Bioinformatic Analyses of Broad H3K79me2 Domains in Different Leukemia Cell Line Data Sets
Abstract
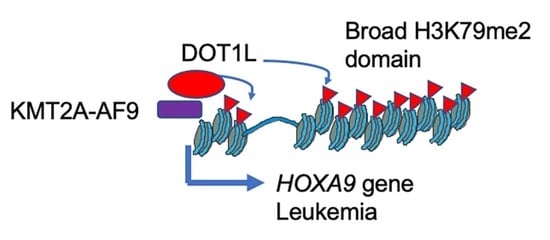
:1. Introduction
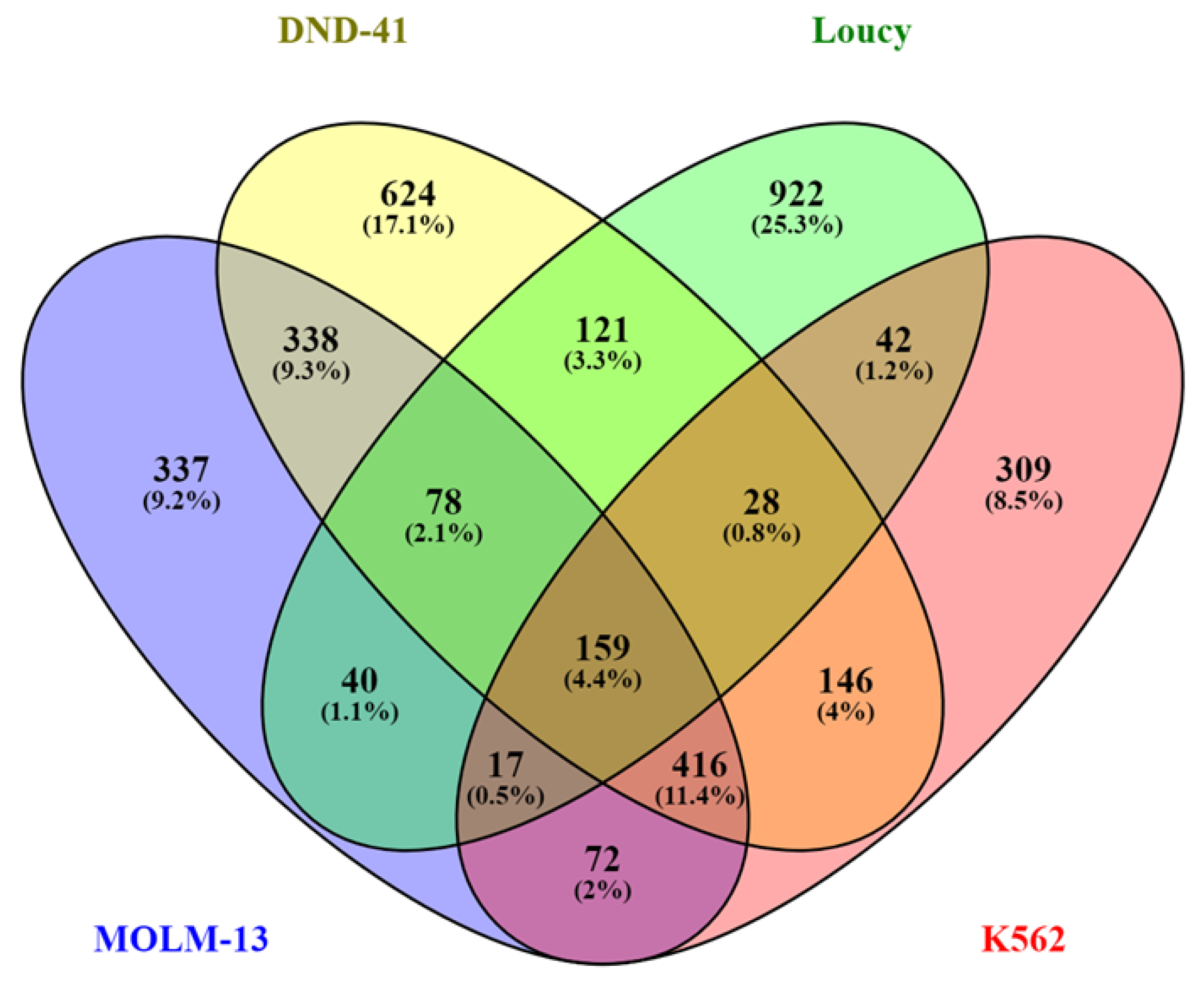
2. Results
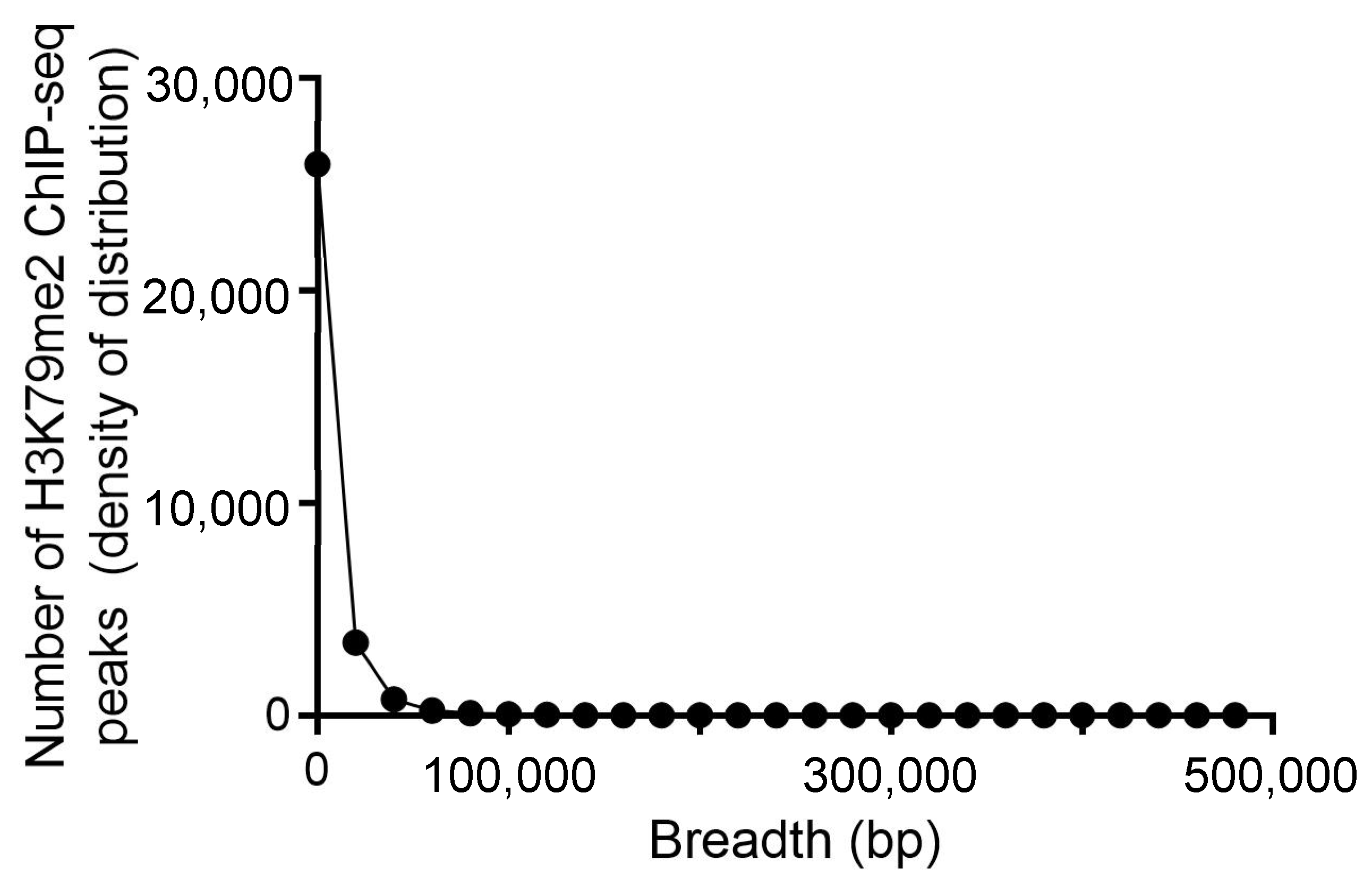
2.1. Identification of Genes with a Broad H3K79me2 Domain in Leukemic Cells
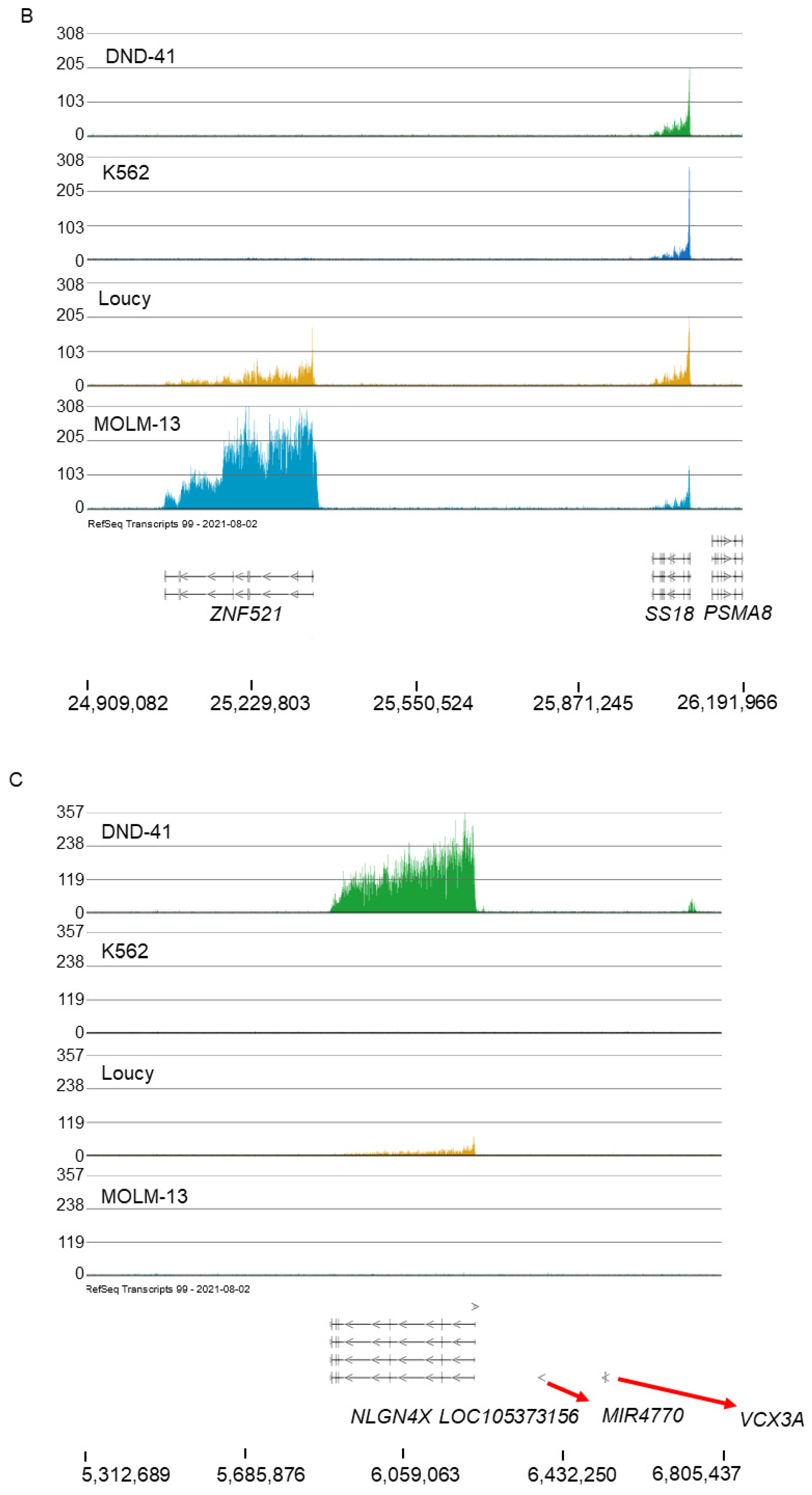
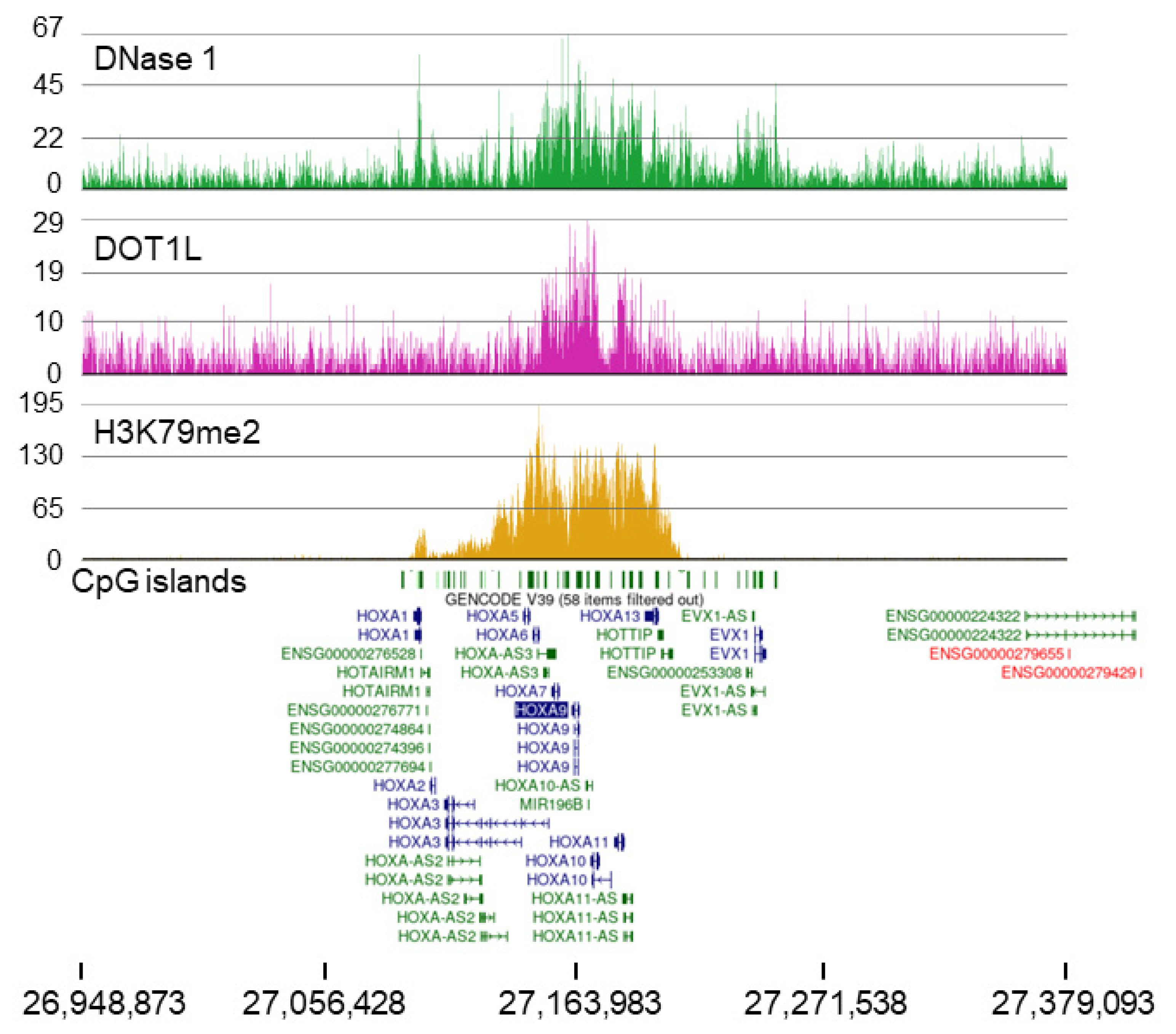
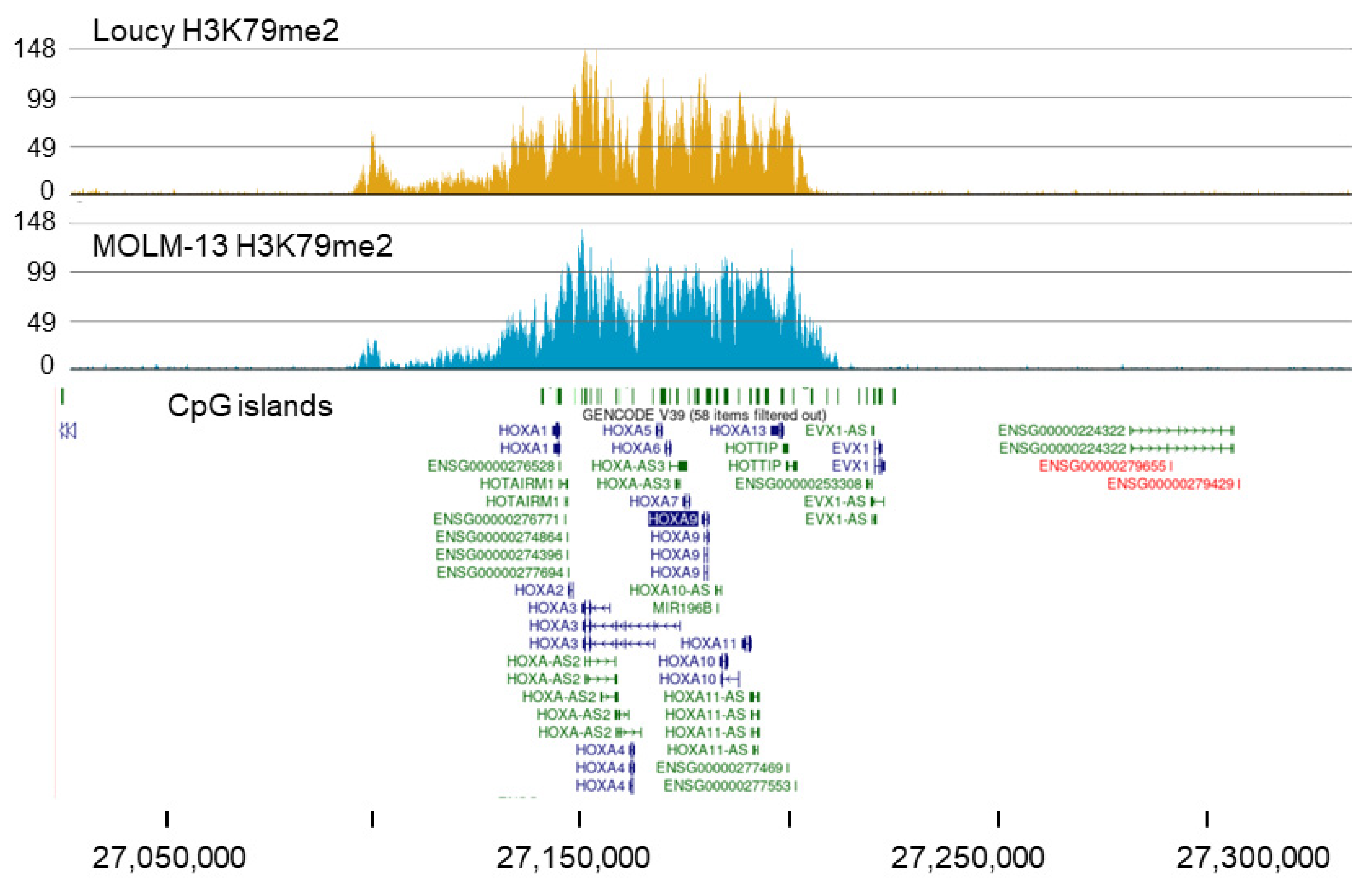
2.2. MOLM-13 and HOXA9 Cluster
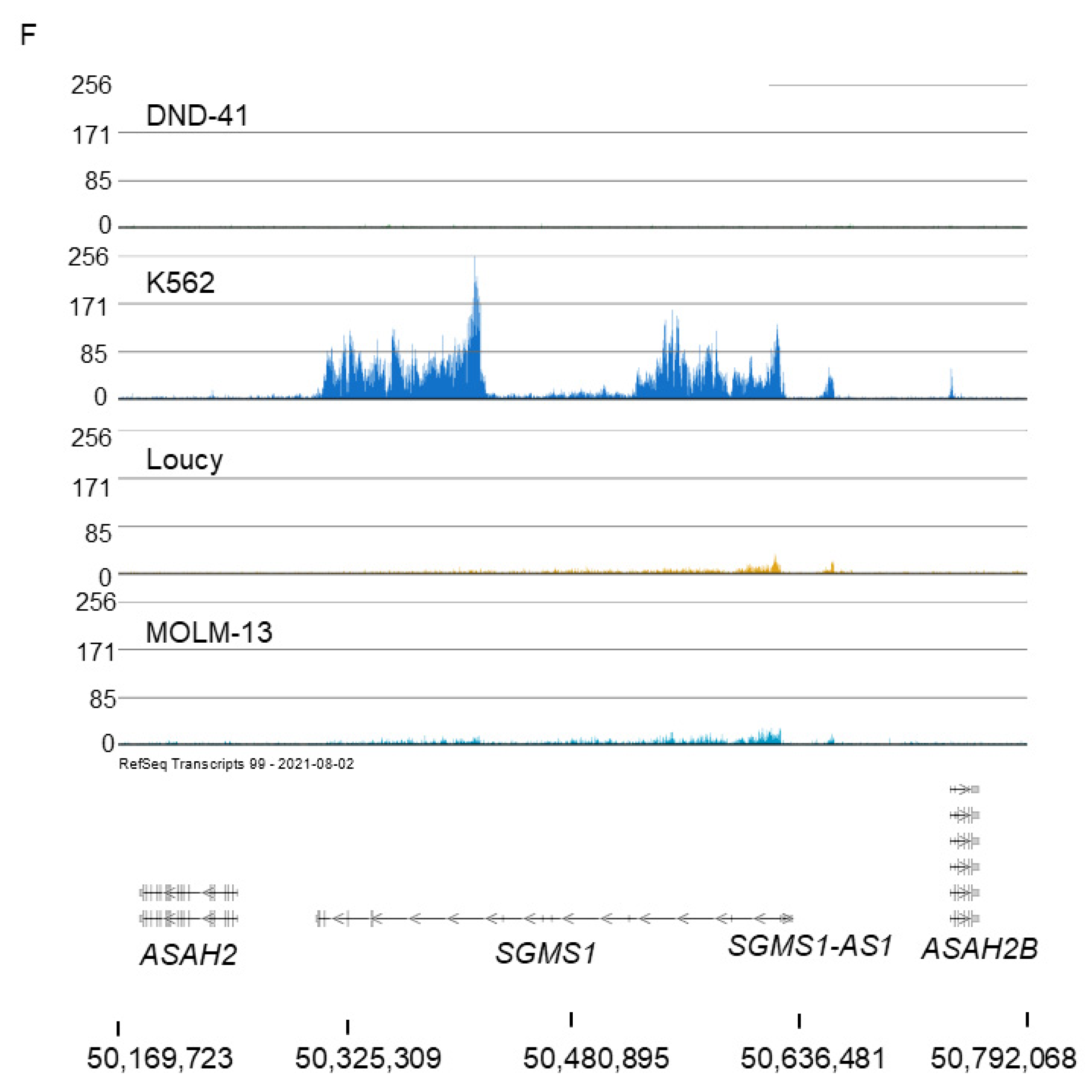
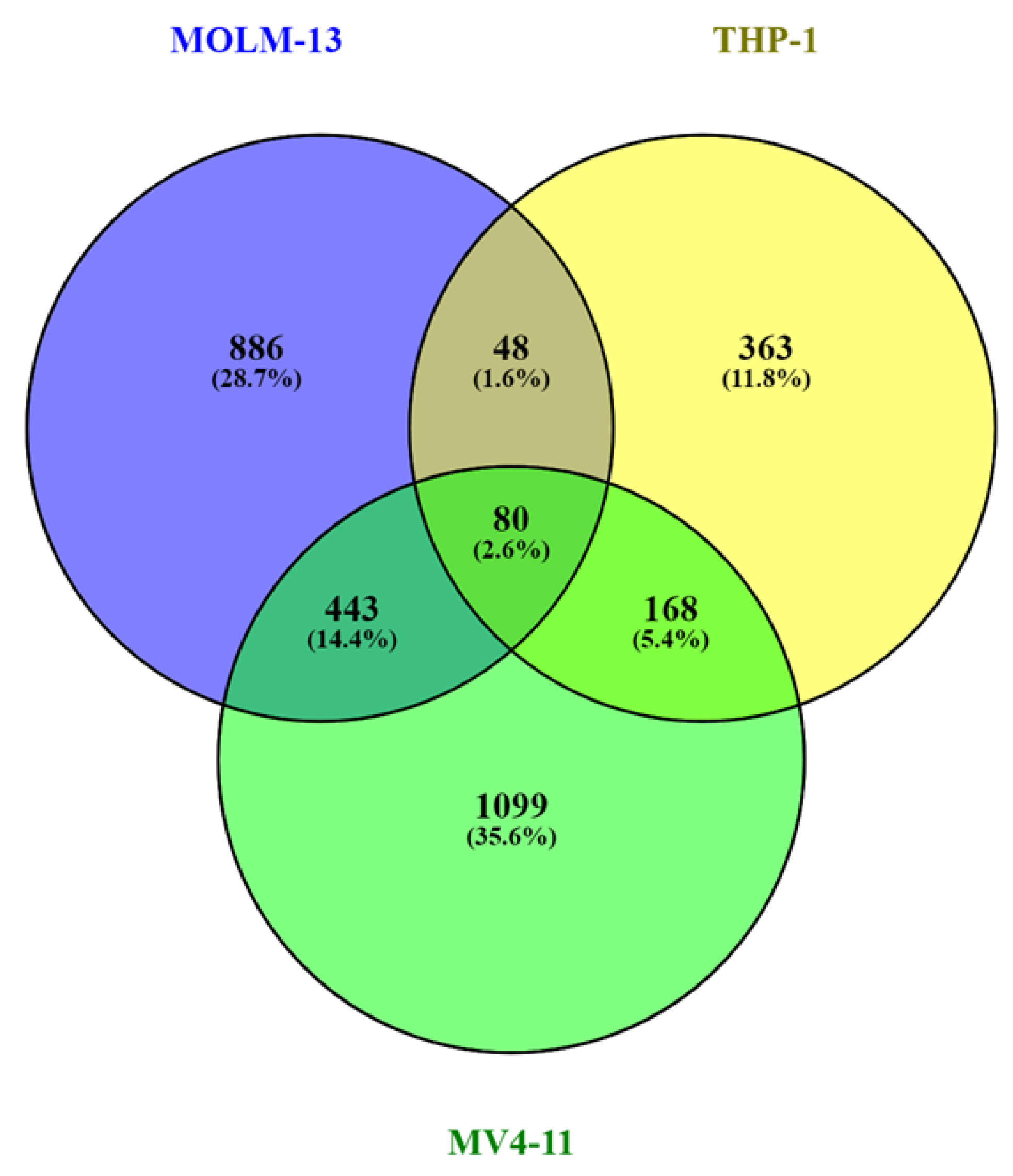
2.3. Broad H3K79me2 Domain in AML Cell Lines
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Data Collection
5.2. ChIP Sequence Analyses (MACS2)
5.3. Broad H3K79me2 Domain
5.4. RNA Sequence Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Muntean, A.G.; Hess, J.L. The Pathogenesis of Mixed-Lineage Leukemia. Annu. Rev. Pathol. Mech. Dis. 2012, 7, 283–301. [Google Scholar] [CrossRef] [PubMed]
- Marschalek, R. Systematic Classification of Mixed-Lineage Leukemia Fusion Partners Predicts Additional Cancer Pathways. Ann. Lab. Med. 2016, 36, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Marschalek, R. MLL Leukemia and Future Treatment Strategies. Arch. Pharm. 2015, 348, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Slany, R.K. The molecular biology of mixed lineage leukemia. Haematologica 2009, 94, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Castiglioni, S.; Di Fede, E.; Bernardelli, C.; Lettieri, A.; Parodi, C.; Grazioli, P.; Colombo, E.A.; Ancona, S.; Milani, D.; Ottaviano, E.; et al. KMT2A: Umbrella Gene for Multiple Diseases. Genes 2022, 13, 514. [Google Scholar] [CrossRef]
- Britten, O.; Ragusa, D.; Tosi, S.; Kamel, Y.M. MLL-Rearranged Acute Leukemia with t(4;11)(Q21;Q23)-Current Treatment Options. Is There a Role for CAR-T Cell Therapy? Cells 2019, 8, 1341. [Google Scholar] [CrossRef]
- Yokoyama, A. Leukemogenesis via aberrant self-renewal by the MLL/AEP-mediated transcriptional activation system. Cancer Sci. 2021, 112, 3935–3944. [Google Scholar] [CrossRef]
- Collins, C.T.; Hess, J.L. Role of HOXA9 in leukemia: Dysregulation, cofactors and essential targets. Oncogene 2015, 35, 1090–1098. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; Armstrong, S.A. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 2007, 7, 823–833. [Google Scholar] [CrossRef]
- Matsuo, Y.; MacLeod, R.; Uphoff, C.; Drexler, H.; Nishizaki, C.; Katayama, Y.; Kimura, G.; Fujii, N.; Omoto, E.; Harada, M.; et al. Two acute monocytic leukemia (AML-M5a) cell lines (MOLM-13 and MOLM-14) with interclonal phenotypic heterogeneity showing MLL-AF9 fusion resulting from an occult chromosome insertion, ins(11;9)(q23;p22p23). Leukemia 1997, 11, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Min, J.; Feng, Q.; Li, Z.; Zhang, Y.; Xu, R.-M.; Keck, W.M. Structure of the Catalytic Domain of Human DOT1L, a Non-SET Domain Nucleosomal Histone Methyltransferase Mosomes during the Cell Division (Grewal and Elgin, 2002). A Recent Addition to the Histone Modification Reper-Toire Is Histone H3 Lysine-79 (Lys79) M. Cell 2003, 112, 711–723. [Google Scholar] [CrossRef]
- Wood, K.; Tellier, M.; Murphy, S. DOT1L and H3K79 Methylation in Transcription and Genomic Stability. Biomolecules 2018, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Daigle, S.R.; Olhava, E.J.; Therkelsen, C.A.; Basavapathruni, A.; Jin, L.; Boriack-Sjodin, P.A.; Allain, C.J.; Klaus, C.R.; Raimondi, A.; Scott, M.P.; et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood 2013, 122, 1017–1025. [Google Scholar] [CrossRef]
- Sarno, F.; Nebbioso, A.; Altucci, L. DOT1L: A key target in normal chromatin remodelling and in mixed-lineage leukaemia treatment. Epigenetics 2019, 15, 439–453. [Google Scholar] [CrossRef]
- Li, Y.; Wen, H.; Xi, Y.; Tanaka, K.; Wang, H.; Peng, D.; Ren, Y.; Jin, Q.; Dent, S.Y.; Li, W.; et al. AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation. Cell 2014, 159, 558–571. [Google Scholar] [CrossRef]
- Wan, L.; Wen, H.; Li, Y.; Lyu, J.; Xi, Y.; Hoshii, T.; Joseph, J.K.; Wang, X.; Loh, Y.H.E.; Erb, M.A.; et al. ENL Links Histone Acetylation to Oncogenic Gene Expression in Acute Myeloid Leukaemia. Nature 2017, 543, 265–269. [Google Scholar] [CrossRef]
- Wang, X.; Chen, C.-W.; Armstrong, S.A. The role of DOT1L in the maintenance of leukemia gene expression. Curr. Opin. Genet. Dev. 2016, 36, 68–72. [Google Scholar] [CrossRef]
- Faber, J.; Krivtsov, A.V.; Stubbs, M.C.; Wright, R.; Davis, T.N.; Van Den Heuvel-Eibrink, M.; Zwaan, C.M.; Kung, A.L.; Armstrong, S.A. HOXA9 Is Required for Survival in Human MLL-Rearranged Acute Leukemias. Blood 2009, 113, 2375–2385. [Google Scholar] [CrossRef]
- Wu, A.; Zhi, J.; Tian, T.; Cihan, A.; Cevher, M.A.; Liu, Z.; David, Y.; Muir, T.W.; Roeder, R.G.; Yu, M. DOT1L complex regulates transcriptional initiation in human erythroleukemic cells. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Beacon, T.H.; Delcuve, G.P.; López, C.; Nardocci, G.; Kovalchuk, I.; van Wijnen, A.J.; Davie, J.R. The dynamic broad epigenetic (H3K4me3, H3K27ac) domain as a mark of essential genes. Clin. Epigenetics 2021, 13, 138. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Chen, Z.; Wu, D.; Zhang, L.; Lin, X.; Su, J.; Rodriguez, B.; Xi, Y.; Xia, Z.; Chen, X.; et al. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat. Genet. 2015, 47, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Benayoun, B.A.A.; Pollina, E.A.A.; Ucar, D.; Mahmoudi, S.; Karra, K.; Wong, E.D.D.; Devarajan, K.; Daugherty, A.C.C.; Kundaje, A.B.B.; Mancini, E.; et al. H3K4me3 Breadth Is Linked to Cell Identity and Transcriptional Consistency. Cell 2014, 158, 673–688. [Google Scholar] [CrossRef]
- López, C.; Barnon, M.T.; Beacon, T.H.; Nardocci, G.; Davie, J.R. The key role of differential broad H3K4me3 and H3K4ac domains in breast cancer. Gene 2022, 826. [Google Scholar] [CrossRef] [PubMed]
- Belhocine, M.; Simonin, M.; Flores, J.D.A.; Cieslak, A.; Manosalva, I.; Pradel, L.; Smith, C.; Mathieu, E.-L.; Charbonnier, G.; Martens, J.H.; et al. Dynamics of broad H3K4me3 domains uncover an epigenetic switch between cell identity and cancer-related genes. Genome Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Chen, M.; Eng, R.; DeJong, J.; Sinha, A.U.; Rahnamay, N.F.; Koche, R.; Al-Shahrour, F.; Minehart, J.; Chen, C.-W.; et al. MLL-AF9– and HOXA9-mediated acute myeloid leukemia stem cell self-renewal requires JMJD1C. J. Clin. Investig. 2016, 126, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Lozzio, B.B.; Lozzio, C.B. Properties and usefulness of the original K-562 human myelogenous leukemia cell line. Leuk. Res. 1979, 3, 363–370. [Google Scholar] [CrossRef]
- Gribble, S.M.; Roberts, I.; Grace, C.; Andrews, K.M.; Green, A.R.; Nacheva, E.P. Cytogenetics of the Chronic Myeloid Leukemia-Derived Cell Line K562. Cancer Genet. Cytogenet. 2000, 118, 1–8. [Google Scholar] [CrossRef]
- Naumann, S.; Reutzel, D.; Speicher, M.; Decker, H.-J. Complete karyotype characterization of the K562 cell line by combined application of G-banding, multiplex-fluorescence in situ hybridization, fluorescence in situ hybridization, and comparative genomic hybridization. Leuk. Res. 2001, 25, 313–322. [Google Scholar] [CrossRef]
- Prokocimer, M.; Peller, S.; Ben-Bassat, H.; Goldfinger, N.; Rotter, V. P53 Gene Mutation in a T-Acute Lymphoblastic Leukemia Cell Line (Loucy) with t(16:20) and 5q-Chromosomal Aberrations. Leuk. Lymphoma 1998, 29, 607–611. [Google Scholar] [CrossRef]
- Minowada, J.; Tsubota, T.; Nakazawa, S.; Srivastava, B.I.S.; Huang, C.C.; Oshimura, M.; Sonta, S.; Han, T.; Sinks, L.F.; Sandberg, A.A. Establishment and Characterization of Leukemic T-Cell Lines, B-Cell Lines, and Null-Cell Line: A Progress Report on Surface Antigen Study of Fresh Lymphatic Leukemias in Man. In Immunological Diagnosis of Leukemias and Lymphomas; Springer: Berlin/Heidelberg, Germany, 1977; Volume 20, pp. 241–251. [Google Scholar] [CrossRef]
- Aziz, N.A.A.; Mokhtar, N.M.; Harun, R.; Mollah, M.M.H.; Rose, I.M.; Sagap, I.; Tamil, A.M.; Ngah, W.Z.W.; Jamal, R. A 19-Gene expression signature as a predictor of survival in colorectal cancer. BMC Med. Genom. 2016, 9, 58. [Google Scholar] [CrossRef]
- Al Barashdi, M.A.; Ali, A.; McMullin, M.F.; Mills, K. Protein tyrosine phosphatase receptor type C (PTPRC or CD45). J. Clin. Pathol. 2021, 74, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Henderson, H.J.; Karanam, B.; Samant, R.; Vig, K.; Singh, S.R.; Yates, C.; Bedi, D. Neuroligin 4X overexpression in human breast cancer is associated with poor relapse-free survival. PLoS ONE 2017, 12, e0189662. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Kim, H.S.; Hwang, J.; Kim, Y.H.; Lim, G.Y.; Sohn, W.J.; Yoon, S.R.; Kim, J.Y.; Park, T.S.; Oh, S.H.; et al. Regulation of HOXA9 Activity by Predominant Expression of DACH1 against C/EBPα and GATA-1 in Myeloid Leukemia with MLL-AF9. Biochem. Biophys. Res. Commun. 2012, 426, 299–305. [Google Scholar] [CrossRef]
- Germano, G.; Morello, G.; Aveic, S.; Pinazza, M.; Minuzzo, S.; Frasson, C.; Persano, L.; Bonvini, P.; Viola, G.; Bresolin, S.; et al. ZNF521 sustains the differentiation block in MLL-rearranged acute myeloid leukemia. Oncotarget 2017, 8, 26129–26141. [Google Scholar] [CrossRef]
- NCBI. Genes & Expression. Gene. Available online: https://www.ncbi.nlm.nih.gov/gene/ (accessed on 23 April 2021).
- Gatford, N.J.F.; Deans, P.J.M.; Duarte, R.R.R.; Chennell, G.; Sellers, K.J.; Raval, P.; Srivastava, D.P. Neuroligin-3 and neuroligin-4X form nanoscopic clusters and regulate growth cone organization and size. Hum. Mol. Genet. 2021, 31, 674–691. [Google Scholar] [CrossRef]
- Allen, C.E.; Mak, C.-H.; Wu, L.-C. The κB transcriptional enhancer motif and signal sequences of V(D)J recombination are targets for the zinc finger protein HIVEP3/KRC: A site selection amplification binding study. BMC Immunol. 2002, 3, 10. [Google Scholar] [CrossRef]
- Krovi, S.H.; Zhang, J.; Michaels-Foster, M.J.; Brunetti, T.; Loh, L.; Scott-Browne, J.; Gapin, L. Thymic iNKT single cell analyses unmask the common developmental program of mouse innate T cells. Nat. Commun. 2020, 11, 6238. [Google Scholar] [CrossRef]
- Juric, D.; Lacayo, N.J.; Ramsey, M.C.; Racevskis, J.; Wiernik, P.H.; Rowe, J.M.; Goldstone, A.H.; O’Dwyer, P.J.; Paietta, E.; Sikic, B.I. Differential Gene Expression Patterns and Interaction Networks in BCR-ABL–Positive and –Negative Adult Acute Lymphoblastic Leukemias. J. Clin. Oncol. 2007, 25, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Lv, G.; Wang, Y.; Ji, C.; Shi, C.; Li, Y. SPRY1 promotes cell proliferation and inhibits apoptosis by activating Hedgehog pathway in acute myeloid leukemia. Hematology 2021, 27, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bilal, F.; Montfort, A.; Gilhodes, J.; Garcia, V.; Riond, J.; Carpentier, S.; Filleron, T.; Colacios, C.; Levade, T.; Daher, A.; et al. Sphingomyelin Synthase 1 (SMS1) Downregulation Is Associated With Sphingolipid Reprogramming and a Worse Prognosis in Melanoma. Front. Pharmacol. 2019, 10, 443. [Google Scholar] [CrossRef] [PubMed]
- Argiropoulos, B.; Humphries, R.K. Hox genes in hematopoiesis and leukemogenesis. Oncogene 2007, 26, 6766–6776. [Google Scholar] [CrossRef] [PubMed]
- Rezsohazy, R.; Saurin, A.; Maurel-Zaffran, C.; Graba, Y. Cellular and molecular insights into Hox protein action. Development 2015, 142, 1212–1227. [Google Scholar] [CrossRef] [PubMed]
- Gabra, M.M.; Salmena, L. microRNAs and Acute Myeloid Leukemia Chemoresistance: A Mechanistic Overview. Front. Oncol. 2017, 7, 255. [Google Scholar] [CrossRef]
- Chen, L.; Hu, N.; Wang, C.; Zhao, H. HOTAIRM1 knockdown enhances cytarabine-induced cytotoxicity by suppression of glycolysis through the Wnt/β-catenin/PFKP pathway in acute myeloid leukemia cells. Arch. Biochem. Biophys. 2020, 680, 108244. [Google Scholar] [CrossRef]
- Van Vlierberghe, P.; Van Grotel, M.; Tchinda, J.; Lee, C.; Beverloo, H.B.; Van Der Spek, P.J.; Stubbs, A.; Cools, J.; Nagata, K.; Fornerod, M.; et al. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood 2008, 111, 4668–4680. [Google Scholar] [CrossRef]
- Mendes, A.; Fahrenkrog, B. NUP214 in Leukemia: It’s More than Transport. Cells 2019, 8, 76. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Liu, X.; Jiang, H.; Chen, B. A Promising Glycolysis- and Immune-Related Prognostic Signature for Glioblastoma. World Neurosurg. 2022, 161, e363–e375. [Google Scholar] [CrossRef]
- Dabiri, A.; Sharifi, M.; Sarmadi, A. Knockdown of SOX12 Expression Induced Apoptotic Factors Is Associated with TWIST1 and CTNNB1 Expression in Human Acute Myeloid Leukemia Cells. Int. J. Mol. Cell. Med. 2021, 10, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, W.; Yan, X.J.; Lin, X.Q.; Li, W.; Mi, J.Q.; Li, J.M.; Zhu, J.; Chen, Z.; Chen, S.J. DNMT3A Mutation Leads to Leukemic Extramedullary Infiltration Mediated by TWIST1. J. Hematol. Oncol. 2016, 9, 106. [Google Scholar] [CrossRef] [PubMed]
- Jukonen, J.; Moyano-Galceran, L.; Höpfner, K.; Pietilä, E.A.; Lehtinen, L.; Huhtinen, K.; Gucciardo, E.; Hynninen, J.; Hietanen, S.; Grénman, S.; et al. Aggressive and recurrent ovarian cancers upregulate ephrinA5, a non-canonical effector of EphA2 signaling duality. Sci. Rep. 2021, 11, 8856. [Google Scholar] [CrossRef]
- Xie, J.; Xing, S.; Shen, B.-Y.; Chen, H.-T.; Sun, B.; Wang, Z.-T.; Wang, J.-W.; Lu, X.-X. PIWIL1 interacting RNA piR-017061 inhibits pancreatic cancer growth via regulating EFNA5. Hum. Cell 2021, 34, 550–563. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Kakadia, P.M.; Chen, Y.; Li, Y.-Q.; Deshpande, A.J.; Buske, C.; Zhang, K.-L.; Zhang, Y.; Xu, G.-L.; Bohlander, S.K. Global reduction of the epigenetic H3K79 methylation mark and increased chromosomal instability in CALM-AF10–positive leukemias. Blood 2009, 114, 651–658. [Google Scholar] [CrossRef]
- Vlaming, H.; McLean, C.M.; Korthout, T.; Alemdehy, M.F.; Hendriks, S.; Lancini, C.; Palit, S.; Klarenbeek, S.; Kwesi-Maliepaard, E.M.; Molenaar, T.M.; et al. Conserved crosstalk between histone deacetylation and H3K79 methylation generates DOT1L-dose dependency in HDAC1-deficient thymic lymphoma. EMBO J. 2019, 38, e101564. [Google Scholar] [CrossRef]
- Zhang, K.; Siino, J.S.; Jones, P.R.; Yau, P.M.; Bradbury, E.M. A mass spectrometric ?Western blot? to evaluate the correlations between histone methylation and histone acetylation. PROTEOMICS 2004, 4, 3765–3775. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, G.; Hollander, D.; Voichek, Y.; Ast, G.; Oren, M. Cotranscriptional histone H2B monoubiquitylation is tightly coupled with RNA polymerase II elongation rate. Genome Res. 2014, 24, 1572–1583. [Google Scholar] [CrossRef]
- Davie, J.R.; Murphy, L.C. Level of ubiquitinated histone H2B in chromatin is coupled to ongoing transcription. Biochemistry 1990, 29, 4752–4757. [Google Scholar] [CrossRef]
- Martin, B.J.E.; Brind’Amour, J.; Kuzmin, A.; Jensen, K.N.; Liu, Z.C.; Lorincz, M.; Howe, L.J. Transcription shapes genome-wide histone acetylation patterns. Nat. Commun. 2021, 12, 210. [Google Scholar] [CrossRef]
- Zhang, L.Q.; Fan, G.L.; Liu, J.J.; Liu, L.; Li, Q.Z.; Lin, H. Identification of Key Histone Modifications and Their Regulatory Regions on Gene Expression Level Changes in Chronic Myelogenous Leukemia. Front. Cell Dev. Biol. 2021, 8, 621578. [Google Scholar] [CrossRef] [PubMed]
- Richter, W.F.; Shah, R.N.; Ruthenburg, A.J. Non-canonical H3K79me2-dependent pathways promote the survival of MLL-rearranged leukemia. eLife 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Kang, C.; Yang, H.-S.; Jung, T.; Hebert, H.; Chung, K.Y.; Kim, S.J.; Hohng, S.; Song, J.-J. Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase. Genes Dev. 2019, 33, 620–625. [Google Scholar] [CrossRef] [PubMed]
- Parker, S.C.J.; Stitzel, M.L.; Taylor, D.L.; Orozco, J.M.; Erdos, M.R.; Akiyama, J.A.; van Bueren, K.L.; Chines, P.S.; Narisu, N.; Black, B.L.; et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. USA 2013, 110, 17921–17926. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Liu, Q.; Garza, N.; Kornblau, S.; Jin, V.X. Integrative analysis reveals functional and regulatory roles of H3K79me2 in mediating alternative splicing. Genome Med. 2018, 10, 30. [Google Scholar] [CrossRef]
- Izaguirre-Carbonell, J.; Christiansen, L.; Burns, R.; Schmitz, J.; Li, C.; Mokry, R.L.; Bluemn, T.; Zheng, Y.; Shen, J.; Carlson, K.-S.; et al. Critical role of Jumonji domain of JMJD1C in MLL-rearranged leukemia. Blood Adv. 2019, 3, 1499–1511. [Google Scholar] [CrossRef]
- Chen, M.; Zhu, N.; Liu, X.; Laurent, B.; Tang, Z.; Eng, R.; Shi, Y.; Armstrong, S.A.; Roeder, R.G. JMJD1C is required for the survival of acute myeloid leukemia by functioning as a coactivator for key transcription factors. Genes Dev. 2015, 29, 2123–2139. [Google Scholar] [CrossRef]
- Wang, F.; Li, Z.; Wang, G.; Tian, X.; Zhou, J.; Yu, W.; Fan, Z.; Dong, L.; Lu, J.; Xu, J.; et al. Integrated transcriptomic and epigenetic data analysis identifiesaberrant expression of genes in acute myeloid leukemia with MLL-AF9 translocation. Mol. Med. Rep. 2019, 21, 883–893. [Google Scholar] [CrossRef]
- Shah, R.; Grzybowski, A.; Cornett, E.; Johnstone, A.L.; Dickson, B.M.; Boone, B.A.; Cheek, M.A.; Cowles, M.W.; Maryanski, D.; Meiners, M.J.; et al. Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies. Mol. Cell 2018, 72, 162–177.e7. [Google Scholar] [CrossRef] [Green Version]
- Cierpicki, T.; Risner, L.E.; Grembecka, J.; Lukasik, S.M.; Popovic, R.; Omonkowska, M.; Shultis, D.D.; Zeleznik-Le, N.J.; Bushweller, J.H. Structure of the MLL CXXC domain–DNA complex and its functional role in MLL-AF9 leukemia. Nat. Struct. Mol. Biol. 2009, 17, 62–68. [Google Scholar] [CrossRef] [Green Version]


| MOLM-13 Top 5% | |||||
|---|---|---|---|---|---|
| Term | Overlap | p-Value | Adjusted p-Value | Odds Ratio | Combined Score |
| Phosphorylation (GO:0016310) | 70/400 | 5.35 × 10−12 | 2.09 × 10−8 | 2.785 | 72.295 |
| Positive regulation of transcription, DNA-templated (GO:0045893) | 144/1183 | 4.52 × 10−10 | 8.81 × 10−7 | 1.848 | 39.758 |
| Protein phosphorylation (GO:0006468) | 75/496 | 1.13 × 10−9 | 1.47 × 10−6 | 2.336 | 48.130 |
| Protein dephosphorylation (GO:0006470) | 32/139 | 4.05 × 10−9 | 3.95 × 10−6 | 3.869 | 74.773 |
| DND-41 Top 5% | |||||
| Term | Overlap | p-value | Adjusted p-value | Odds Ratio | Combined Score |
| Mitotic cell-cycle phase transition (GO:0044772) | 59/209 | 1.06 × 10−14 | 4.49 × 10−11 | 3.812 | 122.675 |
| G1/S transition of mitotic cell cycle (GO:0000082) | 31/85 | 1.65 × 10−11 | 3.50 × 10−8 | 5.510 | 136.810 |
| Ubiquitin-dependent protein catabolic process (GO:0006511) | 71/354 | 1.10 × 10−9 | 1.30 × 10−6 | 2.429 | 49.941 |
| Protein dephosphorylation (GO:0006470) | 38/139 | 1.54 × 10−9 | 1.30 × 10−6 | 3.615 | 73.357 |
| K562 Top 5% | |||||
| Term | Overlap | p-value | Adjusted p-value | Odds Ratio | Combined Score |
| Negative regulation of translation (GO:0017148) | 26/90 | 7.53 × 10−12 | 2.78 × 10−8 | 6.549 | 167.727 |
| Regulation of translation (GO:0006417) | 36/178 | 7.56 × 10−11 | 1.39 × 10−7 | 4.105 | 95.670 |
| Phosphorylation (GO:0016310) | 57/400 | 7.38 × 10−10 | 9.08 × 10−7 | 2.711 | 57.009 |
| Negative regulation of gene expression (GO:0010629) | 48/322 | 3.72 × 10−9 | 3.43 × 10−6 | 2.846 | 55.243 |
| Loucy Top 5% | |||||
| Term | Overlap | p-value | Adjusted p-value | Odds Ratio | Combined Score |
| mRNA splicing, via spliceosome (GO:0000398) | 55/274 | 1.11 × 10−12 | 2.52 × 10−9 | 3.413 | 93.961 |
| RNA splicing via transesterification reactions with bulged adenosine as nucleophile (GO:0000377) | 52/251 | 1.28 × 10−12 | 2.52 × 10−9 | 3.547 | 97.139 |
| mRNA processing (GO:0006397) | 57/300 | 4.58 × 10−12 | 6.01 × 10−9 | 3.188 | 83.248 |
| RNA metabolic process (GO:0016070) | 34/133 | 2.55 × 10−11 | 2.51 × 10−8 | 4.626 | 112.835 |
| MOLM-13 | |||||
|---|---|---|---|---|---|
| Term | Overlap | p-Value | Adjusted p-Value | Odds Ratio | Combined Score |
| Negative regulation of myeloid-cell differentiation (GO:0045638) | 4/22 | 4.56 × 10−4 | 0.403777 | 13.10978 | 100.8647 |
| Phosphorylation (GO:0016310) | 17/400 | 4.81 × 10−4 | 0.403777 | 2.674282 | 20.42911 |
| Regulation of cellular senescence (GO:2000772) | 4/31 | 0.001739 | 0.573737 | 8.735847 | 55.50902 |
| Negative regulation of NF-kappaB transcription-factor activity (GO:0032088) | 6/79 | 0.002167 | 0.573737 | 4.864462 | 29.83979 |
| DND-41 | |||||
| Term | Overlap | p-value | Adjusted p-value | Odds Ratio | Combined Score |
| Mitotic-chromosome condensation (GO:0007076) | 6/27 | 1.52 × 10−4 | 0.134681 | 8.94822 | 78.65524 |
| DNA dealkylation (GO:0035510) | 4/10 | 1.70 × 10−4 | 0.134681 | 20.82796 | 180.8268 |
| DNA dealkylation involved in DNA repair (GO:0006307) | 4/10 | 1.70 × 10−4 | 0.134681 | 20.82796 | 180.8268 |
| Mitotic-cell-cycle phase transition (GO:0044772) | 17/209 | 3.14 × 10−4 | 0.18717 | 2.798325 | 22.56889 |
| K562 | |||||
| Term | Overlap | p-value | Adjusted p-value | Odds Ratio | Combined Score |
| Regulation of organelle organization (GO:0033043) | 7/80 | 2.38 × 10−4 | 0.207771 | 6.229066 | 51.9768 |
| Regulation of cytoskeleton organization (GO:0051493) | 8/112 | 3.51 × 10−4 | 0.207771 | 5.005622 | 39.81809 |
| Negative regulation of stress-fiber assembly (GO:0051497) | 4/24 | 4.65 × 10−4 | 0.207771 | 12.89902 | 98.97629 |
| Establishment of endothelial intestinal barrier (GO:0090557) | 3/11 | 5.50 × 10−4 | 0.207771 | 24.12132 | 181.0597 |
| Loucy | |||||
| Term | Overlap | p-value | Adjusted p-value | Odds Ratio | Combined Score |
| RNA splicing, via transesterification reactions with bulged adenosine as nucleophile (GO:0000377) | 31/251 | 6.03 × 10−7 | 0.001095 | 2.982339 | 42.71137 |
| Mitotic-spindle organization (GO:0007052) | 23/157 | 8.91 × 10−7 | 0.001095 | 3.616888 | 50.38532 |
| mRNA splicing via spliceosome (GO:0000398) | 32/274 | 1.39 × 10−6 | 0.001095 | 2.798551 | 37.75151 |
| Gene expression (GO:0010467) | 38/356 | 1.40 × 10−6 | 0.001095 | 2.535929 | 34.17959 |
| MV4-11 Top 5% | |||||
|---|---|---|---|---|---|
| Term | Overlap | p-Value | Adjusted p-Value | Odds Ratio | Combined Score |
| mRNA splicing via spliceosome (GO:0000398) | 64/274 | 5.72 × 10−13 | 2.37 × 10−9 | 3.178282 | 89.59665 |
| mRNA processing (GO:0006397) | 66/300 | 4.57 × 10−12 | 9.48 × 10−9 | 2.940925 | 76.79013 |
| RNA processing (GO:0006396) | 46/179 | 3.34 × 10−11 | 4.61 × 10−8 | 3.584974 | 86.48194 |
| RNA splicing via transesterification reactions with bulged adenosine as nucleophile (GO:0000377) | 56/251 | 1.07 × 10−10 | 1.11 × 10−7 | 2.983586 | 68.49088 |
| THP-1 top 5% | |||||
| Term | Overlap | p-value | Adjusted p-value | Odds Ratio | Combined Score |
| mRNA splicing via spliceosome (GO:0000398) | 34/274 | 3.18 × 10−11 | 8.11 × 10−8 | 4.32956 | 104.6518 |
| RNA splicing via transesterification reactions with bulged adenosine as nucleophile (GO:0000377) | 32/251 | 5.98 × 10−11 | 8.11 × 10−8 | 4.456271 | 104.9003 |
| mRNA processing (GO:0006397) | 33/300 | 1.47 × 10−9 | 1.33 × 10−6 | 3.765912 | 76.5817 |
| RNA metabolic process (GO:0016070) | 19/133 | 7.67 × 10−8 | 5.20 × 10−5 | 5.007031 | 82.0337 |
| MV4-11 | |||||
|---|---|---|---|---|---|
| Term | Overlap | p-Value | Adjusted p-Value | Odds Ratio | Combined Score |
| Import into nucleus (GO:0051170) | 14/77 | 6.81 × 10−5 | 0.147625 | 3.858269 | 36.98274 |
| Nucleotide-excision repair (GO:0006289) | 16/105 | 1.92 × 10−4 | 0.147625 | 3.122745 | 26.72998 |
| Positive regulation of establishment of protein localization to mitochondrion (GO:1903749) | 11/56 | 1.99 × 10−4 | 0.147625 | 4.236438 | 36.0943 |
| mRNA splicing via spliceosome (GO:0000398) | 30/274 | 2.59 × 10−4 | 0.147625 | 2.145831 | 17.72484 |
| THP-1 | |||||
| Term | Overlap | p-value | Adjusted p-value | Odds Ratio | Combined Score |
| Microtubule cytoskeleton organization involved in mitosis (GO:1902850) | 10/128 | 1.17 × 10−4 | 0.097514 | 4.685985 | 42.43785 |
| Mitotic-spindle organization (GO:0007052) | 11/157 | 1.43 × 10−4 | 0.097514 | 4.171875 | 36.93698 |
| Positive regulation of double-strand break repair via nonhomologous end joining (GO:2001034) | 4/16 | 1.63 × 10−4 | 0.097514 | 18.22191 | 158.8791 |
| Nuclear-membrane reassembly (GO:0031468) | 6/54 | 4.25 × 10−4 | 0.164331 | 6.858894 | 53.24899 |
| MOLM-13 | |||||
| Term | Overlap | p-value | Adjusted p-value | Odds Ratio | Combined Score |
| Phosphorylation (GO:0016310) | 45/400 | 9.90 × 10−9 | 2.98 × 10−5 | 2.827469 | 52.11112 |
| Protein phosphorylation (GO:0006468) | 44/496 | 1.02 × 10−5 | 0.015305 | 2.157547 | 24.80196 |
| Regulation of intracellular-signal transduction (GO:1902531) | 38/437 | 6.19 × 10−5 | 0.062065 | 2.101864 | 20.36704 |
| Transmembrane-receptor protein tyrosine kinase-signaling pathway (GO:0007169) | 35/404 | 1.28 × 10−4 | 0.096379 | 2.089284 | 18.72459 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, P.; Sattarifard, H.; Fatemiyan, N.; Lakowski, T.M.; Davie, J.R. Bioinformatic Analyses of Broad H3K79me2 Domains in Different Leukemia Cell Line Data Sets. Cells 2022, 11, 2830. https://doi.org/10.3390/cells11182830
Sharma P, Sattarifard H, Fatemiyan N, Lakowski TM, Davie JR. Bioinformatic Analyses of Broad H3K79me2 Domains in Different Leukemia Cell Line Data Sets. Cells. 2022; 11(18):2830. https://doi.org/10.3390/cells11182830
Chicago/Turabian StyleSharma, Prerna, Hedieh Sattarifard, Narges Fatemiyan, Ted M. Lakowski, and James R. Davie. 2022. "Bioinformatic Analyses of Broad H3K79me2 Domains in Different Leukemia Cell Line Data Sets" Cells 11, no. 18: 2830. https://doi.org/10.3390/cells11182830