Fission Impossible (?)—New Insights into Disorders of Peroxisome Dynamics
Abstract
:1. Introduction
2. Growth and Division of Peroxisomes
2.1. Membrane Deformation and Elongation
2.2. Membrane Constriction and Assembly of Fission Sites
2.3. Membrane Scission
2.4. Pulling Forces, ER Contacts and Lipid Transfer
2.5. Multiple Roles of PO Membrane Dynamics
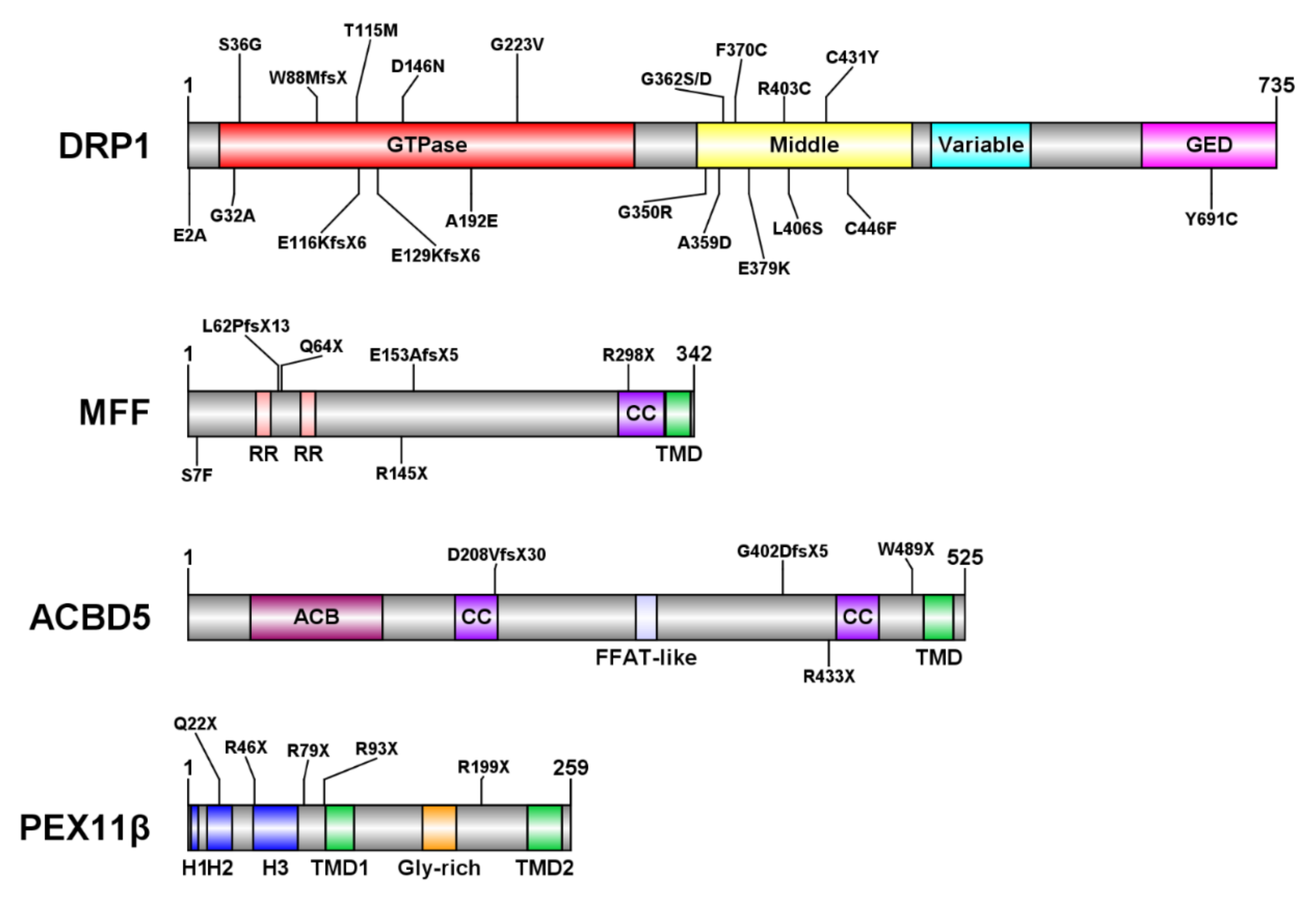
3. Disorders of Peroxisome Dynamics and Plasticity
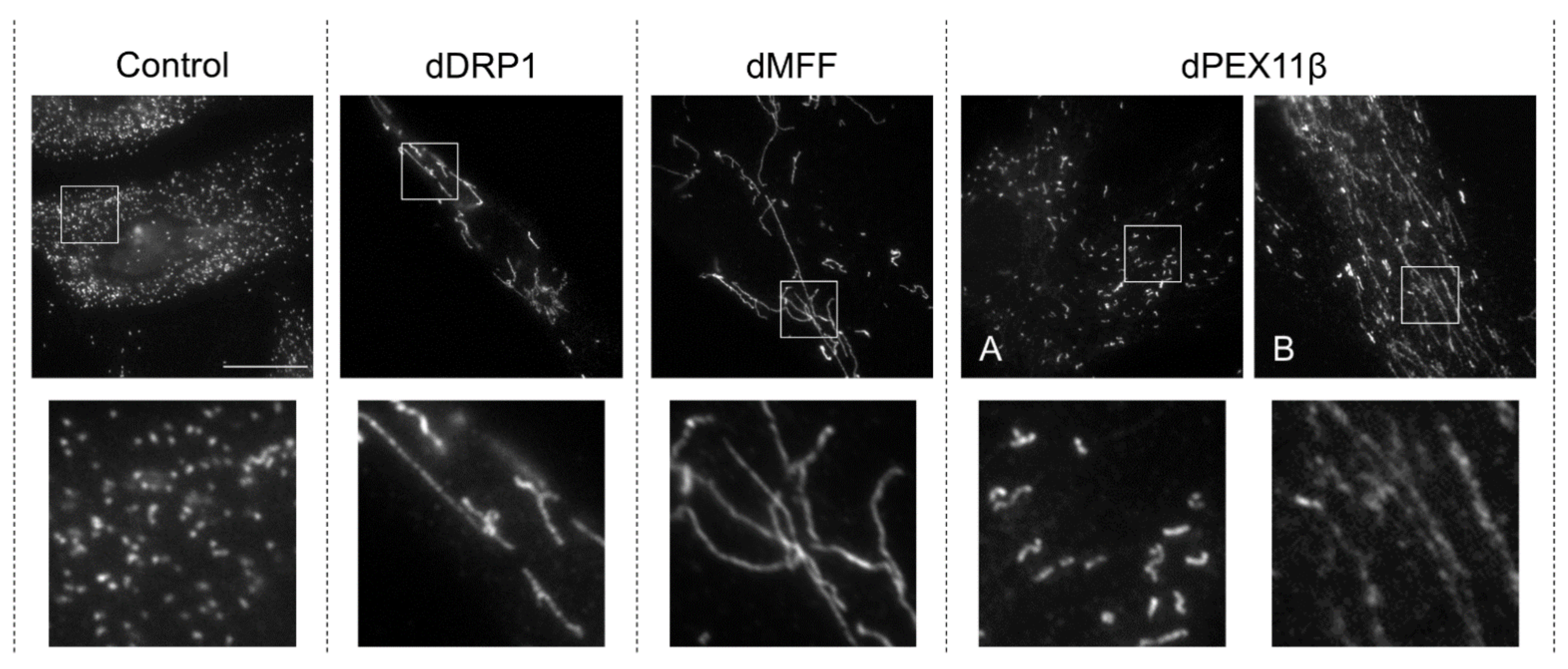
3.1. DRP1 Deficiency
3.2. Mitochondrial Fission Factor (MFF) Deficiency
3.3. ACBD5 Deficiency
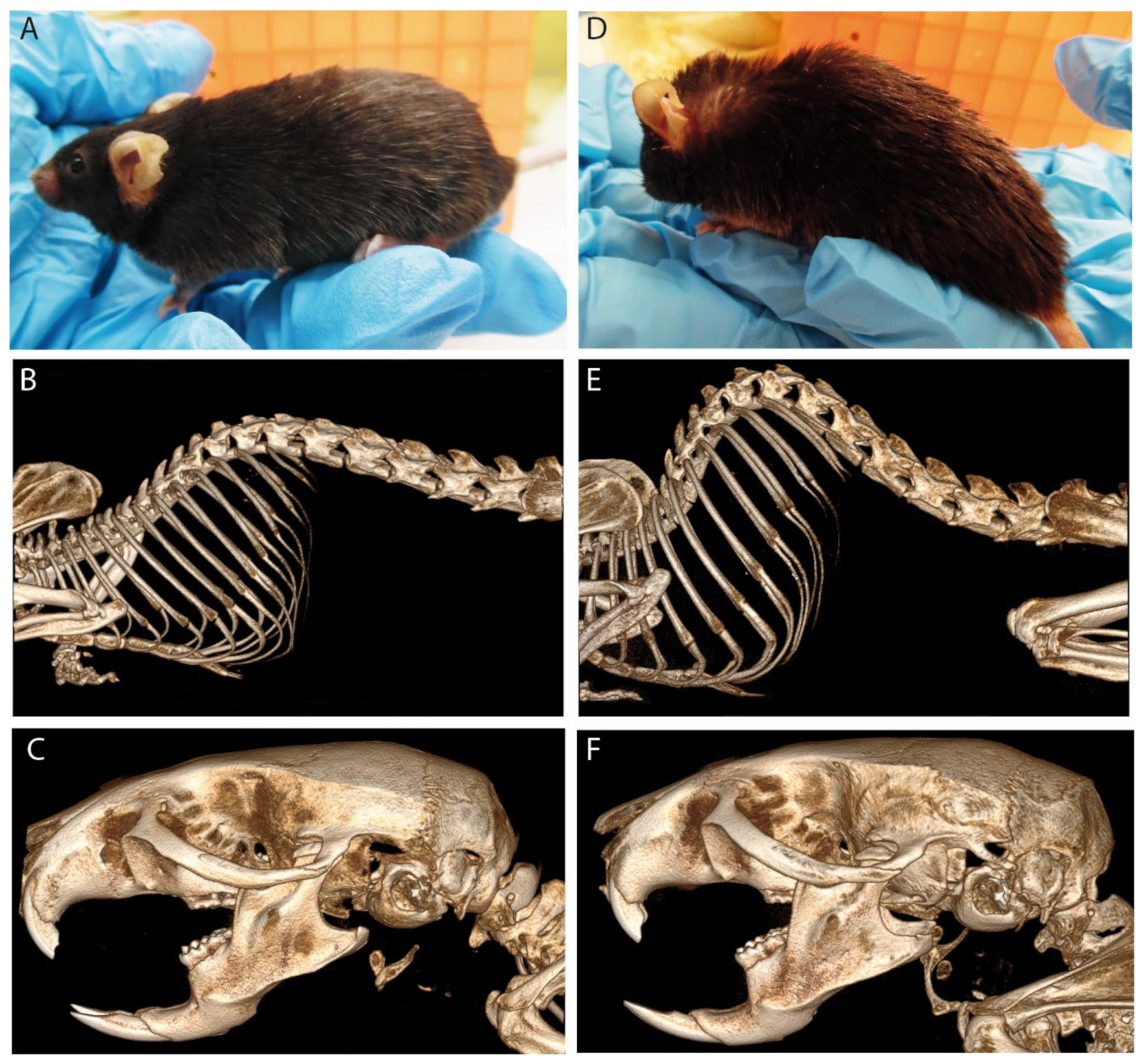
3.4. PEX11β Deficiency
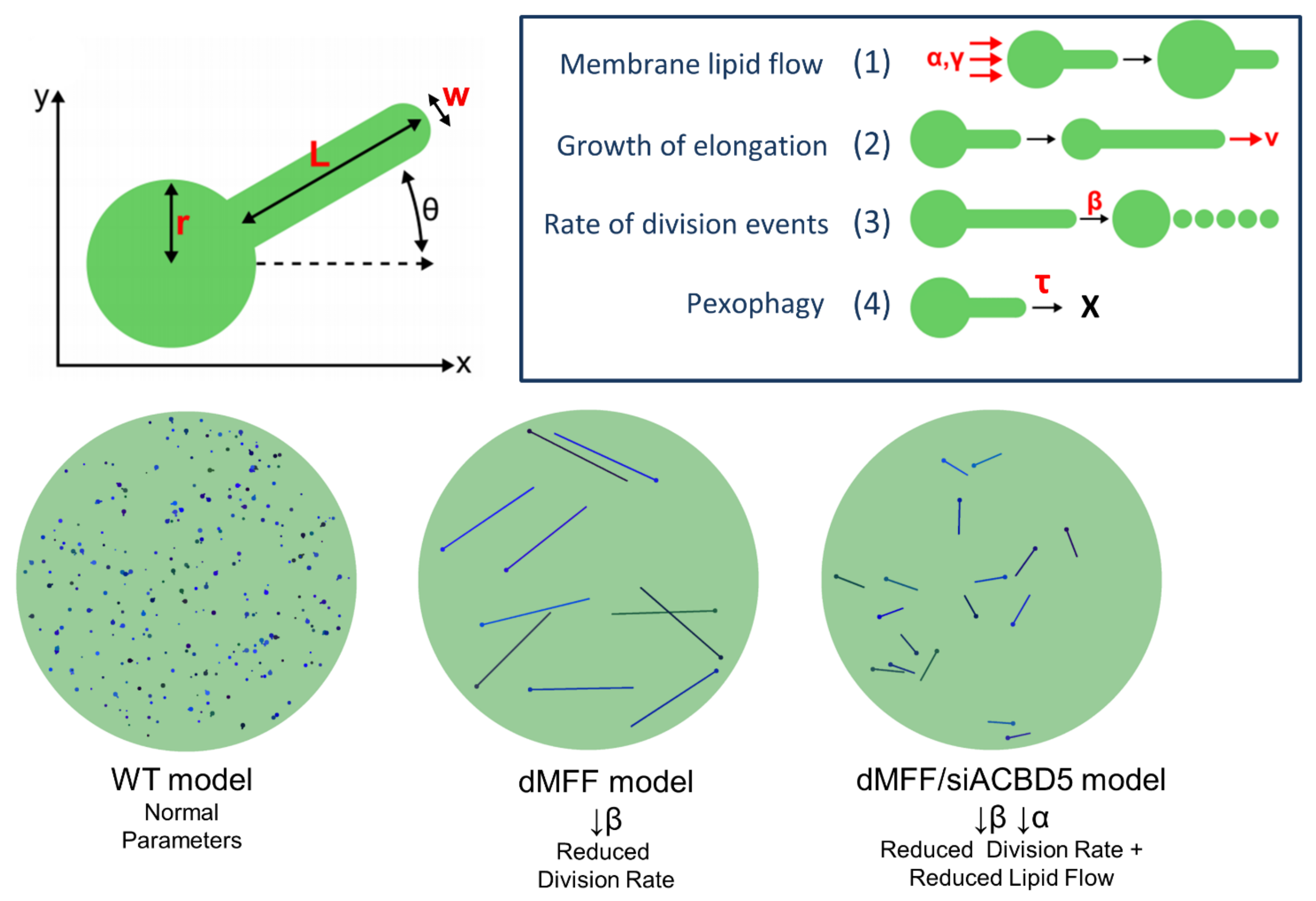
4. Modelling and Prediction of Peroxisomal Dynamics
5. Potential Strategies for Treatment
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Overview of DRP1, MFF, PEX11β, and ACBD5-Deficient Patients
| Clinical Features | Genotype | Mutation | Organelle Alterations * | Reference |
|---|---|---|---|---|
| Microcephaly, abnormal brain development, optic atrophy hypoplasia, lactic acidosis, | c.1184C>A (p.A395D) De novo heterozygous | Missense mutation in middle domain | Defective fission of mitochondria and peroxisomes, slightly elevated VLCFA and lactate levels but no alterations to mitochondrial metabolism | [96] |
| (Mutant overexpression in HeLa/in vitro assays) Elongated mitochondria, reduced DRP1 levels at mitochondria, reduced formation of higher-order structures and compromised GTPase activity | [194] | |||
| Infantile encephalopathy, lactic acidosis, poor feeding, global developmental delay, hypotonia and status epilepticus | c.1048G>A (p.G350R) or c.1135G>A (p.E379K) De novo heterozygous | Missense mutation in middle domain | (Expression in Drosophila DRP1-deficient cells): (p.G350R): Increase in peroxisomal size and decreased number. Reduced mitochondrial size and defect in mitochondrial trafficking. (p.E379K): No notable alterations to peroxisomes or mitochondria | [195] |
| Refractory epilepsy with prolonged survival | c.1085G>A (p.G362D) De novo heterozygous | Missense mutation in middle domain | Hyperfusion of the mitochondrial network | [97] |
| Postnatal microcephaly, developmental delay and pain insensitivity | c.1084G>A (p.G362S) De novo heterozygous | Missense mutation in middle domain | Decreased respiratory chain complex IV activity. Impaired fission with elongated mitochondrial structure | [98] |
| Epileptic encephalopathy | c.1207C>T (p.R403C) De novo heterozygous | Missense mutation in middle domain | (Expression in mouse embryonic fibroblasts): Dominant-negative effect, with reduced DRP1 oligomerization and mitochondrial fission activity | [196] |
| Mild developmental delay and intellectual disability, paroxysmal dystonia, acute status epilepticus, progressive global cerebral atrophy | Normal mitochondrial and peroxisomal metabolism, elongated mitochondria | [99] | ||
| Hyperfused elongated mitochondria with abnormal cristae, reduced efficiency of oxidative phosphorylation, increased mitochondrial membrane potential, elongated peroxisomes | [105] | |||
| Encephalopathy in infancy | c.1217T>C (p.L406S) De novo heterozygous | Missense mutation in middle domain | Elongation of peroxisomes and mitochondria | [100] |
| Hypotonia and absent respiratory effort. Demyelination and reduction of the number of axons in the sural nerve | c.261dup (p.W88MfsX) + c.385_386del (p.E129KfsX6) compound heterozygous | Frame shift, truncation No detectable DRP1 protein | Giant mitochondria with abnormal cristae in hippocampal and Purkinje neurons; normal mitochondrial morphology in glial and non-neuronal cells | [102] |
| Slowly progressive infantile encephalopathy | c.106A>G (p.S36G) + c.346_347delGA (p.E116KfsX6) compound heterozygous | Missense mutation in GTPase domain | Elongated and constricted mitochondria, elongated peroxisomes with abnormal distribution | [101] |
| Isolated dominant optic atrophy | c.5A>C (p.E2A) or c.575C>A (p.A192E) heterozygous, dominantly inherited | Missense mutation in GTPase domain | Dominant negative effect with elongated mitochondrial network | [94] |
| Severe infantile parkinsonism, global encephalopathy, hypomyelination | c.1337G>T (p.C446F) De novo heterozygous | Missense mutation in middle domain | Elongated mitochondria, elongated and constricted peroxisomes. Reduced number of mitochondria and peroxisomes. Increased lactate levels | [95] |
| Hypotonia, developmental delay, seizures (p.C431Y), optic atrophy (p.G32A) | c.1292G>A (p.C431Y) or c.95G>C (p.G32A) De novo heterozygous | Missense mutation in middle domain (p.C431Y) or GTPase domain (p.G32A) | (p.G32A): Dominant negative effect with elongated, highly connected mitochondrial network. No alterations to peroxisomes | [107] |
| (p.G32A): Hyperfused elongated mitochondria with abnormal cristae, reduced efficiency of oxidative phosphorylation, increased mitochondrial membrane potential, elongated peroxisomes | [105] | |||
| Hypotonia, developmental delay, abnormal movement | c.305C>T (p.T115M) homozygous | Missense mutation in GTPase domain | Elongated mitochondria, reduced mtDNA content, reduced mitochondrial respiration, impaired cell growth, reduced DRP1 oligomerization Normal peroxisome morphology and VLCFA levels | [106] |
| Static encephalopathy, developmental delay, seizures, nystagmus | c.2072A>G, (p.Y691C) De novo heterozygous | Missense mutation in GTPase effector domain | Normal peroxisomal metabolism (Expression in Drosophila DRP1-deficient cells): Dominant negative effect with enlarged peroxisomes showing abnormal distribution, increased mitochondrial connectivity and abnormal distribution | [108] |
| Severe, early-onset epileptic encephalopathy, developmental delay, progressive cerebral atrophy | c.668G>T (p.G223V) or c.1109T>G (p.F370C) De novo heterozygous | Missense mutation in GTPase domain (p.G223V) or middle domain (p.F370C) | (p.G223V): mixed population of hyperfused and swollen/rod-shaped mitochondria (p.F370C): elongated mitochondria Both: uneven mitochondrial distribution, normal cellular respiration, mixed population of spherical and elongated peroxisomes | [103] |
| Psychomotor developmental delay, axonal sensory neuropathy leading to global hypotonia and severe ataxia | c.436G>A (p.D146N) De novo heterozygous | Missense mutation in GTPase domain | Dominant negative effect with hyperfused ‘balloon-like’ mitochondrial network, reduced mitochondrial turnover; elongation of peroxisomes, increase in peroxisomal mass | [104] |
| Clinical Features | Genotype | Mutation | Organelle Alterations * | Reference |
|---|---|---|---|---|
| Developmental delay, abnormal intensity on brain MRI of globus pallidus, motor and speech deficits, mild hypertonia, borderline microcephaly and pale optic disc | c.190C>T (p.Q64X) homozygous | Nonsense mutation, truncation before TMD | Elongated peroxisomes and mitochondria | [119] |
| Elongated peroxisomes, normal peroxisome metabolism, altered peroxisome redox environment | [55] | |||
| Developmental delay, peripheral neuropathy, optic atrophy, and Leigh-like encephalopathy | c.184dup 892C>T (p.L62PfsX13) (p.R298X) compound heterozygous | Nonsense mutation/frame shift, truncation before TMD | Elongation of peroxisomes and mitochondria, increased mitochondrial branching, normal mitochondrial and peroxisomal metabolism | [120] |
| c.453_454del (p.E153AfsX5) homozygous | Frame shift, truncation before TMD | |||
| Epileptic encephalopathy, neurological regression, severe intellectual disability, microcephaly, tetraparesis, optic atrophy | c.892C>T (p.R298X) homozygous | Nonsense mutation, truncation before TMD Low levels of MFF proteins and truncated forms detected | Elongation of peroxisomes, constricted elongated mitochondria, normal mitochondrial and peroxisomal metabolism | [121] |
| Developmental delay, neuroregression, microcephaly, optic atrophy, hearing defects | c.433C>T (p.R145X) | Nonsense mutation, truncation before TMD | Rounded, swollen mitochondria in lymphoblastoid cells | [122] |
| Spastic cerebral palsy, global developmental delay, bilateral thalamic lesions. (Homozygous mother and sibling asymptomatic) | c.19_20AG>TT (p.S7F) homozygous | Missense mutation in cytoplasmic N-terminus | Normal lactate levels in serum | [123] |
| Clinical Features | Genotype | Mutation | Organelle Alterations * | Reference |
|---|---|---|---|---|
| Mild intellectual disability, congenital cataracts, progressive hearing loss and polyneuropathy | c.64C>T (p.Q22X) homozygous | Nonsense mutation, truncation No detectable PEX11β protein | Enlarged and slightly elongated peroxisomes, normal peroxisomal metabolism, matrix protein import compromised in ~10% of cells. Partial rescue of peroxisome phenotype by PEX11γ expression | [130] |
| Mild intellectual disability, congenital cataracts, developmental delay, short stature, hearing defects | c.235C>T (p.R79X) homozygous or c.136C>T (p.R46X) homozygous or c.595C>T (p.R199X) heterozygous + ex1-3 del heterozygous | Nonsense mutation, truncation | Normal peroxisomal metabolism except (p.R79X): very low plasmalogen levels in blood plasma (p.R46X): elevated C24:0/C22:0 in blood plasma | [131] |
| Bilateral nystagmus, congenital cataracts with myopia, strabismus, high muscle tone, mental retardation | c.277C>T (p.R93X) homozygous | Nonsense mutation, truncation | Normal peroxisome metabolism | [156] |
| Clinical Features | Genotype | Mutation | Organelle Alterations * | Reference |
|---|---|---|---|---|
| Cone-rod dystrophy, spastic paraparesis, leukodystrophy | c.1205 + 1G>A (p.G402DfsX5) homozygous | Frame shift, truncation No detectable ACBD5 protein | Impaired β-oxidation of VLCFA in peroxisomes, normal plasmalogen synthesis | [137,143] |
| Retinal dystrophy, progressive leukodystrophy and microcephaly, ataxia, dysarthria, hypomyelination with diffuse abnormality in deep white matter | c.626-689_937-234delins936+1075_c.936+1230inv (p.D208VfsX30) homozygous | Exon 7/8 deletion No detectable ACBD5 protein | Normal presence of import-competent peroxisomes. Increased VLCFA levels, reduced C26:0 β-oxidation, reduced plasmalogen biosynthesis | [76,140,142] |
| Leukodystrophy, nystagmus, cone-rod dystrophy, spastic paraparesis, psychomotor developmental regression | c.1467G>A, (p.W489X) homozygous | Nonsense mutation, truncation | Elevated C26:0, C24:0/C22:0, and C26:0/C22:0 in plasma, decreased C22:0, C24:0 levels and phytanic acid in plasma | [138] |
| Leukodystrophy, retinal dystrophy, nystagmus | c.1297C>T, (p.R433X) homozygous | Nonsense mutation, truncation Very little detectable ACBD5 protein | Elevated C26:0 in plasma (patient 1), elevated C24:0 and C22:0 in plasma (patient 2) | [139] |
References
- Korobova, F.; Ramabhadran, V.; Higgs, H.N. An Actin-Dependent Step in Mitochondrial Fission Mediated by the ER-Associated Formin INF2. Science 2013, 339, 464–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, J.R.; Lackner, L.L.; West, M.; DiBenedetto, J.R.; Nunnari, J.; Voeltz, G.K. ER Tubules Mark Sites of Mitochondrial Division. Science 2011, 334, 358–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, S.C.; Uchiyama, L.F.; Nunnari, J. ER-Mitochondria Contacts Couple MtDNA Synthesis with Mitochondrial Division in Human Cells. Science 2016, 353, aaf5549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraus, F.; Ryan, M.T. The Constriction and Scission Machineries Involved in Mitochondrial Fission. J. Cell Sci. 2017, 130, 2953–2960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakrabarti, R.; Ji, W.-K.; Stan, R.V.; de Juan Sanz, J.; Ryan, T.A.; Higgs, H.N. INF2-Mediated Actin Polymerization at the ER Stimulates Mitochondrial Calcium Uptake, Inner Membrane Constriction, and Division. J. Cell Biol. 2018, 217, 251–268. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Chan, D.C. Mitochondrial Dynamics—Fusion, Fission, Movement, and Mitophagy—In Neurodegenerative Diseases. Hum. Mol. Genet. 2009, 18, R169–R176. [Google Scholar] [CrossRef]
- Yapa, N.M.B.; Lisnyak, V.; Reljic, B.; Ryan, M.T. Mitochondrial Dynamics in Health and Disease. FEBS Lett. 2021, 595, 1184–1204. [Google Scholar] [CrossRef]
- Schrader, M.; Costello, J.L.; Godinho, L.F.; Azadi, A.S.; Islinger, M. Proliferation and Fission of Peroxisomes—An Update. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 971–983. [Google Scholar] [CrossRef]
- Carmichael, R.E.; Schrader, M. Determinants of Peroxisome Membrane Dynamics. Front. Physiol. 2022, 13, 834411. [Google Scholar] [CrossRef]
- Schrader, M.; Yoon, Y. Mitochondria and Peroxisomes: Are the “Big Brother” and the “Little Sister” Closer than Assumed? BioEssays 2007, 29, 1105–1114. [Google Scholar] [CrossRef]
- Schrader, M.; Costello, J.; Godinho, L.F.; Islinger, M. Peroxisome-Mitochondria Interplay and Disease. J. Inherit. Metab. Dis. 2015, 38, 681–702. [Google Scholar] [CrossRef] [PubMed]
- Imoto, Y.; Itoh, K.; Fujiki, Y. Molecular Basis of Mitochondrial and Peroxisomal Division Machineries. Int. J. Mol. Sci. 2020, 21, 5452. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, C.; Grabiger, S.; Schwefel, D.; Faelber, K.; Rosenbaum, E.; Mears, J.; Rocks, O.; Daumke, O. Structural Insights into Oligomerization and Mitochondrial Remodelling of Dynamin 1-like Protein. EMBO J. 2013, 32, 1280–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalia, R.; Wang, R.Y.-R.; Yusuf, A.; Thomas, P.V.; Agard, D.A.; Shaw, J.M.; Frost, A. Structural Basis of Mitochondrial Receptor Binding and Constriction by DRP1. Nature 2018, 558, 401–405. [Google Scholar] [CrossRef]
- Delille, H.K.; Alves, R.; Schrader, M. Biogenesis of Peroxisomes and Mitochondria: Linked by Division. Histochem. Cell Biol. 2009, 131, 441–446. [Google Scholar] [CrossRef]
- Bonekamp, N.A.; Sampaio, P.; de Abreu, F.V.; Lüers, G.H.; Schrader, M. Transient Complex Interactions of Mammalian Peroxisomes without Exchange of Matrix or Membrane Marker Proteins. Traffic 2012, 13, 960–978. [Google Scholar] [CrossRef]
- Huybrechts, S.J.; Van Veldhoven, P.P.; Brees, C.; Mannaerts, G.P.; Los, G.V.; Fransen, M. Peroxisome Dynamics in Cultured Mammalian Cells. Traffic 2009, 10, 1722–1733. [Google Scholar] [CrossRef]
- Tilokani, L.; Nagashima, S.; Paupe, V.; Prudent, J. Mitochondrial Dynamics: Overview of Molecular Mechanisms. Essays Biochem. 2018, 62, 341–360. [Google Scholar] [CrossRef] [Green Version]
- Palmer, C.S.; Elgass, K.D.; Parton, R.G.; Osellame, L.D.; Stojanovski, D.; Ryan, M.T. Adaptor Proteins MiD49 and MiD51 Can Act Independently of Mff and Fis1 in Drp1 Recruitment and Are Specific for Mitochondrial Fission. J. Biol. Chem. 2013, 288, 27584–27593. [Google Scholar] [CrossRef] [Green Version]
- Sugiura, A.; Mattie, S.; Prudent, J.; McBride, H.M. Newly Born Peroxisomes Are a Hybrid of Mitochondrial and ER-Derived Pre-Peroxisomes. Nature 2017, 542, 251–254. [Google Scholar] [CrossRef]
- Aleksic, M.; Golic, I.; Kalezic, A.; Jankovic, A.; Korac, B.; Korac, A. Hypothyroidism Intensifies Both Canonic and the De Novo Pathway of Peroxisomal Biogenesis in Rat Brown Adipocytes in a Time-Dependent Manner. Cells 2021, 10, 2248. [Google Scholar] [CrossRef] [PubMed]
- Neuspiel, M.; Schauss, A.C.; Braschi, E.; Zunino, R.; Rippstein, P.; Rachubinski, R.A.; Andrade-Navarro, M.A.; McBride, H.M. Cargo-Selected Transport from the Mitochondria to Peroxisomes Is Mediated by Vesicular Carriers. Curr. Biol. 2008, 18, 102–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanders, R.J.A.; Waterham, H.R. Biochemistry of Mammalian Peroxisomes Revisited. Annu. Rev. Biochem. 2006, 75, 295–332. [Google Scholar] [CrossRef] [PubMed]
- Fransen, M.; Lismont, C.; Walton, P. The Peroxisome-Mitochondria Connection: How and Why? Int. J. Mol. Sci. 2017, 18, 1126. [Google Scholar] [CrossRef]
- Costello, J.L.; Passmore, J.B.; Islinger, M.; Schrader, M. Multi-Localized Proteins: The Peroxisome-Mitochondria Connection. In Subcellular Biochemistry; Springer: New York, NY, USA, 2018; Volume 89, pp. 383–415. [Google Scholar]
- Shai, N.; Yifrach, E.; van Roermund, C.W.T.; Cohen, N.; Bibi, C.; IJlst, L.; Cavellini, L.; Meurisse, J.; Schuster, R.; Zada, L.; et al. Systematic Mapping of Contact Sites Reveals Tethers and a Function for the Peroxisome-Mitochondria Contact. Nat. Commun. 2018, 9, 1761. [Google Scholar] [CrossRef]
- Kustatscher, G.; Grabowski, P.; Schrader, T.A.; Passmore, J.B.; Schrader, M.; Rappsilber, J. Co-Regulation Map of the Human Proteome Enables Identification of Protein Functions. Nat. Biotechnol. 2019, 37, 1361–1371. [Google Scholar] [CrossRef]
- Silva, B.S.C.; DiGiovanni, L.; Kumar, R.; Carmichael, R.E.; Kim, P.K.; Schrader, M. Maintaining Social Contacts: The Physiological Relevance of Organelle Interactions. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118800. [Google Scholar] [CrossRef]
- Huber, N.; Guimaraes, S.; Schrader, M.; Suter, U.; Niemann, A. Charcot-Marie-Tooth Disease-Associated Mutants of GDAP1 Dissociate Its Roles in Peroxisomal and Mitochondrial Fission. EMBO Rep. 2013, 14, 545–552. [Google Scholar] [CrossRef] [Green Version]
- Dixit, E.; Boulant, S.; Zhang, Y.; Lee, A.S.Y.Y.; Odendall, C.; Shum, B.; Hacohen, N.; Chen, Z.J.; Whelan, S.P.; Fransen, M.; et al. Peroxisomes Are Signaling Platforms for Antiviral Innate Immunity. Cell 2010, 141, 668–681. [Google Scholar] [CrossRef] [Green Version]
- Castro, I.G.; Richards, D.M.; Metz, J.; Costello, J.L.; Passmore, J.B.; Schrader, T.A.; Gouveia, A.; Ribeiro, D.; Schrader, M. A Role for Mitochondrial Rho GTPase 1 (MIRO1) in Motility and Membrane Dynamics of Peroxisomes. Traffic 2018, 19, 229–242. [Google Scholar] [CrossRef] [Green Version]
- Okumoto, K.; Ono, T.; Toyama, R.; Shimomura, A.; Nagata, A.; Fujiki, Y. New Splicing Variants of Mitochondrial Rho GTPase-1 (Miro1) Transport Peroxisomes. J. Cell Biol. 2018, 217, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Covill-Cooke, C.; Toncheva, V.S.; Drew, J.; Birsa, N.; López-Doménech, G.; Kittler, J.T. Peroxisomal Fission Is Modulated by the Mitochondrial Rho-GTPases, Miro1 and Miro2. EMBO Rep. 2020, 21, e49865. [Google Scholar] [CrossRef] [PubMed]
- Borgese, N.; Coy-Vergara, J.; Colombo, S.F.; Schwappach, B. The Ways of Tails: The GET Pathway and More. Protein J. 2019, 38, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Pieuchot, L.; Loh, R.A.; Yang, J.; Kari, T.M.A.; Wong, J.Y.; Jedd, G. Hydrophobic Handoff for Direct Delivery of Peroxisome Tail-Anchored Proteins. Nat. Commun. 2014, 5, 5790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costello, J.L.; Castro, I.G.; Camões, F.; Schrader, T.A.; McNeall, D.; Yang, J.; Giannopoulou, E.-A.; Gomes, S.; Pogenberg, V.; Bonekamp, N.A.; et al. Predicting the Targeting of Tail-Anchored Proteins to Subcellular Compartments in Mammalian Cells. J. Cell Sci. 2017, 130, 1675–1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yagita, Y.; Hiromasa, T.; Fujiki, Y. Tail-Anchored PEX26 Targets Peroxisomes via a PEX19-Dependent and TRC40-Independent Class I Pathway. J. Cell Biol. 2013, 200, 651–666. [Google Scholar] [CrossRef] [Green Version]
- Lazarow, P.B.; Fujiki, Y. Biogenesis of Peroxisomes. Annu. Rev. Cell Biol. 1985, 1, 489–530. [Google Scholar] [CrossRef]
- Schrader, M.; Fahimi, H.D. Growth and Division of Peroxisomes. Int. Rev. Cytol. 2006, 255, 237–290. [Google Scholar]
- Delille, H.K.; Agricola, B.; Guimaraes, S.C.; Borta, H.; Lüers, G.H.; Fransen, M.; Schrader, M. Pex11pbeta-Mediated Growth and Division of Mammalian Peroxisomes Follows a Maturation Pathway. J. Cell Sci. 2010, 123, 2750–2762. [Google Scholar] [CrossRef] [Green Version]
- Bonekamp, N.A.; Grille, S.; Cardoso, M.J.; Almeida, M.; Aroso, M.; Gomes, S.; Magalhaes, A.C.; Ribeiro, D.; Islinger, M.; Schrader, M. Self-Interaction of Human Pex11pbeta during Peroxisomal Growth and Division. PLoS ONE 2013, 8, e53424. [Google Scholar] [CrossRef] [Green Version]
- Koch, J.; Brocard, C. PEX11 Proteins Attract Mff and Human Fis1 to Coordinate Peroxisomal Fission. J. Cell Sci. 2012, 125, 3813–3826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opaliński, Ł.; Kiel, J.A.K.W.; Williams, C.; Veenhuis, M.; van der Klei, I.J. Membrane Curvature during Peroxisome Fission Requires Pex11. EMBO J. 2011, 30, 5–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, J.; Thomas, A.S.; Grabietz, T.; Landgraf, C.; Volkmer, R.; Marrink, S.J.; Williams, C.; Melo, M.N. The N-Terminal Amphipathic Helix of Pex11p Self-Interacts to Induce Membrane Remodelling during Peroxisome Fission. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1292–1300. [Google Scholar] [CrossRef] [PubMed]
- Itoyama, A.; Honsho, M.; Abe, Y.; Moser, A.; Yoshida, Y.; Fujiki, Y.; Gould, S.J. Docosahexaenoic Acid Mediates Peroxisomal Elongation, a Prerequisite for Peroxisome Division. J. Cell Sci. 2012, 125, 589–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, S.; Tanaka, A.; Fujiki, Y. Fis1, DLP1, and Pex11p Coordinately Regulate Peroxisome Morphogenesis. Exp. Cell Res. 2007, 313, 1675–1686. [Google Scholar] [CrossRef]
- Koch, A.; Schneider, G.; Lüers, G.H.; Schrader, M. Peroxisome Elongation and Constriction but Not Fission Can Occur Independently of Dynamin-like Protein 1. J. Cell Sci. 2004, 117, 3995–4006. [Google Scholar] [CrossRef] [Green Version]
- Schrader, M.; Reuber, B.E.; Morrell, J.C.; Jimenez-Sanchez, G.; Obie, C.; Stroh, T.A.; Valle, D.; Schroer, T.A.; Gould, S.J. Expression of PEX11beta Mediates Peroxisome Proliferation in the Absence of Extracellular Stimuli. J. Biol. Chem. 1998, 273, 29607–29614. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, Y.; Niwa, H.; Honsho, M.; Itoyama, A.; Fujiki, Y. Pex11 Mediates Peroxisomal Proliferation by Promoting Deformation of the Lipid Membrane. Biol. Open 2015, 4, 710–721. [Google Scholar] [CrossRef] [Green Version]
- Feng, Q.; Kornmann, B. Mechanical Forces on Cellular Organelles. J. Cell Sci. 2018, 131, jcs218479. [Google Scholar] [CrossRef] [Green Version]
- Mahecic, D.; Carlini, L.; Kleele, T.; Colom, A.; Goujon, A.; Matile, S.; Roux, A.; Manley, S. Mitochondrial Membrane Tension Governs Fission. Cell Rep. 2021, 35, 108947. [Google Scholar] [CrossRef]
- Costello, J.L.; Castro, I.G.; Hacker, C.; Schrader, T.A.; Metz, J.; Zeuschner, D.; Azadi, A.S.; Godinho, L.F.; Costina, V.; Findeisen, P.; et al. ACBD5 and VAPB Mediate Membrane Associations between Peroxisomes and the ER. J. Cell Biol. 2017, 216, 331–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, R.; Cheng, D.; Coyaud, É.; Freeman, S.; Di Pietro, E.; Wang, Y.; Vissa, A.; Yip, C.M.; Fairn, G.D.; Braverman, N.; et al. VAPs and ACBD5 Tether Peroxisomes to the ER for Peroxisome Maintenance and Lipid Homeostasis. J. Cell Biol. 2017, 216, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Bishop, A.; Kamoshita, M.; Passmore, J.B.; Hacker, C.; Schrader, T.A.; Waterham, H.R.; Costello, J.L.; Schrader, M. Fluorescent Tools to Analyse Peroxisome-ER Interactions in Mammalian Cells. Contact 2019, 2, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passmore, J.B.; Carmichael, R.E.; Schrader, T.A.; Godinho, L.F.; Ferdinandusse, S.; Lismont, C.; Wang, Y.; Hacker, C.; Islinger, M.; Fransen, M.; et al. Mitochondrial Fission Factor (MFF) Is a Critical Regulator of Peroxisome Maturation. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118709. [Google Scholar] [CrossRef]
- Koch, A.; Yoon, Y.; Bonekamp, N.A.; Mcniven, M.A.; Schrader, M. A Role for Fis1 in Both Mitochondrial and Peroxisomal Fission in Mammalian Cells. Mol. Biol. Cell 2005, 16, 5077–5086. [Google Scholar] [CrossRef]
- Itoyama, A.; Michiyuki, S.; Honsho, M.; Yamamoto, T.; Moser, A.; Yoshida, Y.; Fujiki, Y. Mff Functions with Pex11p and DLP1 in Peroxisomal Fission. Biol. Open 2013, 2, 998–1006. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.; Opalinski, L.; Landgraf, C.; Costello, J.; Schrader, M.; Krikken, A.M.; Knoops, K.; Kram, A.M.; Volkmer, R.; van der Klei, I.J. The Membrane Remodeling Protein Pex11p Activates the GTPase Dnm1p during Peroxisomal Fission. Proc. Natl. Acad. Sci. USA 2015, 112, 6377–6382. [Google Scholar] [CrossRef] [Green Version]
- Imoto, Y.; Abe, Y.; Honsho, M.; Okumoto, K.; Ohnuma, M.; Kuroiwa, H.; Kuroiwa, T.; Fujiki, Y. Onsite GTP Fuelling via DYNAMO1 Drives Division of Mitochondria and Peroxisomes. Nat. Commun. 2018, 9, 4634. [Google Scholar] [CrossRef]
- Honsho, M.; Abe, Y.; Imoto, Y.; Chang, Z.-F.; Mandel, H.; Falik-Zaccai, T.C.; Fujiki, Y. Mammalian Homologue NME3 of DYNAMO1 Regulates Peroxisome Division. Int. J. Mol. Sci. 2020, 21, 8040. [Google Scholar] [CrossRef]
- Otera, H.; Wang, C.; Cleland, M.M.; Setoguchi, K.; Yokota, S.; Youle, R.J.; Mihara, K. Mff Is an Essential Factor for Mitochondrial Recruitment of Drp1 during Mitochondrial Fission in Mammalian Cells. J. Cell Biol. 2010, 191, 1141–1158. [Google Scholar] [CrossRef] [Green Version]
- Ihenacho, U.K.; Meacham, K.A.; Harwig, M.C.; Widlansky, M.E.; Hill, R.B. Mitochondrial Fission Protein 1: Emerging Roles in Organellar Form and Function in Health and Disease. Front. Endocrinol. 2021, 12, 660095. [Google Scholar] [CrossRef] [PubMed]
- Schrader, T.A.; Carmichael, R.E.; Islinger, M.; Costello, J.L.; Hacker, C.; Bonekamp, N.A.; Weishaupt, J.H.; Andersen, P.M.; Schrader, M. PEX11β and FIS1 Cooperate in Peroxisome Division Independent of Mitochondrial Fission Factor. J. Cell Sci. 2022, 259924. [Google Scholar] [CrossRef] [PubMed]
- Schrader, M.; Thiemann, M.; Fahimi, H.D. Peroxisomal Motility and Interaction with Microtubules. Microsc. Res. Tech. 2003, 61, 171–178. [Google Scholar] [CrossRef]
- Neuhaus, A.; Eggeling, C.; Erdmann, R.; Schliebs, W. Why Do Peroxisomes Associate with the Cytoskeleton? Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 1019–1026. [Google Scholar] [CrossRef]
- Covill-Cooke, C.; Toncheva, V.S.; Kittler, J.T. Regulation of Peroxisomal Trafficking and Distribution. Cell. Mol. Life Sci. 2021, 78, 1929–1941. [Google Scholar] [CrossRef]
- Zinsmaier, K.E. Mitochondrial Miro GTPases Coordinate Mitochondrial and Peroxisomal Dynamics. Small GTPases 2021, 12, 372–398. [Google Scholar] [CrossRef]
- Schrader, M.; Burkhardt, J.K.; Baumgart, E.; Lüers, G.H.; Spring, H.; Volkl, A.; Fahimi, H.D.; Völkl, A.; Fahimi, H.D. Interaction of Microtubules with Peroxisomes. Tubular and Spherical Peroxisomes in HepG2 Cells and Their Alteractions Induced by Microtubule-Active Drugs. Eur. J. Cell Biol. 1996, 69, 24–35. [Google Scholar]
- Passmore, J.B.; Pinho, S.; Gomez-Lazaro, M.; Schrader, M. The Respiratory Chain Inhibitor Rotenone Affects Peroxisomal Dynamics via Its Microtubule-Destabilising Activity. Histochem. Cell Biol. 2017, 148, 331–341. [Google Scholar] [CrossRef] [Green Version]
- Dodt, G.; Braverman, N.; Wong, C.; Moser, A.; Moser, H.W.; Watkins, P.; Valle, D.; Gould, S.J. Mutations in the PTS1 Receptor Gene, PXR1, Define Complementation Group 2 of the Peroxisome Biogenesis Disorders. Nat. Genet. 1995, 9, 115–125. [Google Scholar] [CrossRef]
- Miyata, N.; Fujiki, Y. Shuttling Mechanism of Peroxisome Targeting Signal Type 1 Receptor Pex5: ATP-Independent Import and ATP-Dependent Export. Mol. Cell. Biol. 2005, 25, 10822–10832. [Google Scholar] [CrossRef] [Green Version]
- Steinberg, S.J.; Raymond, G.V.; Braverman, N.E.; Moser, A.B. Zellweger Spectrum Disorder. In GeneReviews® [Internet]; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Mirzaa, G.M., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2003. [Google Scholar]
- Waterham, H.R.; Ferdinandusse, S.; Wanders, R.J.A. Human Disorders of Peroxisome Metabolism and Biogenesis. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.J.; Imanaka, T.; Shio, H.; Small, G.M.; Lazarow, P.B. Peroxisomal Membrane Ghosts in Zellweger Syndrome—Aberrant Organelle Assembly. Science 1988, 239, 1536–1538. [Google Scholar] [CrossRef] [PubMed]
- Schrader, M.; Kamoshita, M.; Islinger, M. Organelle Interplay—Peroxisome Interactions in Health and Disease. J. Inherit. Metab. Dis. 2020, 43, 71–89. [Google Scholar] [CrossRef] [PubMed]
- Ferdinandusse, S.; Falkenberg, K.D.; Koster, J.; Mooyer, P.A.; Jones, R.; van Roermund, C.W.T.; Pizzino, A.; Schrader, M.; Wanders, R.J.A.; Vanderver, A.; et al. ACBD5 Deficiency Causes a Defect in Peroxisomal Very Long-Chain Fatty Acid Metabolism. J. Med. Genet. 2017, 54, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldwin, H.A.; Wang, C.; Kanfer, G.; Shah, H.V.; Velayos-Baeza, A.; Dulovic-Mahlow, M.; Brüggemann, N.; Anding, A.; Baehrecke, E.H.; Maric, D.; et al. VPS13D Promotes Peroxisome Biogenesis. J. Cell Biol. 2021, 220, e202001188. [Google Scholar] [CrossRef] [PubMed]
- Guillén-Samander, A.; Leonzino, M.; Hanna, M.G.; Tang, N.; Shen, H.; De Camilli, P. VPS13D Bridges the ER to Mitochondria and Peroxisomes via Miro. J. Cell Biol. 2021, 220, e202010004. [Google Scholar] [CrossRef]
- Kumar, N.; Leonzino, M.; Hancock-Cerutti, W.; Horenkamp, F.A.; Li, P.Q.; Lees, J.A.; Wheeler, H.; Reinisch, K.M.; De Camilli, P. VPS13A and VPS13C Are Lipid Transport Proteins Differentially Localized at ER Contact Sites. J. Cell Biol. 2018, 217, 3625–3639. [Google Scholar] [CrossRef] [Green Version]
- Mathur, J.; Mammone, A.; Barton, K.A. Organelle Extensions in Plant Cells. J. Integr. Plant Biol. 2012, 54, 851–867. [Google Scholar] [CrossRef]
- Jaipargas, E.-A.; Mathur, N.; Bou Daher, F.; Wasteneys, G.O.; Mathur, J. High Light Intensity Leads to Increased Peroxule-Mitochondria Interactions in Plants. Front. Cell Dev. Biol. 2016, 4, 6. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Sanz-Fernández, M.; Hu, J.; Sandalio, L.M. Peroxisomes Extend Peroxules in a Fast Response to Stress via a Reactive Oxygen Species-Mediated Induction of the Peroxin PEX11a. Plant Physiol. 2016, 171, 1665–1674. [Google Scholar] [CrossRef] [Green Version]
- Koch, A.; Thiemann, M.; Grabenbauer, M.; Yoon, Y.; McNiven, M.A.; Schrader, M. Dynamin-like Protein 1 Is Involved in Peroxisomal Fission. J. Biol. Chem. 2003, 278, 8597–8605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smirnova, E.; Shurland, D.-L.; Ryazantsev, S.N.; van der Bliek, A.M. A Human Dynamin-Related Protein Controls the Distribution of Mitochondria. J. Cell Biol. 1998, 143, 351–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamerkar, S.C.; Kraus, F.; Sharpe, A.J.; Pucadyil, T.J.; Ryan, M.T. Dynamin-Related Protein 1 Has Membrane Constricting and Severing Abilities Sufficient for Mitochondrial and Peroxisomal Fission. Nat. Commun. 2018, 9, 5239. [Google Scholar] [CrossRef] [PubMed]
- Sesaki, H.; Adachi, Y.; Kageyama, Y.; Itoh, K.; Iijima, M. In Vivo Functions of Drp1: Lessons Learned from Yeast Genetics and Mouse Knockouts. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 1179–1185. [Google Scholar] [CrossRef] [Green Version]
- Francy, C.A.; Alvarez, F.J.D.; Zhou, L.; Ramachandran, R.; Mears, J.A. The Mechanoenzymatic Core of Dynamin-Related Protein 1 Comprises the Minimal Machinery Required for Membrane Constriction. J. Biol. Chem. 2015, 290, 11692–11703. [Google Scholar] [CrossRef] [Green Version]
- Wasiak, S.; Zunino, R.; McBride, H.M. Bax/Bak Promote Sumoylation of DRP1 and Its Stable Association with Mitochondria during Apoptotic Cell Death. J. Cell Biol. 2007, 177, 439–450. [Google Scholar] [CrossRef] [Green Version]
- Cribbs, J.T.; Strack, S. Reversible Phosphorylation of Drp1 by Cyclic AMP-Dependent Protein Kinase and Calcineurin Regulates Mitochondrial Fission and Cell Death. EMBO Rep. 2007, 8, 939–944. [Google Scholar] [CrossRef] [Green Version]
- Taguchi, N.; Ishihara, N.; Jofuku, A.; Oka, T.; Mihara, K. Mitotic Phosphorylation of Dynamin-Related GTPase Drp1 Participates in Mitochondrial Fission. J. Biol. Chem. 2007, 282, 11521–11529. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Hildick, K.L.; Luo, J.; Dearden, L.; Wilkinson, K.A.; Henley, J.M. SENP3-Mediated DeSUMOylation of Dynamin-Related Protein 1 Promotes Cell Death Following Ischaemia. EMBO J. 2013, 32, 1514–1528. [Google Scholar] [CrossRef]
- Banerjee, R.; Mukherjee, A.; Nagotu, S. Mitochondrial Dynamics and Its Impact on Human Health and Diseases: Inside the DRP1 Blackbox. J. Mol. Med. 2022, 100, 1–21. [Google Scholar] [CrossRef]
- Parikh, S. The Neurologic Manifestations of Mitochondrial Disease. Dev. Disabil. Res. Rev. 2010, 16, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Gerber, S.; Charif, M.; Chevrollier, A.; Chaumette, T.; Angebault, C.; Kane, M.S.; Paris, A.; Alban, J.; Quiles, M.; Delettre, C.; et al. Mutations in DNM1L, as in OPA1, Result in Dominant Optic Atrophy despite Opposite Effects on Mitochondrial Fusion and Fission. Brain 2017, 140, 2586–2596. [Google Scholar] [CrossRef] [PubMed]
- Díez, H.; Cortès-Saladelafont, E.; Ormazábal, A.; Marmiese, A.F.; Armstrong, J.; Matalonga, L.; Bravo, M.; Briones, P.; Emperador, S.; Montoya, J.; et al. Severe Infantile Parkinsonism Because of a de Novo Mutation on DLP1 Mitochondrial-Peroxisomal Protein. Mov. Disord. 2017, 32, 1108–1110. [Google Scholar] [CrossRef] [PubMed]
- Waterham, H.R.; Koster, J.; van Roermund, C.W.T.; Mooyer, P.A.W.; Wanders, R.J.A.; Leonard, J.V. A Lethal Defect of Mitochondrial and Peroxisomal Fission. N. Engl. J. Med. 2007, 356, 1736–1741. [Google Scholar] [CrossRef]
- Vanstone, J.R.; Smith, A.M.; McBride, S.; Naas, T.; Holcik, M.; Antoun, G.; Harper, M.-E.; Michaud, J.; Sell, E.; Chakraborty, P.; et al. DNM1L-Related Mitochondrial Fission Defect Presenting as Refractory Epilepsy. Eur. J. Hum. Genet. 2016, 24, 1084–1088. [Google Scholar] [CrossRef] [Green Version]
- Sheffer, R.; Douiev, L.; Edvardson, S.; Shaag, A.; Tamimi, K.; Soiferman, D.; Meiner, V.; Saada, A. Postnatal Microcephaly and Pain Insensitivity Due to a de Novo Heterozygous DNM1L Mutation Causing Impaired Mitochondrial Fission and Function. Am. J. Med. Genet. A 2016, 170, 1603–1607. [Google Scholar] [CrossRef]
- Ryan, C.S.; Fine, A.L.; Cohen, A.L.; Schiltz, B.M.; Renaud, D.L.; Wirrell, E.C.; Patterson, M.C.; Boczek, N.J.; Liu, R.; Babovic-Vuksanovic, D.; et al. De Novo DNM1L Variant in a Teenager with Progressive Paroxysmal Dystonia and Lethal Super-Refractory Myoclonic Status Epilepticus. J. Child Neurol. 2018, 33, 651–658. [Google Scholar] [CrossRef]
- Zaha, K.; Matsumoto, H.; Itoh, M.; Saitsu, H.; Kato, K.; Kato, M.; Ogata, S.; Murayama, K.; Kishita, Y.; Mizuno, Y.; et al. DNM1L-Related Encephalopathy in Infancy with Leigh Syndrome-like Phenotype and Suppression-Burst. Clin. Genet. 2016, 90, 472–474. [Google Scholar] [CrossRef]
- Nasca, A.; Legati, A.; Baruffini, E.; Nolli, C.; Moroni, I.; Ardissone, A.; Goffrini, P.; Ghezzi, D. Biallelic Mutations in DNM1L Are Associated with a Slowly Progressive Infantile Encephalopathy. Hum. Mutat. 2016, 37, 898–903. [Google Scholar] [CrossRef] [Green Version]
- Yoon, G.; Malam, Z.; Paton, T.; Marshall, C.R.; Hyatt, E.; Ivakine, Z.; Scherer, S.W.; Lee, K.-S.; Hawkins, C.; Cohn, R.D.; et al. Lethal Disorder of Mitochondrial Fission Caused by Mutations in DNM1L. J. Pediatr. 2016, 171, 313–316.e2. [Google Scholar] [CrossRef]
- Verrigni, D.; Di Nottia, M.; Ardissone, A.; Baruffini, E.; Nasca, A.; Legati, A.; Bellacchio, E.; Fagiolari, G.; Martinelli, D.; Fusco, L.; et al. Clinical-Genetic Features and Peculiar Muscle Histopathology in Infantile DNM1L-Related Mitochondrial Epileptic Encephalopathy. Hum. Mutat. 2019, 40, 601–618. [Google Scholar] [CrossRef] [PubMed]
- Longo, F.; Benedetti, S.; Zambon, A.A.; Sora, M.G.N.; Di Resta, C.; De Ritis, D.; Quattrini, A.; Maltecca, F.; Ferrari, M.; Previtali, S.C. Impaired Turnover of Hyperfused Mitochondria in Severe Axonal Neuropathy Due to a Novel DRP1 Mutation. Hum. Mol. Genet. 2020, 29, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Robertson, G.L.; Riffle, S.N.; Patel, M.; Marshall, A.; Beasley, H.; Lopez, E.G.; Shao, J.; Vue, Z.; Hinton, A.; Mears, J.A.; et al. DRP1-Mediated Mitochondrial Fission Is Essential to Maintain Cristae Morphology and Bioenergetics. bioRxiv 2022. [Google Scholar] [CrossRef]
- Hogarth, K.A.; Costford, S.R.; Yoon, G.; Sondheimer, N.; Maynes, J.T. DNM1L Variant Alters Baseline Mitochondrial Function and Response to Stress in a Patient with Severe Neurological Dysfunction. Biochem. Genet. 2018, 56, 56–77. [Google Scholar] [CrossRef] [PubMed]
- Whitley, B.N.; Lam, C.; Cui, H.; Haude, K.; Bai, R.; Escobar, L.; Hamilton, A.; Brady, L.; Tarnopolsky, M.A.; Dengle, L.; et al. Aberrant Drp1-Mediated Mitochondrial Division Presents in Humans with Variable Outcomes. Hum. Mol. Genet. 2018, 27, 3710–3719. [Google Scholar] [CrossRef] [PubMed]
- Assia Batzir, N.; Bhagwat, P.K.; Eble, T.N.; Liu, P.; Eng, C.M.; Elsea, S.H.; Robak, L.A.; Scaglia, F.; Goldman, A.M.; Dhar, S.U.; et al. De Novo Missense Variant in the GTPase Effector Domain (GED) of DNM1L Leads to Static Encephalopathy and Seizures. Cold Spring Harb. Mol. Case Stud. 2019, 5, 3673. [Google Scholar] [CrossRef] [Green Version]
- Wakabayashi, J.; Zhang, Z.; Wakabayashi, N.; Tamura, Y.; Fukaya, M.; Kensler, T.W.; Iijima, M.; Sesaki, H. The Dynamin-Related GTPase Drp1 Is Required for Embryonic and Brain Development in Mice. J. Cell Biol. 2009, 186, 805–816. [Google Scholar] [CrossRef] [Green Version]
- Gandre-Babbe, S.; van der Bliek, A.M. The Novel Tail-Anchored Membrane Protein Mff Controls Mitochondrial and Peroxisomal Fission in Mammalian Cells. Mol. Biol. Cell 2008, 19, 2402–2412. [Google Scholar] [CrossRef] [Green Version]
- Clinton, R.W.; Francy, C.A.; Ramachandran, R.; Qi, X.; Mears, J.A. Dynamin-Related Protein 1 Oligomerization in Solution Impairs Functional Interactions with Membrane-Anchored Mitochondrial Fission Factor. J. Biol. Chem. 2016, 291, 478–492. [Google Scholar] [CrossRef] [Green Version]
- Koirala, S.; Guo, Q.; Kalia, R.; Bui, H.T.; Eckert, D.M.; Frost, A.; Shaw, J.M. Interchangeable Adaptors Regulate Mitochondrial Dynamin Assembly for Membrane Scission. Proc. Natl. Acad. Sci. USA 2013, 110, E1342–E1351. [Google Scholar] [CrossRef] [Green Version]
- Toyama, E.Q.; Herzig, S.; Courchet, J.; Lewis, T.L.; Losón, O.C.; Hellberg, K.; Young, N.P.; Chen, H.; Polleux, F.; Chan, D.C.; et al. AMP-Activated Protein Kinase Mediates Mitochondrial Fission in Response to Energy Stress. Science 2016, 351, 275–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, A.; Kage, F.; Higgs, H.N. Mff Oligomerization Is Required for Drp1 Activation and Synergy with Actin Filaments during Mitochondrial Division. Mol. Biol. Cell 2021, 32, ar5. [Google Scholar] [CrossRef] [PubMed]
- Losón, O.C.; Song, Z.; Chen, H.; Chan, D.C. Fis1, Mff, MiD49, and MiD51 Mediate Drp1 Recruitment in Mitochondrial Fission. Mol. Biol. Cell 2013, 24, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, P.J.; Francy, C.A.; Stepanyants, N.; Lehman, L.; Baglio, A.; Mears, J.A.; Qi, X.; Ramachandran, R. Distinct Splice Variants of Dynamin-Related Protein 1 Differentially Utilize Mitochondrial Fission Factor as an Effector of Cooperative GTPase Activity. J. Biol. Chem. 2016, 291, 493–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osellame, L.D.; Singh, A.P.; Stroud, D.A.; Palmer, C.S.; Stojanovski, D.; Ramachandran, R.; Ryan, M.T. Cooperative and Independent Roles of the Drp1 Adaptors Mff, MiD49 and MiD51 in Mitochondrial Fission. J. Cell Sci. 2016, 129, 2170–2181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, W.-K.; Chakrabarti, R.; Fan, X.; Schoenfeld, L.; Strack, S.; Higgs, H.N. Receptor-Mediated Drp1 Oligomerization on Endoplasmic Reticulum. J. Cell Biol. 2017, 216, 4123–4139. [Google Scholar] [CrossRef] [Green Version]
- Shamseldin, H.E.; Alshammari, M.; Al-Sheddi, T.; Salih, M.A.; Alkhalidi, H.; Kentab, A.; Repetto, G.M.; Hashem, M.; Alkuraya, F.S. Genomic Analysis of Mitochondrial Diseases in a Consanguineous Population Reveals Novel Candidate Disease Genes. J. Med. Genet. 2012, 49, 234–241. [Google Scholar] [CrossRef]
- Koch, J.; Feichtinger, R.G.; Freisinger, P.; Pies, M.; Schrödl, F.; Iuso, A.; Sperl, W.; Mayr, J.A.; Prokisch, H.; Haack, T.B. Disturbed Mitochondrial and Peroxisomal Dynamics Due to Loss of MFF Causes Leigh-like Encephalopathy, Optic Atrophy and Peripheral Neuropathy. J. Med. Genet. 2016, 53, 270–278. [Google Scholar] [CrossRef]
- Nasca, A.; Nardecchia, F.; Commone, A.; Semeraro, M.; Legati, A.; Garavaglia, B.; Ghezzi, D.; Leuzzi, V. Clinical and Biochemical Features in a Patient with Mitochondrial Fission Factor Gene Alteration. Front. Genet. 2018, 9, 625. [Google Scholar] [CrossRef] [Green Version]
- Panda, I.; Ahmad, I.; Sagar, S.; Zahra, S.; Shamim, U.; Sharma, S.; Faruq, M. Encephalopathy Due to Defective Mitochondrial and Peroxisomal Fission 2 Caused by a Novel MFF Gene Mutation in a Young Child. Clin. Genet. 2020, 97, 933–937. [Google Scholar] [CrossRef]
- Sharma, C.; Saini, A.; Gothwal, M.; Jhirwal, M.; Patwa, P. Mitochondrial Fission Factor Gene Mutation: A Dilemma for Prenatal Diagnosis. Int. J. Appl. Basic Med. Res. 2021, 11, 114–116. [Google Scholar] [CrossRef] [PubMed]
- Lewis, T.L.J.; Kwon, S.-K.; Lee, A.; Shaw, R.; Polleux, F. MFF-Dependent Mitochondrial Fission Regulates Presynaptic Release and Axon Branching by Limiting Axonal Mitochondria Size. Nat. Commun. 2018, 9, 5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khacho, M.; Slack, R.S. Mitochondrial Dynamics in the Regulation of Neurogenesis: From Development to the Adult Brain. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2018, 247, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Onoue, K.; Jofuku, A.; Ban-Ishihara, R.; Ishihara, T.; Maeda, M.; Koshiba, T.; Itoh, T.; Fukuda, M.; Otera, H.; Oka, T.; et al. Fis1 Acts as a Mitochondrial Recruitment Factor for TBC1D15 That Is Involved in Regulation of Mitochondrial Morphology. J. Cell Sci. 2013, 126, 176–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burté, F.; Carelli, V.; Chinnery, P.F.; Yu-Wai-Man, P. Disturbed Mitochondrial Dynamics and Neurodegenerative Disorders. Nat. Rev. Neurol. 2015, 11, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Z.-H.; Cai, Q. Mitochondrial Transport in Neurons: Impact on Synaptic Homeostasis and Neurodegeneration. Nat. Rev. Neurosci. 2012, 13, 77–93. [Google Scholar] [CrossRef] [Green Version]
- Berger, J.; Dorninger, F.; Forss-Petter, S.; Kunze, M. Peroxisomes in Brain Development and Function. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 934–955. [Google Scholar] [CrossRef] [Green Version]
- Ebberink, M.S.; Koster, J.; Visser, G.; van Spronsen, F.; Stolte-Dijkstra, I.; Smit, G.P.A.; Fock, J.M.; Kemp, S.; Wanders, R.J.A.; Waterham, H.R. A Novel Defect of Peroxisome Division Due to a Homozygous Non-Sense Mutation in the PEX11beta Gene. J. Med. Genet. 2012, 49, 307–313. [Google Scholar] [CrossRef]
- Taylor, R.L.; Handley, M.T.; Waller, S.; Campbell, C.; Urquhart, J.; Meynert, A.M.; Ellingford, J.M.; Donnelly, D.; Wilcox, G.; Lloyd, I.C.; et al. Novel PEX11B Mutations Extend the Peroxisome Biogenesis Disorder 14B Phenotypic Spectrum and Underscore Congenital Cataract as an Early Feature. Investig. Ophthalmol. Vis. Sci. 2017, 58, 594–603. [Google Scholar] [CrossRef] [Green Version]
- Nuebel, E.; Morgan, J.T.; Fogarty, S.; Winter, J.M.; Lettlova, S.; Berg, J.A.; Chen, Y.-C.; Kidwell, C.U.; Maschek, J.A.; Clowers, K.J.; et al. The Biochemical Basis of Mitochondrial Dysfunction in Zellweger Spectrum Disorder. EMBO Rep. 2021, 22, e51991. [Google Scholar] [CrossRef]
- Lismont, C.; Koster, J.; Provost, S.; Baes, M.; Van Veldhoven, P.P.; Waterham, H.R.; Fransen, M. Deciphering the Potential Involvement of PXMP2 and PEX11B in Hydrogen Peroxide Permeation across the Peroxisomal Membrane Reveals a Role for PEX11B in Protein Sorting. Biochim. Biophys. Acta Biomembr. 2019, 1861, 182991. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ren, S.; Clish, C.; Jain, M.; Mootha, V.; McCaffery, J.M.; Chan, D.C. Titration of Mitochondrial Fusion Rescues Mff-Deficient Cardiomyopathy. J. Cell Biol. 2015, 211, 795–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islinger, M.; Costello, J.L.; Kors, S.; Soupene, E.; Levine, T.P.; Kuypers, F.A.; Schrader, M. The Diversity of ACBD Proteins—From Lipid Binding to Protein Modulators and Organelle Tethers. Biochim. Biophys. Acta. Mol. Cell Res. 2020, 1867, 118675. [Google Scholar] [CrossRef] [PubMed]
- Costello, J.L.; Castro, I.G.; Schrader, T.A.; Islinger, M.; Schrader, M. Peroxisomal ACBD4 Interacts with VAPB and Promotes ER-Peroxisome Associations. Cell Cycle 2017, 16, 1039–1045. [Google Scholar] [CrossRef] [PubMed]
- Abu-Safieh, L.; Alrashed, M.; Anazi, S.; Alkuraya, H.; Khan, A.O.; Al-Owain, M.; Al-Zahrani, J.; Al-Abdi, L.; Hashem, M.; Al-Tarimi, S.; et al. Autozygome-Guided Exome Sequencing in Retinal Dystrophy Patients Reveals Pathogenetic Mutations and Novel Candidate Disease Genes. Genome Res. 2013, 23, 236–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartlett, M.; Nasiri, N.; Pressman, R.; Bademci, G.; Forghani, I. First Reported Adult Patient with Retinal Dystrophy and Leukodystrophy Caused by a Novel ACBD5 Variant: A Case Report and Review of Literature. Am. J. Med. Genet. A 2021, 185, 1236–1241. [Google Scholar] [CrossRef]
- Gorukmez, O.; Havalı, C.; Gorukmez, O.; Dorum, S. Newly Defined Peroxisomal Disease with Novel ACBD5 Mutation. J. Pediatr. Endocrinol. Metab. 2022, 35, 11–18. [Google Scholar] [CrossRef]
- Helman, G.; Lajoie, B.R.; Crawford, J.; Takanohashi, A.; Walkiewicz, M.; Dolzhenko, E.; Gross, A.M.; Gainullin, V.G.; Bent, S.J.; Jenkinson, E.M.; et al. Genome Sequencing in Persistently Unsolved White Matter Disorders. Ann. Clin. Transl. Neurol. 2020, 7, 144–152. [Google Scholar] [CrossRef]
- Moser, A.B.; Kreiter, N.; Bezman, L.; Lu, S.; Raymond, G.V.; Naidu, S.; Moser, H.W. Plasma Very Long Chain Fatty Acids in 3000 Peroxisome Disease Patients and 29,000 Controls. Ann. Neurol. 1999, 45, 100–110. [Google Scholar] [CrossRef]
- Herzog, K.; Pras-Raves, M.L.; Ferdinandusse, S.; Vervaart, M.A.T.; Luyf, A.C.M.; van Kampen, A.H.C.; Wanders, R.J.A.; Waterham, H.R.; Vaz, F.M. Functional Characterisation of Peroxisomal β-Oxidation Disorders in Fibroblasts Using Lipidomics. J. Inherit. Metab. Dis. 2017, 41, 479–487. [Google Scholar] [CrossRef] [Green Version]
- Yagita, Y.; Shinohara, K.; Abe, Y.; Nakagawa, K.; Al-Owain, M.; Alkuraya, F.S.; Fujiki, Y. Deficiency of a Retinal Dystrophy Protein, Acyl-CoA Binding Domain-Containing 5 (ACBD5), Impairs Peroxisomal β-Oxidation of Very-Long-Chain Fatty Acids. J. Biol. Chem. 2017, 292, 691–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darwisch, W.; von Spangenberg, M.; Lehmann, J.; Singin, Ö.; Deubert, G.; Kühl, S.; Roos, J.; Horstmann, H.; Körber, C.; Hoppe, S.; et al. Cerebellar and Hepatic Alterations in ACBD5-Deficient Mice Are Associated with Unexpected, Distinct Alterations in Cellular Lipid Homeostasis. Commun. Biol. 2020, 3, 713. [Google Scholar] [CrossRef] [PubMed]
- Yeboah, G.K.; Lobanova, E.S.; Brush, R.S.; Agbaga, M.-P. Very Long Chain Fatty Acid-Containing Lipids: A Decade of Novel Insights from the Study of ELOVL4. J. Lipid Res. 2021, 62, 100030. [Google Scholar] [CrossRef] [PubMed]
- Wanders, R.J.A.; Waterham, H.R.; Ferdinandusse, S. Peroxisomes and Their Central Role in Metabolic Interaction Networks in Humans. Subcell. Biochem. 2018, 89, 345–365. [Google Scholar] [CrossRef] [PubMed]
- Tvrdik, P.; Westerberg, R.; Silve, S.; Asadi, A.; Jakobsson, A.; Cannon, B.; Loison, G.; Jacobsson, A. Role of a New Mammalian Gene Family in the Biosynthesis of Very Long Chain Fatty Acids and Sphingolipids. J. Cell Biol. 2000, 149, 707–718. [Google Scholar] [CrossRef] [Green Version]
- Agbaga, M.-P.; Brush, R.S.; Mandal, M.N.A.; Henry, K.; Elliott, M.H.; Anderson, R.E. Role of Stargardt-3 Macular Dystrophy Protein (ELOVL4) in the Biosynthesis of Very Long Chain Fatty Acids. Proc. Natl. Acad. Sci. USA 2008, 105, 12843–12848. [Google Scholar] [CrossRef] [Green Version]
- Kassmann, C.M.; Quintes, S.; Rietdorf, J.; Möbius, W.; Sereda, M.W.; Nientiedt, T.; Saher, G.; Baes, M.; Nave, K.-A. A Role for Myelin-Associated Peroxisomes in Maintaining Paranodal Loops and Axonal Integrity. FEBS Lett. 2011, 585, 2205–2211. [Google Scholar] [CrossRef]
- Beard, M.E.; Novikoff, A.B. Distribution of Peroxisomes (Microbodies) in the Nephron of the Rat: A Cytochemical Study. J. Cell Biol. 1969, 42, 501–518. [Google Scholar] [CrossRef]
- Robison, W.G.J.; Kuwabara, T. Vitamin A Storage and Peroxisomes in Retinal Pigment Epithelium and Liver. Investig. Ophthalmol. Vis. Sci. 1977, 16, 1110–1117. [Google Scholar]
- Wang, Y.; Metz, J.; Costello, J.L.; Passmore, J.; Schrader, M.; Schultz, C.; Islinger, M. Intracellular Redistribution of Neuronal Peroxisomes in Response to ACBD5 Expression. PLoS ONE 2018, 13, e0209507. [Google Scholar] [CrossRef]
- Ilacqua, N.; Anastasia, I.; Raimondi, A.; Lemieux, P.; de Aguiar Vallim, T.Q.; Toth, K.; Koonin, E.V.; Pellegrini, L. A Three-Organelle Complex Made by WrappER Contacts with Peroxisome and Mitochondria Responds to Liver Lipid Flux Changes. J. Cell Sci. 2021, 135, jcs259091. [Google Scholar] [CrossRef] [PubMed]
- Mattiazzi Ušaj, M.; Brložnik, M.; Kaferle, P.; Žitnik, M.; Wolinski, H.; Leitner, F.; Kohlwein, S.D.; Zupan, B.; Petrovič, U. Genome-Wide Localization Study of Yeast Pex11 Identifies Peroxisome–Mitochondria Interactions through the ERMES Complex. J. Mol. Biol. 2015, 427, 2072–2087. [Google Scholar] [CrossRef] [PubMed]
- Mindthoff, S.; Grunau, S.; Steinfort, L.L.; Girzalsky, W.; Hiltunen, J.K.; Erdmann, R.; Antonenkov, V.D. Peroxisomal Pex11 Is a Pore-Forming Protein Homologous to TRPM Channels. Biochim. Biophys. Acta 2016, 1863, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhang, L.; Li, Y.; Gao, J.; Yu, H.; Guo, Y.; Jia, L. Variant Analysis of PEX11B Gene from a Family with Peroxisome Biogenesis Disorder 14B by Whole Exome Sequencing. Mol. Genet. Genomic Med. 2020, 8, e1042. [Google Scholar] [CrossRef] [Green Version]
- Azadi, A.S.; Carmichael, R.E.; Kovacs, W.J.; Koster, J.; Kors, S.; Waterham, H.R.; Schrader, M. A Functional SMAD2/3 Binding Site in the PEX11β Promoter Identifies a Role for TGFβ in Peroxisome Proliferation in Humans. Front. Cell Dev. Biol. 2020, 8, 577637. [Google Scholar] [CrossRef]
- Li, X.; Baumgart, E.; Morrell, J.C.; Jimenez-Sanchez, G.; Valle, D.; Gould, S.J. PEX11beta Deficiency Is Lethal and Impairs Neuronal Migration but Does Not Abrogate Peroxisome Function. Mol. Cell. Biol. 2002, 22, 4358–4365. [Google Scholar] [CrossRef] [Green Version]
- Ahlemeyer, B.; Gottwald, M.; Baumgart-Vogt, E. Deletion of a Single Allele of the Pex11β Gene Is Sufficient to Cause Oxidative Stress, Delayed Differentiation and Neuronal Death in Mouse Brain. Dis. Model. Mech. 2012, 5, 125–140. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Baumgart, E.; Dong, G.-X.; Morrell, J.C.; Jimenez-Sanchez, G.; Valle, D.; Smith, K.D.; Gould, S.J. PEX11alpha Is Required for Peroxisome Proliferation in Response to 4-Phenylbutyrate but Is Dispensable for Peroxisome Proliferator-Activated Receptor Alpha-Mediated Peroxisome Proliferation. Mol. Cell. Biol. 2002, 22, 8226–8240. [Google Scholar] [CrossRef] [Green Version]
- Weng, H.; Ji, X.; Naito, Y.; Endo, K.; Ma, X.; Takahashi, R.; Shen, C.; Hirokawa, G.; Fukushima, Y.; Iwai, N. Pex11α Deficiency Impairs Peroxisome Elongation and Division and Contributes to Nonalcoholic Fatty Liver in Mice. Am. J. Physiol. Metab. 2012, 304, E187–E196. [Google Scholar] [CrossRef] [Green Version]
- Garikapati, V.; Colasante, C.; Baumgart-Vogt, E.; Spengler, B. Sequential Lipidomic, Metabolomic, and Proteomic Analyses of Serum, Liver, and Heart Tissue Specimens from Peroxisomal Biogenesis Factor 11α Knockout Mice. Anal. Bioanal. Chem. 2022, 414, 2235–2250. [Google Scholar] [CrossRef]
- Chen, C.; Wang, H.; Chen, B.; Chen, D.; Lu, C.; Li, H.; Qian, Y.; Tan, Y.; Weng, H.; Cai, L. Pex11a Deficiency Causes Dyslipidaemia and Obesity in Mice. J. Cell. Mol. Med. 2019, 23, 2020–2031. [Google Scholar] [CrossRef] [PubMed]
- Delmaghani, S.; Defourny, J.; Aghaie, A.; Beurg, M.; Dulon, D.; Thelen, N.; Perfettini, I.; Zelles, T.; Aller, M.; Meyer, A.; et al. Hypervulnerability to Sound Exposure through Impaired Adaptive Proliferation of Peroxisomes. Cell 2015, 163, 894–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrader, M.; Castro, I.; Fahimi, H.D.; Islinger, M. Peroxisome Morphology in Pathologies. In Molecular Machines Involved in Peroxisome Biogenesis and Maintenance; Brocard, C., Hartig, A., Eds.; Springer: Vienna, Austria, 2014; pp. 125–151. ISBN 3709117879. [Google Scholar]
- Mast, F.D.; Aitchison, J.D. Characterization of Peroxisomal Regulation Networks. In Proteomics of Peroxisomes; Subcellular Biochemistry; Del Río, L.A., Schrader, M., Eds.; Springer: Singapore, 2018; Volume 89, pp. 367–382. ISBN 978-981-13-2233-4. [Google Scholar]
- Hong, F.; Pan, S.; Guo, Y.; Xu, P.; Zhai, Y. PPARs as Nuclear Receptors for Nutrient and Energy Metabolism. Molecules 2019, 24, 2545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, D.; Castro, I.; Fahimi, H.D.; Schrader, M. Peroxisome Morphology in Pathology. Histol. Histopathol. 2012, 27, 661–676. [Google Scholar] [PubMed]
- He, A.; Dean, J.M.; Lodhi, I.J. Peroxisomes as Cellular Adaptors to Metabolic and Environmental Stress. Trends Cell Biol. 2021, 31, 656–670. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Fahimi, H.D. Biogenesis of Peroxisomes in Regenerating Rat Liver. I. Sequential Changes of Catalase and Urate Oxidase Detected by Ultrastructural Cytochemistry. Eur. J. Cell Biol. 1987, 43, 293–300. [Google Scholar]
- Fringes, B.; Gorgas, K.; Reith, A. Clofibrate Increases the Number of Peroxisomes and of Lamellar Bodies in Alveolar Cells Type II of the Rat Lung. Eur. J. Cell Biol. 1988, 46, 136–143. [Google Scholar]
- Roels, F.; Espeel, M.; Pauwels, M.; De Craemer, D.; Egberts, H.J.; van der Spek, P. Different Types of Peroxisomes in Human Duodenal Epithelium. Gut 1991, 32, 858–865. [Google Scholar] [CrossRef] [Green Version]
- Schrader, M.; Baumgart, E.; Volkl, A.; Fahimi, H.D. Heterogeneity of Peroxisomes in Human Hepatoblastoma Cell Line HepG2. Evidence of Distinct Subpopulations. Eur. J. Cell Biol. 1994, 64, 281–294. [Google Scholar]
- Schrader, M.; King, S.J.; Stroh, T.A.; Schroer, T.A. Real Time Imaging Reveals a Peroxisomal Reticulum in Living Cells. J. Cell Sci. 2000, 113, 3663–3671. [Google Scholar] [CrossRef]
- Litwin, J.A.; Bilińska, B. Morphological Heterogeneity of Peroxisomes in Cultured Mouse Leydig Cells. Folia Histochem. Cytobiol. 1995, 33, 255–258. [Google Scholar] [PubMed]
- Schrader, M.; Krieglstein, K.; Fahimi, H.D. Tubular Peroxisomes in HepG2 Cells: Selective Induction by Growth Factors and Arachidonic Acid. Eur. J. Cell Biol. 1998, 75, 87–96. [Google Scholar] [CrossRef]
- Schrader, M.; Wodopia, R.; Fahimi, H.D. Induction of Tubular Peroxisomes by UV Irradiation and Reactive Oxygen Species in HepG2 Cells. J. Histochem. Cytochem. 1999, 47, 1141–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kors, S.; Hacker, C.; Bolton, C.; Maier, R.; Reimann, L.; Kitchener, E.J.A.; Warscheid, B.; Costello, J.L.; Schrader, M. Regulating Peroxisome-ER Contacts via the ACBD5-VAPB Tether by FFAT Motif Phosphorylation and GSK3β. J. Cell Biol. 2022, 221, e202003143. [Google Scholar] [CrossRef]
- Waterham, H.R.; Ebberink, M.S. Genetics and Molecular Basis of Human Peroxisome Biogenesis Disorders. Biochim. Biophys. Acta 2012, 1822, 1430–1441. [Google Scholar] [CrossRef] [Green Version]
- Parikh, S.; Saneto, R.; Falk, M.J.; Anselm, I.; Cohen, B.H.; Haas, R.; The Mitochondrial Medicine Society. A Modern Approach to the Treatment of Mitochondrial Disease. Curr. Treat. Options Neurol. 2009, 11, 414–430. [Google Scholar] [CrossRef] [Green Version]
- Douiev, L.; Sheffer, R.; Horvath, G.; Saada, A. Bezafibrate Improves Mitochondrial Fission and Function in DNM1L-Deficient Patient Cells. Cells 2020, 9, 301. [Google Scholar] [CrossRef] [Green Version]
- Jamwal, S.; Blackburn, J.K.; Elsworth, J.D. PPARγ/PGC1α Signaling as a Potential Therapeutic Target for Mitochondrial Biogenesis in Neurodegenerative Disorders. Pharmacol. Ther. 2021, 219, 107705. [Google Scholar] [CrossRef]
- Argyriou, C.; Polosa, A.; Song, J.Y.; Omri, S.; Steele, B.; Cécyre, B.; McDougald, D.S.; Di Pietro, E.; Bouchard, J.-F.; Bennett, J.; et al. AAV-Mediated PEX1 Gene Augmentation Improves Visual Function in the PEX1-Gly844Asp Mouse Model for Mild Zellweger Spectrum Disorder. Mol. Ther. Methods Clin. Dev. 2021, 23, 225–240. [Google Scholar] [CrossRef]
- Anderson, G.R.; Wardell, S.E.; Cakir, M.; Yip, C.; Ahn, Y.; Ali, M.; Yllanes, A.P.; Chao, C.A.; McDonnell, D.P.; Wood, K.C. Dysregulation of Mitochondrial Dynamics Proteins Are a Targetable Feature of Human Tumors. Nat. Commun. 2018, 9, 1677. [Google Scholar] [CrossRef] [Green Version]
- Itoh, K.; Nakamura, K.; Iijima, M.; Sesaki, H. Mitochondrial Dynamics in Neurodegeneration. Trends Cell Biol. 2013, 23, 64–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, J.; Wei, X.; Zhi, X.; Wang, X.; Meng, D. Drp1-Dependent Mitochondrial Fission in Cardiovascular Disease. Acta Pharmacol. Sin. 2021, 42, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Manczak, M.; Calkins, M.J.; Reddy, P.H. Impaired Mitochondrial Dynamics and Abnormal Interaction of Amyloid Beta with Mitochondrial Protein Drp1 in Neurons from Patients with Alzheimer’s Disease: Implications for Neuronal Damage. Hum. Mol. Genet. 2011, 20, 2495–2509. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.U.; Saw, N.L.; Shamloo, M.; Mochly-Rosen, D. Drp1/Fis1 Interaction Mediates Mitochondrial Dysfunction, Bioenergetic Failure and Cognitive Decline in Alzheimer’s Disease. Oncotarget 2018, 9, 6128–6143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bordt, E.A.; Clerc, P.; Roelofs, B.A.; Saladino, A.J.; Tretter, L.; Adam-Vizi, V.; Cherok, E.; Khalil, A.; Yadava, N.; Ge, S.X.; et al. The Putative Drp1 Inhibitor Mdivi-1 Is a Reversible Mitochondrial Complex I Inhibitor That Modulates Reactive Oxygen Species. Dev. Cell 2017, 40, 583–594.e6. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Yin, J.; Ma, X.; Zhao, F.; Siedlak, S.L.; Wang, Z.; Torres, S.; Fujioka, H.; Xu, Y.; Perry, G.; et al. Inhibition of Mitochondrial Fragmentation Protects against Alzheimer’s Disease in Rodent Model. Hum. Mol. Genet. 2017, 26, 4118–4131. [Google Scholar] [CrossRef] [Green Version]
- Rappold, P.M.; Cui, M.; Grima, J.C.; Fan, R.Z.; de Mesy-Bentley, K.L.; Chen, L.; Zhuang, X.; Bowers, W.J.; Tieu, K. Drp1 Inhibition Attenuates Neurotoxicity and Dopamine Release Deficits in Vivo. Nat. Commun. 2014, 5, 5244. [Google Scholar] [CrossRef]
- Disatnik, M.-H.; Ferreira, J.C.B.; Campos, J.C.; Gomes, K.S.; Dourado, P.M.M.; Qi, X.; Mochly-Rosen, D. Acute Inhibition of Excessive Mitochondrial Fission after Myocardial Infarction Prevents Long-Term Cardiac Dysfunction. J. Am. Heart Assoc. 2013, 2, e000461. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Cai, D.; Zhong, H.; Liu, F.; Jiang, Q.; Liang, J.; Li, P.; Song, Y.; Ji, A.; Jiao, W.; et al. Mitochondrial Dynamics and Biogenesis Indicators May Serve as Potential Biomarkers for Diagnosis of Myasthenia Gravis. Exp. Ther. Med. 2022, 23, 307. [Google Scholar] [CrossRef]
- Chang, C.-R.; Manlandro, C.M.; Arnoult, D.; Stadler, J.; Posey, A.E.; Hill, R.B.; Blackstone, C. A Lethal de Novo Mutation in the Middle Domain of the Dynamin-Related GTPase Drp1 Impairs Higher Order Assembly and Mitochondrial Division. J. Biol. Chem. 2010, 285, 32494–32503. [Google Scholar] [CrossRef] [Green Version]
- Chao, Y.-H.; Robak, L.A.; Xia, F.; Koenig, M.K.; Adesina, A.; Bacino, C.A.; Scaglia, F.; Bellen, H.J.; Wangler, M.F. Missense Variants in the Middle Domain of DNM1L in Cases of Infantile Encephalopathy Alter Peroxisomes and Mitochondria When Assayed in Drosophila. Hum. Mol. Genet. 2016, 25, 1846–1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahrner, J.A.; Liu, R.; Perry, M.S.; Klein, J.; Chan, D.C. A Novel de Novo Dominant Negative Mutation in DNM1L Impairs Mitochondrial Fission and Presents as Childhood Epileptic Encephalopathy. Am. J. Med. Genet. A 2016, 170, 2002–2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carmichael, R.E.; Islinger, M.; Schrader, M. Fission Impossible (?)—New Insights into Disorders of Peroxisome Dynamics. Cells 2022, 11, 1922. https://doi.org/10.3390/cells11121922
Carmichael RE, Islinger M, Schrader M. Fission Impossible (?)—New Insights into Disorders of Peroxisome Dynamics. Cells. 2022; 11(12):1922. https://doi.org/10.3390/cells11121922
Chicago/Turabian StyleCarmichael, Ruth E., Markus Islinger, and Michael Schrader. 2022. "Fission Impossible (?)—New Insights into Disorders of Peroxisome Dynamics" Cells 11, no. 12: 1922. https://doi.org/10.3390/cells11121922






