cells11111783








