Non-Coding RNAs and Prediction of Preeclampsia in the First Trimester of Pregnancy
Abstract
:1. Introduction
1.1. Preeclampsia
1.2. Pathogenesis of PE
1.3. Prediction and Prevention of PE
1.4. Non-Coding RNA
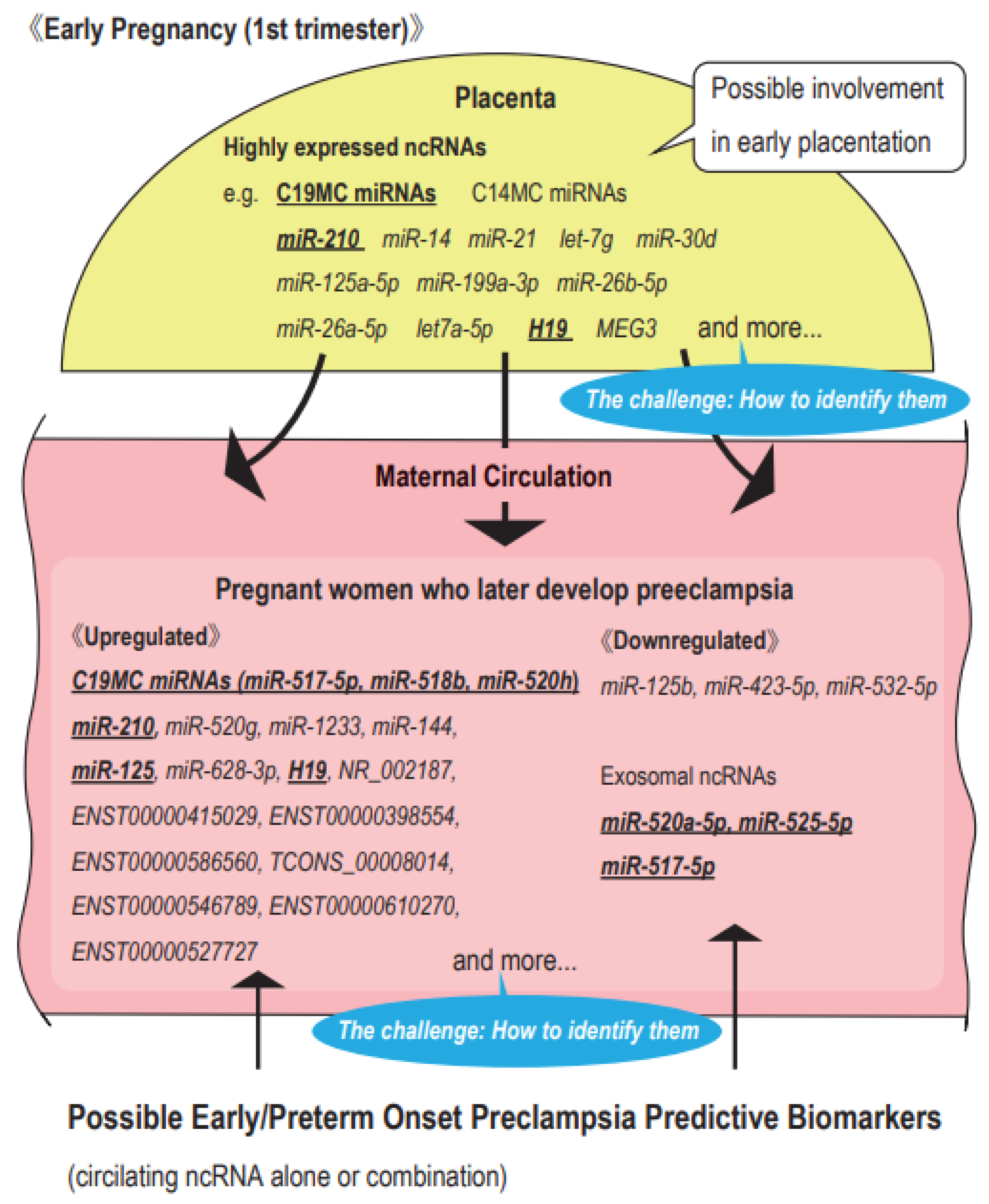
1.5. Possible ncRNAs as Predictive Markers of PE
2. Highly Expressed ncRNAs in the First Trimester Placenta
2.1. miRNAs
2.1.1. Placenta-Specific miRNA Clusters
2.1.2. Hypoxia-Responsive miRNA: miR-210
2.1.3. miRNAs That May Be Involved in Early Placentation Based on an Expression Analysis Using Human Early Placentas
2.2. lncRNAs
3. Notable lncRNAs for Early Placentation
3.1. H19
3.2. MALAT1: Metastasis-Associated Lung Adenocarcinoma Transcript 1
3.3. MEG3: Maternally Expressed Gene 3
4. Possible Circulating ncRNA Biomarkers for PE in the First Trimester
4.1. miRNAs
4.1.1. Placenta-Specific miRNA Clusters
| No | Author | Country | Study Outcome | Race | Sampling Time (GW) | Sample | Case (n)/Control (n) | Predictive ncRNAs | Type of Predictive ncRNA | Quantitative Method | Expression Level | Accuracy | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | AUC | |||||||||||||
| 1 | Hromadnikova (2017) | Czech | PE | Caucasian | 10–13 w | Plasma | PE (21)/Normal (58) | miR-517-5p (C19MC) | miRNA | RT-PCR | Up in PE | 42.9 | 86.2 | 52.9 | 80.6 | 0.70 | [113] |
| miR-518b (C19MC) | Up in PE | 52.4 | 63.8 | 34.4 | 78.7 | 0.55 | |||||||||||
| miR-520h (C19MC) | Up in PE | 14.3 | 96.6 | 60.0 | 75.7 | 0.45 | |||||||||||
| 2 | Hromadnikova (2019) | Czech | PE | Caucasian | 10–13 w | Exosomes from plasma | PE (43)/Normal (102) | miR-520a-5p (C19MC) | miRNA | RT-PCR | Down in PE | 60.5 | 84.0 | 26.0 | 42.0 | 0.69 | [114] |
| miR-525-5p (C19MC) | Down in PE | 51.2 | 84.0 | 22.0 | 42.0 | 0.69 | |||||||||||
| miR-517-5p (C19MC) | Down in PE | 60.4 | 70.0 | 26.0 | 35.0 | 0.63 | |||||||||||
| 3 miRNAs | Down in PE | 65.1 | 78.0 | 28.0 | 39.0 | 0.71 | |||||||||||
| 3 | Jiang L (2017) | China | PE | NA | 8–10 w | Serum | PE (19)/Normal (19) | miR-520g | miRNA | RT-PCR | Up in PE | NA | NA | NA | NA | NA | [115] |
| 4 | Ura (2014) | Italy | PE | Multiracial (Caucasian: > 90%) | 12–14 w | Serum | PE (24)/Normal (24) | miR-1233 | miRNA | RT-PCR | Up in PE (5.4x) | NA | NA | NA | NA | NA | [116] |
| miR-520a | Up in PE (3.2x) | NA | NA | NA | NA | NA | |||||||||||
| miR-210 | Up in PE (3.3x) | NA | NA | NA | NA | NA | |||||||||||
| miR-144 | Down in PE (0.4x) | NA | NA | NA | NA | NA | |||||||||||
| 5 | Licini (2021) | Italy | PE | NA | 12 w | Plasma | PE (13)/Normal (18) | miR-125b | miRNA | RT-PCR | Down in PE | 0.85 | [117] | ||||
| 6 | Martinez-Fierro (2021) | Mexico | PE | NA | 12 w | Serum | PE (6)/Normal (6) | miR-628-3p | miRNA | Taqman low density array | Up in PE (7.7x) | NA | NA | NA | NA | NA | [118] |
| 7 | Martinez-Fierro (2019) | Mexico | PE | NA | 12 w | Serum | PE (16)/Normal (18) | miR-628-3p | miRNA | RT-PCR | Up in PE (7.7x) | NA | NA | NA | NA | NA | [119] |
| 8 | Chen (2021) | China | PE | NA | 11–13 w | Whole blood | PE (24)/Normal (30) | has_circ_0025992 | circRNA | RT-PCR | Up in PE | 54.2 | 93.3 | 86.7 | 71.8 | 0.80 | [120] |
| 9 | Timofeeva (2018) | Russia | EOPE | NA | 11–13 w | Plasma | EOPE (6)/Normal (10) | miR-423-5p | miRNA | RT-PCR | Down in PE | NA | NA | NA | NA | NA | [121] |
| miR-532-5p | Down in PE | NA | NA | NA | NA | NA | |||||||||||
| 10 | Winger (2015) | USA | PE | NA | preconception-9 w | PBMC | PE (12)/Normal (19) | Score using 7 miRNAs (miR-1, miR-133b, miR-199a-5p, miR-1267, miR-1229, miR-148a-3p, miR-223) | miRNA | RT-PCR (Scoring) | NA | 83.3 | 89.5 | 83.0 | 89.0 | 0.90 | [122] |
| EOPE | EOPE (5)/Normal (19) | NA | 80.0 | 89.5 | 67.0 | 94.0 | 0.86 | ||||||||||
| 11 | Winger (2018) | UK | PE | Multiracial | 11–13 w | Plasma | PE (4)/Normal (19) | miR-1267 | miRNA | RT-PCR | NA | 75 | 95 | NA | NA | 0.88 | [123] |
| PE (2)/Normal (7) | miR-148a | NA | 100 | 86 | NA | NA | 0.93 | ||||||||||
| PE (2)/Normal (4) | miR-196a | NA | 100 | 100 | NA | NA | 1.00 | ||||||||||
| PE (3)/Normal (8) | miR-33a | NA | 100 | 100 | NA | NA | 1.00 | ||||||||||
| PE (3)/Normal (9) | miR-575 | NA | 100 | 78 | NA | NA | 0.93 | ||||||||||
| PE (7)/Normal (9) | miR-582 | NA | 100 | 100 | NA | NA | 1.00 | ||||||||||
| PE (3)/Normal (14) | miR-210 | NA | 67 | 100 | NA | NA | 0.86 | ||||||||||
| PE (4)/Normal (20) | miR-16 | NA | 100 | 55 | NA | NA | 0.78 | ||||||||||
| PE (8)/Normal (40) | Score using 8 miRNAs | (Scoring) | NA | 75 | 90 | NA | NA | 0.91 | |||||||||
| 12 | Yoffe (2018) | Israel | EOPE | Multiracial | 11–13 w | Plasma | EOPE (35)/Normal (40) | Score using 25 ncRNAs (12 miRNAs, 4 lncRNAs, 1 rRNA, 7 mitochondrial tRNA, 1 processed transcript) | miRNA, lncRNA, rRNA, mitochondrial tRNA, processed transcript | Small RNA sequencing | NA | 72.0 | 80.0 | NA | NA | 0.86 | [124] |
| 13 | Dai (2021) | China | PE | NA | <20 w * | Serum | PE (97)/Normal (97) | NR_002187 | lncRNA (pseudogene) | RT-PCR | Up in PE | NA | NA | NA | NA | 0.66 | [125] |
| ENST00000415029 | lncRNA | Up in PE | NA | NA | NA | NA | 0.69 | ||||||||||
| ENST00000398554 | lncRNA | Up in PE | NA | NA | NA | NA | 0.65 | ||||||||||
| ENST00000586560 | lncRNA | Up in PE | NA | NA | NA | NA | 0.63 | ||||||||||
| TCONS_00008014 | lncRNA | Up in PE | NA | NA | NA | NA | 0.64 | ||||||||||
| ENST00000546789 | lncRNA | Up in PE | NA | NA | NA | NA | 0.68 | ||||||||||
| ENST00000610270 | lncRNA | Up in PE | NA | NA | NA | NA | 0.66 | ||||||||||
| ENST00000527727 | lncRNA | Up in PE | NA | NA | NA | NA | 0.64 | ||||||||||
| 14 | Tarca (2021) | USA | EOPE | Multiracial (African American > 90%) | 11–17 w | Whole blood | EOPE (13)/Normal (49) | H19 | lncRNA (lincRNA) | Z score obtained from microarrays (EOPE vs. Normal) | Up in PE | NA | NA | NA | NA | NA | [126] |
4.1.2. Hypoxia-Responsive miRNA: miR-210
4.1.3. Circulating Predictive miRNAs of PE Based on Gene Expression Analyses Using Human Samples
4.2. lncRNAs
5. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AUC | area under the curve |
| circRNA | circular RNA |
| CVS | chorionic villous sampling |
| CVT | chorionic villous trophoblasts |
| C14MC | chromosome 14 microRNA cluster |
| C19MC | chromosome 19 microRNA cluster |
| dNK cell | decidual natural killer cell |
| EMT | epithelial-to-mesenchymal transition |
| EOPE | early onset preeclampsia |
| EVT | extravillous trophoblasts |
| FC | fold change |
| ISH | in situ hybridization |
| LDA | low dose aspirin |
| LMD | laser microdissection |
| lncRNA | long non-coding RNA |
| MAP | mean arterial pressure |
| miRNA | microRNA |
| MSC | mesenchymal stem cells |
| mRNA | messenger RNA |
| ncRNA | non-coding RNA |
| NK cell | natural killer cell |
| NSCLC | non-small cell lung cell cancer |
| PE | preeclampsia |
| POPE | preterm onset preeclampsia |
| rRNA | ribosomal RNA |
| RT-PCR | real time-polymerase chain reaction |
| SCT | syncytiotrophoblasts |
| UTR | untranslated region |
| VSMC | vascular smooth muscle cell |
| VT | villous trophoblasts |
References
- Redman, C.W.; Sargent, I.L. Latest advances in understanding preeclampsia. Science 2005, 308, 1592–1594. [Google Scholar] [CrossRef] [PubMed]
- Steegers, E.A.; von Dadelszen, P.; Duvekot, J.J.; Pijnenborg, R. Preeclampsia. Lancet 2010, 376, 631–644. [Google Scholar] [CrossRef]
- Sibai, B.; Dekker, G.; Kupferminc, M. Pre-eclampsia. Lancet 2005, 365, 785–799. [Google Scholar] [CrossRef]
- Tanner, M.S.; Davey, M.A.; Mol, B.W.; Rolnik, D.L. The evolution of the diagnostic criteria of preeclampsia-eclampsia. Am. J. Obstet. Gynecol. 2022, 226, S835–S843. [Google Scholar] [CrossRef]
- Scott, G.; Gillon, T.E.; Pels, A.; von Dadelszen, P.; Magee, L.A. Guidelines-similarities and dissimilarities: A systematic review of international clinical practice guidelines for pregnancy hypertension. Am. J. Obstet. Gynecol. 2022, 226, S1222–S1236. [Google Scholar] [CrossRef]
- Platt, M.J. Outcomes in preterm infants. Public Health 2014, 128, 399–403. [Google Scholar] [CrossRef]
- Glass, H.C.; Costarino, A.T.; Stayer, S.A.; Brett, C.M.; Cladis, F.; Davis, P.J. Outcomes for extremely premature infants. Anesth. Analg. 2015, 120, 1337–1351. [Google Scholar] [CrossRef] [Green Version]
- Pollheimer, J.; Vondra, S.; Baltayeva, J.; Beristain, A.G.; Knöfler, M. Regulation of Placental Extravillous Trophoblasts by the Maternal Uterine Environment. Front. Immunol. 2018, 9, 2597. [Google Scholar] [CrossRef]
- Opichka, M.A.; Rappelt, M.W.; Gutterman, D.D.; Grobe, J.L.; McIntosh, J.J. Vascular Dysfunction in Preeclampsia. Cells 2021, 10, 3055. [Google Scholar] [CrossRef]
- Rolnik, D.L.; Wright, D.; Poon, L.C.Y.; Syngelaki, A.; O’Gorman, N.; de Paco Matallana, C.; Akolekar, R.; Cicero, S.; Janga, D.; Singh, M.; et al. ASPRE trial: Performance of screening for preterm pre-eclampsia. Ultrasound Obstet. Gynecol. 2017, 50, 492–495. [Google Scholar] [CrossRef]
- O’Gorman, N.; Wright, D.; Syngelaki, A.; Akolekar, R.; Wright, A.; Poon, L.C.; Nicolaides, K.H. Competing risks model in screening for preeclampsia by maternal factors and biomarkers at 11–13 weeks gestation. Am. J. Obstet. Gynecol. 2016, 214, 103.e1–103.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, D.; Tan, M.Y.; O’Gorman, N.; Poon, L.C.; Syngelaki, A.; Wright, A.; Nicolaides, K.H. Predictive performance of the competing risk model in screening for preeclampsia. Am. J. Obstet. Gynecol. 2019, 220, 199.e1–199.e13. [Google Scholar] [CrossRef] [Green Version]
- Guy, G.P.; Leslie, K.; Diaz Gomez, D.; Forenc, K.; Buck, E.; Khalil, A.; Thilaganathan, B. Implementation of routine first trimester combined screening for pre-eclampsia: A clinical effectiveness study. BJOG 2021, 128, 149–156. [Google Scholar] [CrossRef]
- Cabunac, P.; Karadžov Orlić, N.; Ardalić, D.; Damnjanović Pažin, B.; Stanimirović, S.; Banjac, G.; Stefanović, A.; Spasojević-Kalimanovska, V.; Egić, A.; Rajović, N.; et al. Use of FMF algorithm for prediction of preeclampsia in high risk pregnancies: A single center longitudinal study. Hypertens. Pregnancy 2021, 40, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Skråstad, R.B.; Hov, G.G.; Blaas, H.G.; Romundstad, P.R.; Salvesen, K. Risk assessment for preeclampsia in nulliparous women at 11–13 weeks gestational age: Prospective evaluation of two algorithms. BJOG 2015, 122, 1781–1788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakovschek, I.C.; Ulrich, D.; Jauk, S.; Csapo, B.; Kolovetsiou-Kreiner, V.; Mayer-Pickel, K.; Stern, C.; Lang, U.; Obermayer-Pietsch, B.; Cervar-Zivkovic, M. Risk assessment for preterm preeclampsia in first trimester: Comparison of three calculation algorithms. Eur. J. Obstet. Gynecol. 2018, 231, 241–247. [Google Scholar] [CrossRef]
- Chaemsaithong, P.; Sahota, D.; Pooh, R.K.; Zheng, M.; Ma, R.; Chaiyasit, N.; Koide, K.; Shaw, S.W.; Seshadri, S.; Choolani, M.; et al. First-trimester pre-eclampsia biomarker profiles in Asian population: Multicenter cohort study. Ultrasound Obstet. Gynecol. 2020, 56, 206–214. [Google Scholar] [CrossRef]
- Guizani, M.; Valsamis, J.; Dutemeyer, V.; Kang, X.; Ceccotti, V.; Khalife, J.; Duiella, S.F.; Blavier, F.; Faraca, A.; Cos, T.; et al. First-Trimester Combined Multimarker Prospective Study for the Detection of Pregnancies at a High Risk of Developing Preeclampsia Using the Fetal Medicine Foundation-Algorithm. Fetal Diagn. Ther. 2018, 43, 266–273. [Google Scholar] [CrossRef]
- Lobo, G.A.R.; Nowak, P.M.; Panigassi, A.P.; Lima, A.I.F.; Araujo Júnior, E.; Nardozza, L.M.M.; Pares, D.B.S. Validation of Fetal Medicine Foundation algorithm for prediction of pre-eclampsia in the first trimester in an unselected Brazilian population. J. Matern.-Fetal Neonatal Med. 2019, 32, 286–292. [Google Scholar] [CrossRef]
- Lakshmy, S.; Ziyaulla, T.; Rose, N. The need for implementation of first trimester screening for preeclampsia and fetal growth restriction in low resource settings. J. Matern.-Fetal Neonatal Med. 2021, 34, 4082–4089. [Google Scholar] [CrossRef]
- Sahai, K.; Saraswathy, S.; Yadav, T.P.; Arora, D.; Krishnan, M. Pre-eclampsia: Molecular events to biomarkers. Med. J. Armed Forces India 2017, 73, 167–174. [Google Scholar] [CrossRef] [Green Version]
- Kuc, S.; Koster, M.P.; Pennings, J.L.; Hankemeier, T.; Berger, R.; Harms, A.C.; Dane, A.D.; Schielen, P.C.; Visser, G.H.; Vreeken, R.J. Metabolomics profiling for identification of novel potential markers in early prediction of preeclampsia. PLoS ONE 2014, 9, e98540. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Krishna, I.; Badell, M.; Samuel, A. Can the quantity of cell-free fetal DNA predict preeclampsia: A systematic review. Prenat. Diagn. 2014, 34, 685–691. [Google Scholar] [CrossRef]
- Contro, E.; Bernabini, D.; Farina, A. Cell-Free Fetal DNA for the Prediction of Pre-Eclampsia at the First and Second Trimesters: A Systematic Review and Meta-Analysis. Mol. Diagn. Ther. 2017, 21, 125–135. [Google Scholar] [CrossRef]
- Taft, R.J.; Pang, K.C.; Mercer, T.R.; Dinger, M.; Mattick, J.S. Non-coding RNAs: Regulators of disease. J. Pathol. 2010, 220, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Schmitz, S.U.; Grote, P.; Herrmann, B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016, 73, 2491–2509. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [Green Version]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.; Carninci, P. Expression Specificity of Disease-Associated lncRNAs: Toward Personalized Medicine. Curr. Top. Microbiol. Immunol. 2016, 394, 237–258. [Google Scholar] [PubMed]
- Tafrihi, M.; Hasheminasab, E. MiRNAs: Biology, Biogenesis, their Web-based Tools, and Databases. Microrna 2019, 8, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, J.; Cairns, M.J. Identifying miRNAs, targets and functions. Brief. Bioinform. 2014, 15, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Gude, N.M.; Roberts, C.T.; Kalionis, B.; King, R.G. Growth and function of the normal human placenta. Thromb. Res. 2004, 114, 397–407. [Google Scholar] [CrossRef]
- Lv, Y.; Lu, C.; Ji, X.; Miao, Z.; Long, W.; Ding, H.; Lv, M. Roles of microRNAs in preeclampsia. J. Cell. Physiol. 2019, 234, 1052–1061. [Google Scholar] [CrossRef]
- Munjas, J.; Sopić, M.; Stefanović, A.; Košir, R.; Ninić, A.; Joksić, I.; Antonić, T.; Spasojević-Kalimanovska, V.; Prosenc Zmrzljak, U. Non-Coding RNAs in Preeclampsia-Molecular Mechanisms and Diagnostic Potential. Int. J. Mol. Sci. 2021, 22, 10652. [Google Scholar] [CrossRef]
- Ali, A.; Hadlich, F.; Abbas, M.W.; Iqbal, M.A.; Tesfaye, D.; Bouma, G.J.; Winger, Q.A.; Ponsuksili, S. MicroRNA-mRNA Networks in Pregnancy Complications: A Comprehensive Downstream Analysis of Potential Biomarkers. Int. J. Mol. Sci. 2021, 22, 2313. [Google Scholar] [CrossRef]
- Jairajpuri, D.S.; Almawi, W.Y. MicroRNA expression pattern in pre-eclampsia (Review). Mol. Med. Rep. 2016, 13, 2351–2358. [Google Scholar] [CrossRef] [Green Version]
- Sheikh, A.M.; Small, H.Y.; Currie, G.; Delles, C. Systematic Review of Micro-RNA Expression in Pre-Eclampsia Identifies a Number of Common Pathways Associated with the Disease. PLoS ONE 2016, 11, e0160808. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.; Yu, R.; Jiang, S.; Xia, Y.; Chen, Y. Recent Advances of MicroRNAs, Long Non-coding RNAs, and Circular RNAs in Preeclampsia. Front. Physiol. 2021, 12, 659638. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Ohkuchi, A.; Kuwata, T.; Usui, R.; Baba, Y.; Suzuki, H.; Chaw Kyi, T.T.; Matsubara, S.; Saito, S.; Takizawa, T. Endogenous and exogenous miR-520c-3p modulates CD44-mediated extravillous trophoblast invasion. Placenta 2017, 50, 25–31. [Google Scholar] [CrossRef]
- Xie, L.; Mouillet, J.F.; Chu, T.; Parks, W.T.; Sadovsky, E.; Knöfler, M.; Sadovsky, Y. C19MC microRNAs regulate the migration of human trophoblasts. Endocrinology 2014, 155, 4975–4985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, S.S.; Ishibashi, O.; Ishikawa, G.; Ishikawa, T.; Katayama, A.; Mishima, T.; Takizawa, T.; Shigihara, T.; Goto, T.; Izumi, A.; et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol. Reprod. 2009, 81, 717–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogoyama, M.; Ohkuchi, A.; Takahashi, H.; Zhao, D.; Matsubara, S.; Takizawa, T. LncRNA H19-Derived miR-675-5p Accelerates the Invasion of Extravillous Trophoblast Cells by Inhibiting GATA2 and Subsequently Activating Matrix Metalloproteinases. Int. J. Mol. Sci. 2021, 22, 1237. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, T.L.; Eisman, L.E.; Joshi, N.V.; Flowers, A.E.; Wu, D.; Wang, Y.; Santiskulvong, C.; Tang, J.; Buttle, R.A.; Sauro, E.; et al. High-throughput miRNA sequencing of the human placenta: Expression throughout gestation. Epigenomics 2021, 13, 995–1012. [Google Scholar] [CrossRef]
- Smith, M.D.; Pillman, K.; Jankovic-Karasoulos, T.; McAninch, D.; Wan, Q.; Bogias, K.J.; McCullough, D.; Bianco-Miotto, T.; Breen, J.; Roberts, C.T. Large-scale transcriptome-wide profiling of microRNAs in human placenta and maternal plasma at early to mid gestation. RNA Biol. 2021, 18, 507–520. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, X.; Wang, R.; Lu, X.; Dang, Y.L.; Wang, H.; Lin, H.Y.; Zhu, C.; Ge, H.; Cross, J.C.; et al. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 2018, 28, 819–832. [Google Scholar] [CrossRef] [Green Version]
- Vento-Tormo, R.; Efremova, M.; Botting, R.A.; Turco, M.Y.; Vento-Tormo, M.; Meyer, K.B.; Park, J.E.; Stephenson, E.; Polański, K.; Goncalves, A.; et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018, 563, 347–353. [Google Scholar] [CrossRef] [Green Version]
- Suryawanshi, H.; Morozov, P.; Straus, A.; Sahasrabudhe, N.; Max, K.E.A.; Garzia, A.; Kustagi, M.; Tuschl, T.; Williams, Z. A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 2018, 4, eaau4788. [Google Scholar] [CrossRef] [Green Version]
- Morales-Prieto, D.M.; Chaiwangyen, W.; Ospina-Prieto, S.; Schneider, U.; Herrmann, J.; Gruhn, B.; Markert, U.R. MicroRNA expression profiles of trophoblastic cells. Placenta 2012, 33, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Miura, S.; Yamasaki, K.; Higashijima, A.; Kinoshita, A.; Yoshiura, K.; Masuzaki, H. Identification of pregnancy-associated microRNAs in maternal plasma. Clin. Chem. 2010, 56, 1767–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Song, W.; Na, Q. The emerging roles of placenta-specific microRNAs in regulating trophoblast proliferation during the first trimester. Aust. N. Z. J. Obstet. Gynaecol. 2012, 52, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Mong, E.F.; Yang, Y.; Akat, K.M.; Canfield, J.; VanWye, J.; Lockhart, J.; Tsibris, J.C.M.; Schatz, F.; Lockwood, C.J.; Tuschl, T.; et al. Chromosome 19 microRNA cluster enhances cell reprogramming by inhibiting epithelial-to-mesenchymal transition. Sci. Rep. 2020, 10, 3029. [Google Scholar] [CrossRef] [Green Version]
- Flor, I.; Neumann, A.; Freter, C.; Helmke, B.M.; Langenbuch, M.; Rippe, V.; Bullerdiek, J. Abundant expression and hemimethylation of C19MC in cell cultures from placenta-derived stromal cells. Biochem. Biophys. Res. Commun. 2012, 422, 411–416. [Google Scholar] [CrossRef]
- Hayder, H.; Fu, G.; Nadeem, L.; O’Brien, J.A.; Lye, S.J.; Peng, C. Overexpression of miR-210-3p Impairs Extravillous Trophoblast Functions Associated with Uterine Spiral Artery Remodeling. Int. J. Mol. Sci. 2021, 22, 3961. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, Y.; Luo, R.; Bian, X.; Wang, Y.; Shao, X.; Li, Y.X.; Liu, M.; Wang, Y.L. A positive feedback self-regulatory loop between miR-210 and HIF-1α mediated by CPEB2 is involved in trophoblast syncytialization: Implication of trophoblast malfunction in preeclampsia. Biol. Reprod. 2020, 102, 560–570. [Google Scholar]
- Wang, R.; Zou, L.; Yang, X. microRNA-210/Long non-coding RNA MEG3 axis inhibits trophoblast cell migration and invasion by suppressing EMT process. Placenta 2021, 109, 64–71. [Google Scholar] [CrossRef]
- Gu, Y.; Sun, J.; Groome, L.J.; Wang, Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am. J. Physiol.-Endocrinol. Metab. 2013, 304, E836–E843. [Google Scholar] [CrossRef]
- Singh, K.; Williams, J., III; Brown, J.; Wang, E.T.; Lee, B.; Gonzalez, T.L.; Cui, J.; Goodarzi, M.O.; Pisarska, M.D. Up-regulation of microRNA-202-3p in first trimester placenta of pregnancies destined to develop severe preeclampsia, a pilot study. Pregnancy Hypertens. 2017, 10, 7–9. [Google Scholar] [CrossRef]
- Wu, H.; Wang, H.; Liu, M.; Bai, Y.; Li, Y.X.; Ji, L.; Peng, C.; Yu, Y.; Wang, Y.L. MiR-195 participates in the placental disorder of preeclampsia via targeting activin receptor type-2B in trophoblastic cells. J. Hypertens. 2016, 34, 1371–1379. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; He, D.; Xie, H.; Zhao, Y.; Peng, Z.; Deng, H.; Hu, J.; Jiang, B.; Liu, N. H19 regulates angiogenic capacity of extravillous trophoblasts by H19/miR-106a-5p/VEGFA axis. Arch. Gynecol. Obstet. 2020, 301, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Morales-Prieto, D.M.; Ospina-Prieto, S.; Chaiwangyen, W.; Schoenleben, M.; Markert, U.R. Pregnancy-associated miRNA-clusters. J. Reprod. Immunol. 2013, 97, 51–61. [Google Scholar] [CrossRef]
- Fóthi, Á.; Biró, O.; Erdei, Z.; Apáti, Á.; Orbán, T.I. Tissue-specific and transcription-dependent mechanisms regulate primary microRNA processing efficiency of the human chromosome 19 MicroRNA cluster. RNA Biol. 2021, 18, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Bayer, A.; Lennemann, N.J.; Ouyang, Y.; Sadovsky, E.; Sheridan, M.A.; Roberts, R.M.; Coyne, C.B.; Sadovsky, Y. Chromosome 19 microRNAs exert antiviral activity independent from type III interferon signaling. Placenta 2018, 61, 33–38. [Google Scholar] [CrossRef]
- Mouillet, J.F.; Ouyang, Y.; Bayer, A.; Coyne, C.B.; Sadovsky, Y. The role of trophoblastic microRNAs in placental viral infection. Int. J. Dev. Biol. 2014, 58, 281–289. [Google Scholar] [CrossRef] [Green Version]
- Su, J.L.; Chen, P.S.; Johansson, G.; Kuo, M.L. Function and regulation of let-7 family microRNAs. Microrna 2012, 1, 34–39. [Google Scholar] [CrossRef]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef]
- Li, Y.; Huang, H.; Ye, X.; Huang, Z.; Chen, X.; Wu, F.; Lin, T. miR-202-3p negatively regulates MMP-1 to inhibit the proliferation, migration and invasion of lung adenocarcinoma cells. Cell Cycle 2021, 20, 406–416. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, Y.; Li, L.; Ma, L.; Zhou, C. Dysregulation of miR-202-3p Affects Migration and Invasion of Endometrial Stromal Cells in Endometriosis via Targeting ROCK1. Reprod. Sci. 2020, 27, 731–742. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, C.; Wang, M.; Su, L.; Qu, Y.; Li, J.; Yu, B.; Yan, M.; Yu, Y.; Liu, B.; et al. Decrease of miR-202-3p expression, a novel tumor suppressor, in gastric cancer. PLoS ONE 2013, 8, e69756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nordin, M.; Bergman, D.; Halje, M.; Engström, W.; Ward, A. Epigenetic regulation of the Igf2/H19 gene cluster. Cell Prolif. 2014, 47, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Cullen, B.R. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007, 13, 313–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, Y.; Tayama, C.; Tomikawa, J.; Akaishi, R.; Kamura, H.; Matsuoka, K.; Wake, N.; Minakami, H.; Kato, K.; Yamada, T.; et al. Placenta-specific epimutation at H19-DMR among common pregnancy complications: Its frequency and effect on the expression patterns of H19 and IGF2. Clin. Epigenet. 2019, 11, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecerf, C.; Le Bourhis, X.; Adriaenssens, E. The long non-coding RNA H19: An active player with multiple facets to sustain the hallmarks of cancer. Cell. Mol. Life Sci. 2019, 76, 4673–4687. [Google Scholar] [CrossRef]
- Matouk, I.J.; Halle, D.; Raveh, E.; Gilon, M.; Sorin, V.; Hochberg, A. The role of the oncofetal H19 lncRNA in tumor metastasis: Orchestrating the EMT-MET decision. Oncotarget 2016, 7, 3748–3765. [Google Scholar] [CrossRef] [Green Version]
- Raveh, E.; Matouk, I.J.; Gilon, M.; Hochberg, A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis—A proposed unifying theory. Mol. Cancer 2015, 14, 184. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.; Li, Z.; Wang, W.; Zeng, Y.; Liu, Z.; Qiu, J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013, 333, 213–221. [Google Scholar] [CrossRef]
- Liu, C.; Chen, Z.; Fang, J.; Xu, A.; Zhang, W.; Wang, Z. H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumor Biol. 2016, 37, 263–270. [Google Scholar] [CrossRef] [Green Version]
- Vennin, C.; Spruyt, N.; Dahmani, F.; Julien, S.; Bertucci, F.; Finetti, P.; Chassat, T.; Bourette, R.P.; Le Bourhis, X.; Adriaenssens, E. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget 2015, 6, 29209–29223. [Google Scholar] [CrossRef] [Green Version]
- He, D.; Wang, J.; Zhang, C.; Shan, B.; Deng, X.; Li, B.; Zhou, Y.; Chen, W.; Hong, J.; Gao, Y.; et al. Down-regulation of miR-675-5p contributes to tumor progression and development by targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol. Cancer 2015, 14, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, M.; Chen, Q.; Liu, X.; Sun, Q.; Zhao, X.; Deng, R.; Wang, Y.; Huang, J.; Xu, M.; Yan, J.; et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014, 281, 3766–3775. [Google Scholar] [CrossRef] [PubMed]
- Shih, J.W.; Kung, H.J. Long non-coding RNA and tumor hypoxia: New players ushered toward an old arena. J. Biomed. Sci. 2017, 24, 53. [Google Scholar] [CrossRef]
- Huppertz, B.; Weiss, G.; Moser, G. Trophoblast invasion and oxygenation of the placenta: Measurements versus presumptions. J. Reprod. Immunol. 2014, 101–102, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xia, Y.; Zhang, H.; Guo, H.; Feng, K.; Zhang, C. Overexpression of long non-coding RNA H19 promotes invasion and autophagy via the PI3K/AKT/mTOR pathways in trophoblast cells. Biomed. Pharmacother. 2018, 101, 691–697. [Google Scholar] [CrossRef]
- Harati-Sadegh, M.; Kohan, L.; Teimoori, B.; Mehrabani, M.; Salimi, S. The effects of placental long noncoding RNA H19 polymorphisms and promoter methylation on H19 expression in association with preeclampsia susceptibility. IUBMB Life 2020, 72, 413–425. [Google Scholar] [CrossRef]
- Gao, W.L.; Liu, M.; Yang, Y.; Yang, H.; Liao, Q.; Bai, Y.; Li, Y.X.; Li, D.; Peng, C.; Wang, Y.L. The imprinted H19 gene regulates human placental trophoblast cell proliferation via encoding miR-675 that targets Nodal Modulator 1 (NOMO1). RNA Biol. 2012, 9, 1002–1010. [Google Scholar] [CrossRef] [Green Version]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [Green Version]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 39. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Hao, Q.; Prasanth, K.V. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018, 34, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hamblin, M.H.; Yin, K.J. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017, 14, 1705–1714. [Google Scholar] [CrossRef] [PubMed]
- Puthanveetil, P.; Gutschner, T.; Lorenzen, J. MALAT1: A therapeutic candidate for a broad spectrum of vascular and cardiorenal complications. Hypertens. Res. 2020, 43, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, S.; Li, Q.; Ji, Q.; Guo, P.; Liu, X. MALAT1: A long non-coding RNA highly associated with human cancers. Oncol. Lett. 2018, 16, 19–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okugawa, Y.; Toiyama, Y.; Hur, K.; Toden, S.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Kusunoki, M.; Boland, C.R.; et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis 2014, 35, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.T.; Shi, D.B.; Wang, Y.W.; Li, X.X.; Xu, Y.; Tripathi, P.; Gu, W.L.; Cai, G.X.; Cai, S.J. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 3174–3181. [Google Scholar]
- Hirata, H.; Hinoda, Y.; Shahryari, V.; Deng, G.; Nakajima, K.; Tabatabai, Z.L.; Ishii, N.; Dahiya, R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015, 75, 1322–1331. [Google Scholar] [CrossRef] [Green Version]
- Lai, M.C.; Yang, Z.; Zhou, L.; Zhu, Q.Q.; Xie, H.Y.; Zhang, F.; Wu, L.M.; Chen, L.M.; Zheng, S.S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 2012, 29, 1810–1816. [Google Scholar] [CrossRef]
- Fan, Y.; Shen, B.; Tan, M.; Mu, X.; Qin, Y.; Zhang, F.; Liu, Y. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 2014, 20, 1531–1541. [Google Scholar] [CrossRef] [Green Version]
- Vassallo, I.; Zinn, P.; Lai, M.; Rajakannu, P.; Hamou, M.F.; Hegi, M.E. WIF1 re-expression in glioblastoma inhibits migration through attenuation of non-canonical WNT signaling by downregulating the lncRNA MALAT1. Oncogene 2016, 35, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Sui, S.; Zhang, J.; Bai, N.; Shi, Q.; Zhang, G.; Gao, S.; You, Z.; Zhan, C.; Liu, F.; et al. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 4881–4891. [Google Scholar] [PubMed]
- Chen, H.; Meng, T.; Liu, X.; Sun, M.; Tong, C.; Liu, J.; Wang, H.; Du, J. Long non-coding RNA MALAT-1 is downregulated in preeclampsia and regulates proliferation, apoptosis, migration and invasion of JEG-3 trophoblast cells. Int. J. Clin. Exp. Pathol. 2015, 8, 12718–12727. [Google Scholar] [PubMed]
- Li, X.; Song, Y.; Liu, F.; Liu, D.; Miao, H.; Ren, J.; Xu, J.; Ding, L.; Hu, Y.; Wang, Z.; et al. Long Non-Coding RNA MALAT1 Promotes Proliferation, Angiogenesis, and Immunosuppressive Properties of Mesenchymal Stem Cells by Inducing VEGF and IDO. J. Cell. Biochem. 2017, 118, 2780–2791. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Y.; Yu, M.S.; Li, X.; Zhang, Z.; Han, C.R.; Yan, B. Overexpression of long non-coding RNA MEG3 suppresses breast cancer cell proliferation, invasion, and angiogenesis through AKT pathway. Tumor Biol. 2017, 39, 1010428317701311. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007, 282, 24731–24742. [Google Scholar] [CrossRef] [Green Version]
- Al-Rugeebah, A.; Alanazi, M.; Parine, N.R. MEG3: An Oncogenic Long Non-coding RNA in Different Cancers. Pathol. Oncol. Res. 2019, 25, 859–874. [Google Scholar] [CrossRef]
- Zhang, W.; Shi, S.; Jiang, J.; Li, X.; Lu, H.; Ren, F. LncRNA MEG3 inhibits cell epithelial-mesenchymal transition by sponging miR-421 targeting E-cadherin in breast cancer. Biomed. Pharmacother. 2017, 91, 312–319. [Google Scholar] [CrossRef]
- Xu, G.; Meng, L.; Yuan, D.; Li, K.; Zhang, Y.; Dang, C.; Zhu, K. MEG3/miR-21 axis affects cell mobility by suppressing epithelial-mesenchymal transition in gastric cancer. Oncol. Rep. 2018, 40, 39–48. [Google Scholar] [CrossRef] [Green Version]
- Dan, J.; Wang, J.; Wang, Y.; Zhu, M.; Yang, X.; Peng, Z.; Jiang, H.; Chen, L. LncRNA-MEG3 inhibits proliferation and metastasis by regulating miRNA-21 in gastric cancer. Biomed. Pharmacother. 2018, 99, 931–938. [Google Scholar] [CrossRef]
- Zhang, Y.; Zou, Y.; Wang, W.; Zuo, Q.; Jiang, Z.; Sun, M.; De, W.; Sun, L. Down-regulated long non-coding RNA MEG3 and its effect on promoting apoptosis and suppressing migration of trophoblast cells. J. Cell. Biochem. 2015, 116, 542–550. [Google Scholar] [CrossRef]
- Liu, W.; Liu, X.; Luo, M.; Liu, X.; Luo, Q.; Tao, H.; Wu, D.; Lu, S.; Jin, J.; Zhao, Y.; et al. dNK derived IFN-γ mediates VSMC migration and apoptosis via the induction of LncRNA MEG3: A role in uterovascular transformation. Placenta 2017, 50, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Zhao, D.; Ohkuchi, A.; Kuwata, T.; Yoshitake, H.; Yuge, K.; Takizawa, T.; Matsubara, S.; Suzuki, M.; Saito, S.; et al. Maternal peripheral blood natural killer cells incorporate placenta-associated microRNAs during pregnancy. Int. J. Mol. Med. 2015, 35, 1511–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hromadnikova, I.; Kotlabova, K.; Ivankova, K.; Krofta, L. First trimester screening of circulating C19MC microRNAs and the evaluation of their potential to predict the onset of preeclampsia and IUGR. PLoS ONE 2017, 12, e0171756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hromadnikova, I.; Dvorakova, L.; Kotlabova, K.; Krofta, L. The Prediction of Gestational Hypertension, Preeclampsia and Fetal Growth Restriction via the First Trimester Screening of Plasma Exosomal C19MC microRNAs. Int. J. Mol. Sci. 2019, 20, 2972. [Google Scholar] [CrossRef]
- Jiang, L.; Long, A.; Tan, L.; Hong, M.; Wu, J.; Cai, L.; Li, Q. Elevated microRNA-520g in pre-eclampsia inhibits migration and invasion of trophoblasts. Placenta 2017, 51, 70–75. [Google Scholar] [CrossRef]
- Ura, B.; Feriotto, G.; Monasta, L.; Bilel, S.; Zweyer, M.; Celeghini, C. Potential role of circulating microRNAs as early markers of preeclampsia. Taiwan J. Obstet. Gynecol. 2014, 53, 232–234. [Google Scholar] [CrossRef] [Green Version]
- Licini, C.; Avellini, C.; Picchiassi, E.; Mensà, E.; Fantone, S.; Ramini, D.; Tersigni, C.; Tossetta, G.; Castellucci, C.; Tarquini, F.; et al. Pre-eclampsia predictive ability of maternal miR-125b: A clinical and experimental study. Transl. Res. 2021, 228, 13–27. [Google Scholar] [CrossRef]
- Martinez-Fierro, M.L.; Garza-Veloz, I. Analysis of Circulating microRNA Signatures and Preeclampsia Development. Cells 2021, 10, 1003. [Google Scholar] [CrossRef]
- Martinez-Fierro, M.L.; Carrillo-Arriaga, J.G.; Luevano, M.; Lugo-Trampe, A.; Delgado-Enciso, I.; Rodriguez-Sanchez, I.P.; Garza-Veloz, I. Serum levels of miR-628-3p and miR-628-5p during the early pregnancy are increased in women who subsequently develop preeclampsia. Pregnancy Hypertens. 2019, 16, 120–125. [Google Scholar] [CrossRef]
- Chen, D.; He, B.; Zheng, P.; Wang, S.; Zhao, X.; Liu, J.; Yang, X.; Cheng, W. Identification of mRNA-, circRNA- and lncRNA- Associated ceRNA Networks and Potential Biomarkers for Preeclampsia From Umbilical Vein Endothelial Cells. Front. Mol. Biosci. 2021, 8, 652250. [Google Scholar] [CrossRef]
- Timofeeva, A.V.; Gusar, V.A.; Kan, N.E.; Prozorovskaya, K.N.; Karapetyan, A.O.; Bayev, O.R.; Chagovets, V.V.; Kliver, S.F.; Iakovishina, D.Y.; Frankevich, V.E.; et al. Identification of potential early biomarkers of preeclampsia. Placenta 2018, 61, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Winger, E.E.; Reed, J.L.; Ji, X. First-trimester maternal cell microRNA is a superior pregnancy marker to immunological testing for predicting adverse pregnancy outcome. J. Reprod. Immunol. 2015, 110, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Winger, E.E.; Reed, J.L.; Ji, X.; Nicolaides, K. Peripheral blood cell microRNA quantification during the first trimester predicts preeclampsia: Proof of concept. PLoS ONE 2018, 13, e0190654. [Google Scholar] [CrossRef] [PubMed]
- Yoffe, L.; Gilam, A.; Yaron, O.; Polsky, A.; Farberov, L.; Syngelaki, A.; Nicolaides, K.; Hod, M.; Shomron, N. Early Detection of Preeclampsia Using Circulating Small non-coding RNA. Sci. Rep. 2018, 8, 3401. [Google Scholar] [CrossRef]
- Dai, C.; Zhao, C.; Xu, M.; Sui, X.; Sun, L.; Liu, Y.; Su, M.; Wang, H.; Yuan, Y.; Zhang, S.; et al. Serum lncRNAs in early pregnancy as potential biomarkers for the prediction of pregnancy-induced hypertension, including preeclampsia. Mol. Ther. Nucleic Acids 2021, 24, 416–425. [Google Scholar] [CrossRef]
- Tarca, A.L.; Romero, R.; Erez, O.; Gudicha, D.W.; Than, N.G.; Benshalom-Tirosh, N.; Pacora, P.; Hsu, C.D.; Chaiworapongsa, T.; Hassan, S.S.; et al. Maternal whole blood mRNA signatures identify women at risk of early preeclampsia: A longitudinal study. J. Matern.-Fetal Neonatal Med. 2021, 34, 3463–3474. [Google Scholar] [CrossRef]
- Hromadnikova, I.; Kotlabova, K.; Hympanova, L.; Doucha, J.; Krofta, L. First trimester screening of circulating C19MC microRNAs can predict subsequent onset of gestational hypertension. PLoS ONE 2014, 9, e113735. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Turturici, G.; Tinnirello, R.; Sconzo, G.; Geraci, F. Extracellular membrane vesicles as a mechanism of cell-to-cell communication: Advantages and disadvantages. Am. J. Physiol. Cell Physiol. 2014, 306, C621–C633. [Google Scholar] [CrossRef] [Green Version]
- Kambe, S.; Yoshitake, H.; Yuge, K.; Ishida, Y.; Ali, M.M.; Takizawa, T.; Kuwata, T.; Ohkuchi, A.; Matsubara, S.; Suzuki, M.; et al. Human exosomal placenta-associated miR-517a-3p modulates the expression of PRKG1 mRNA in Jurkat cells. Biol. Reprod. 2014, 91, 129. [Google Scholar] [CrossRef] [Green Version]
- Vashukova, E.S.; Kozyulina, P.Y.; Illarionov, R.A.; Yurkina, N.O.; Pachuliia, O.V.; Butenko, M.G.; Postnikova, T.B.; Ivanova, L.A.; Eremeeva, D.R.; Zainulina, M.S.; et al. High-Throughput Sequencing of Circulating MicroRNAs in Plasma and Serum during Pregnancy Progression. Life 2021, 11, 1055. [Google Scholar] [CrossRef] [PubMed]

| Author | Type of ncRNA | Study Method | Features | Sampling Time | Sample Size | Reference |
|---|---|---|---|---|---|---|
| Morales-Prieto (2012) | miRNA | Microarray analysis (1st vs. 3rd trimester placentas) ↓ Validation by RT-PCR | ・C19MC miRNAs significantly increased from 1st to 3rd trimester trophoblasts. ・C14MC miRNA significantly decreased from 1st to 3rd trimester trophoblasts. ・miR-14, miR-21, and let-7g were highly expressed throughout pregnancy. ・In the clustering analysis, C14MC miRNAs were close to JEG3, a choriocarcinoma cell line, while C19MC miRNAs were close to HTR-8/SVneo, an EVT cell line. | 1st trimester | 3 | [51] |
| 3rd trimester | 3 | |||||
| Luo (2009) | miRNA | Small RNA library sequencing | ・30–40% of the miRNAs highly expressed in 1st trimester placentas were C19MC miRNAs. | 1st trimester (7–11 w) | placenta: 6, plasma: 3 | [44] |
| 3rd trimester (36–38 w) | placenta: 6, plasma: 3 | |||||
| ISH | ・C19MC miRNAs were more highly expressed in SCT than in CVT. | 1st trimester | ||||
| RT-PCR analysis of C19MC miRNAs in adult organs | ・miR-517a, miR-517b, miR-518b, miR-519A, and miR-512-3p were expressed exclusively in the placenta among adult organs. | NA | ||||
| RT-PCR comparative analysis of C19MC miRNAs in maternal plasma (before vs. after delivery) | ・The expression levels of C19MC miRNAs (miR-517a and miR-518b) in maternal plasma markedly decreased after delivery. | 1 day before delivery 3 days after delivery | 6 6 | |||
| Miura (2010) | miRNA | Microarray analysis (1st vs. 3rd trimester placentas/plasma) | ・Among 82 placenta-predominant miRNAs (>100-fold higher in the placenta than in plasma), C19MC miRNAs and C14MC miRNAs accounted for 53.7 and 15.9%, respectively. | 1st trimester (12–13 w) 3rd trimester (38–39 w) | placenta: 2, plasma: 2 placenta: 2, plasma: 2 | [52] |
| RT-PCR comparative analysis using plasma samples (1st vs. 3rd trimester vs. post delivery) | ・Among placenta-predominant miRNAs, 24 miRNAs significantly decreased after delivery, particularly miR-515-3p, miR-517a, miR-517c, miR-518b, miR-526b (C19MC), and miR-323-3p (C14MC). | 1st trimester (12–13 w) 3rd trimester (38–39 w) Day 1 after delivery | 10 10 10 | |||
| Ogoyama (2021) | miRNA | RNA sequencing for EVT and CVT isolated from human early placentas | ・The expression of C14MC miRNAs was significantly lower in EVT than in CVT. | 7 w | 3 | [45] |
| lncRNA | ・The expression level of lncRNA H19 accounted for 70% of all lncRNAs in EVT. | |||||
| Gonzalez (2021) | miRNA | miRNA sequencing (1st vs. 3rd trimester placentas) | ・C19MC miRNAs significantly increased from 1st to 3rd trimester trophoblasts. ・C14MC miRNA significantly decreased from 1st to 3rd trimester trophoblasts. | 1st trimester 3rd trimester | 113 47 | [46] |
| Smith (2021) | miRNA | miRNA sequencing using placentas and plasma from 6–23 w of gestation. ↓ Expression comparison (6–10 w vs. 11–23 w) | ・The top 10 miRNAs with the highest expression levels in the placenta were miR-30d-5p, miR-125a-5p, miR-517a-3p*, miR-199a-3p, miR-26b-5p, miR-26a-5p, let-7a-5p, miR-21-5p, miR-126-3p, and miR-516b-5p* (C19MC*). ・Thirty-four out of 48 C19MC miRNAs (75%) were downregulated in 11–23 w of gestation. ・Fifty-six out of 77 C14MC miRNAs (73%) were up-regulated in 11–23 w of gestation. ・The top highest miRNAs in the placenta were also highly expressed in plasma. | 6–10 w 11–23 w | total 86 | [47] |
| No | Author | ncRNA | Type of ncRNA | Study Method | Features | Sampling Time | Sample Size | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | Wang (2012) | miR-517b | miRNA (C19MC) | ISH and RT-PCR | ・miR-517b and miR-519a were located in the trophoblast layer. | 6–9 w | ISH: 1 in each GW | [53] |
| miR-519a | RT-PCR: 10 in each GW | |||||||
| 2 | Mong (2020) | miR-517a miR-517c | miRNA (C19MC) | ISH | ・miR-517a/c were more highly expressed in VT than in EVT. | 7–8 w | 2 | [54] |
| Functional analysis using EVT-like cells differentiated from iPSC | ・When iPSC differentiated into EVT cells, miR-517a/c expression decreased and EMT-related gene expression increased. | NA | NA | |||||
| 3 | Takahashi (2017) | miR-520c-3p | miRNA (C19MC) | RT-PCR following LMD | ・miR-520c-3p was highly expressed in the maternal decidua stroma. | 7–9 w | 5 | [42] |
| RT-PCR using human EVT cells isolated from 1st trimester placentas | ・miR-520c-3p was more highly expressed in VT than in EVT. | 7–11 w | 6 | |||||
| ・miR-520c-3p directly inhibited CD44. | ||||||||
| 4 | Xie (2014) | miR-517-3p | miRNA (C19MC) | RT-PCR comparison analysis (VT vs. EVT cells obtained from 1st trimester placentas by LMD) | ・C19MC miRNAs were more highly expressed in VT than in EVT. | 6–12 w | 7 | [43] |
| miR-518b | ||||||||
| miR-519d | ||||||||
| miR-520g | Functional analysis using a human EVT cell line transfected with the C19MC cluster by a plasmid vector | ・C19MC miRNAs attenuated the migration of an EVT cell line. | NA | NA | ||||
| miR-515-5p | ||||||||
| miR-1323 | ||||||||
| 5 | Flor (2012) | miR-520-3p miR-519a-3p miR-517a-3p | miRNA (C19MC) | RT-PCR | ・C19MC miRNAs were highly expressed in 1st trimester placentas. | 10–14 w | 4–20 | [55] |
| DNA methylation analysis | ・The CpG island upstream of the C19MC cluster in the paternal allele escaped methylation. | NA | NA | |||||
| 6 | Hayder (2021) | miR-210-3p | miRNA | RT-PCR comparison analysis (among 1st trimester, 2nd trimester, preterm, and term placentas) | ・The expression level of miR-210-3p decreased as gestational weeks increased. | 1st: 5–12 w | 8 | [56] |
| 2nd: 13–25 w | 10 | |||||||
| preterm: 26–36 w | 13 | |||||||
| term: 37–40 w | 17 | |||||||
| Culture of EVT cells isolated from human 1st trimester placentas and a functional analysis | ・miR-210-3p inhibited the migration and outgrowth of EVT cells. | 6–9 w | 10 in the miR-210-3p group 11 in the control group | |||||
| ・miR-210-3p directly inhibited CDX, a major transcription factor responsible for trophoblast self-renewal. | ||||||||
| 7 | Wang (2020) | miR-210 | miRNA | ISH | ・miR-210 and its direct target CPEB2 (HIF1α inhibitor) were co-located in SCT and VT. | 7–8 w | NA | [57] |
| Functional analysis using a human trophoblast cell line | ・Positive feedback was shown between miR-210 and HIFα mediated by CPEB1. | NA | NA | |||||
| 8 | Wang (2021) | miR-210 | miRNA | ISH | ・miR-210 and MEG3 were highly expressed in human 1st trimester trophoblasts. ・As VTs differentiated into EVTs and invaded the maternal decidua, miR-210 expression decreased. | 6–9 w | NA | [58] |
| MEG3 | lncRNA | Functional analysis using a human EVT cell line | ・miR-210 inhibited the EMT of an EVT cell line by suppressing MEG3 expression. | NA | NA | |||
| 9 | Gu (2013) | miR-371-5p | miRNA | Microarray analysis (1st vs. 3rd trimester placentas) ↓ Validation by RT-PCR and ISH | ・The expression levels of miR371-5p, miR-17-3p, and miR-708-5p were significantly higher in 1st trimester placentas | 6–8 w (1st) 30–38 w (3rd) | microarray: 6 in each group RT-PCR: 6 in each group ISH: 1 in each group | [59] |
| miR-17-3p | ||||||||
| miR-708-5p | ||||||||
| 10 | Singh (2017) | miR-202-3p | miRNA | Microarray analysis (placentas of women who later developed severe PE vs. those who did not ) ↓ Validation by RT-PCR | ・Nine miRNAs were differentially expressed in the severe PE onset group at 11–13 w. ・The expression of miR-202-3p was 7-fold higher in the severe PE onset group at 11–13 w. | 11–13 w | 4 in each group | [60] |
| 11 | Wu (2016) | miR-195 | miRNA | ISH | ・miR-195 and its direct target ActR2B were co-located in VT, SCT, and EVT. | 7–9 w | NA | [61] |
| Functional analysis using a human EVT cell line | ・miR-195 accelerated the invasion of an EVT cell line. | NA | NA | |||||
| 12 | Zeng (2020) | H19 | lncRNA | ISH RT-PCR comparison analysis (normal vs. RM group) | ・H19 was located in trophoblasts, particularly EVT. ・The expression level of H19 was significantly lower in the RM group. | 6–12 w | ISH: 1 in each group RT-PCR: 12 in each group | [62] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogoyama, M.; Takahashi, H.; Suzuki, H.; Ohkuchi, A.; Fujiwara, H.; Takizawa, T. Non-Coding RNAs and Prediction of Preeclampsia in the First Trimester of Pregnancy. Cells 2022, 11, 2428. https://doi.org/10.3390/cells11152428
Ogoyama M, Takahashi H, Suzuki H, Ohkuchi A, Fujiwara H, Takizawa T. Non-Coding RNAs and Prediction of Preeclampsia in the First Trimester of Pregnancy. Cells. 2022; 11(15):2428. https://doi.org/10.3390/cells11152428
Chicago/Turabian StyleOgoyama, Manabu, Hironori Takahashi, Hirotada Suzuki, Akihide Ohkuchi, Hiroyuki Fujiwara, and Toshihiro Takizawa. 2022. "Non-Coding RNAs and Prediction of Preeclampsia in the First Trimester of Pregnancy" Cells 11, no. 15: 2428. https://doi.org/10.3390/cells11152428







