Physical Training Chronically Stimulates the Motor Neuron Cell Nucleus in the Ts65Dn Mouse, a Model of Down Syndrome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Physical Activity
2.2. Tissue Processing
2.3. Ultrastructural Morphometry
2.4. Ultrastructural Immunocytochemistry
2.5. Statistics
3. Results
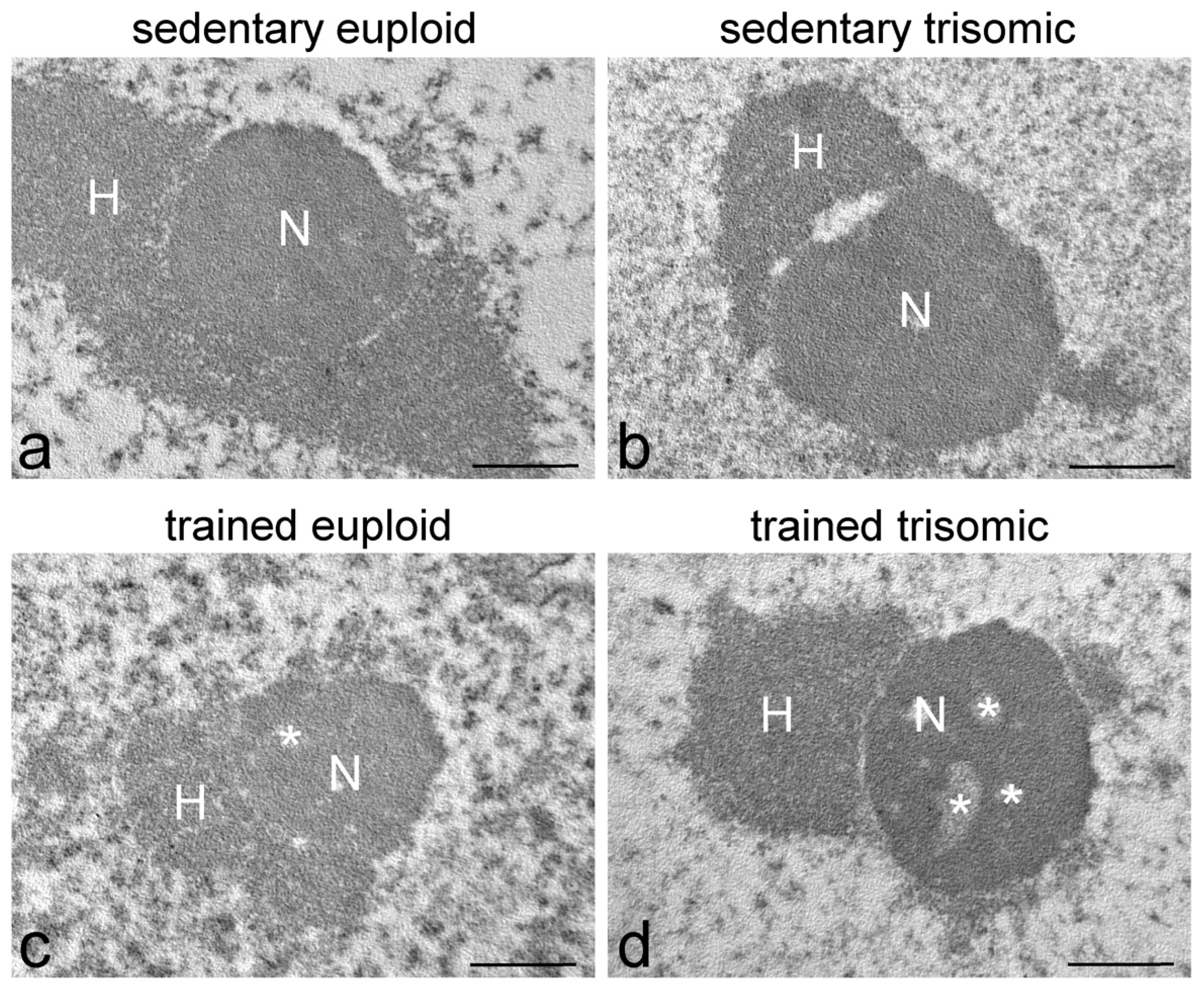
3.1. Ultrastructural Morphology
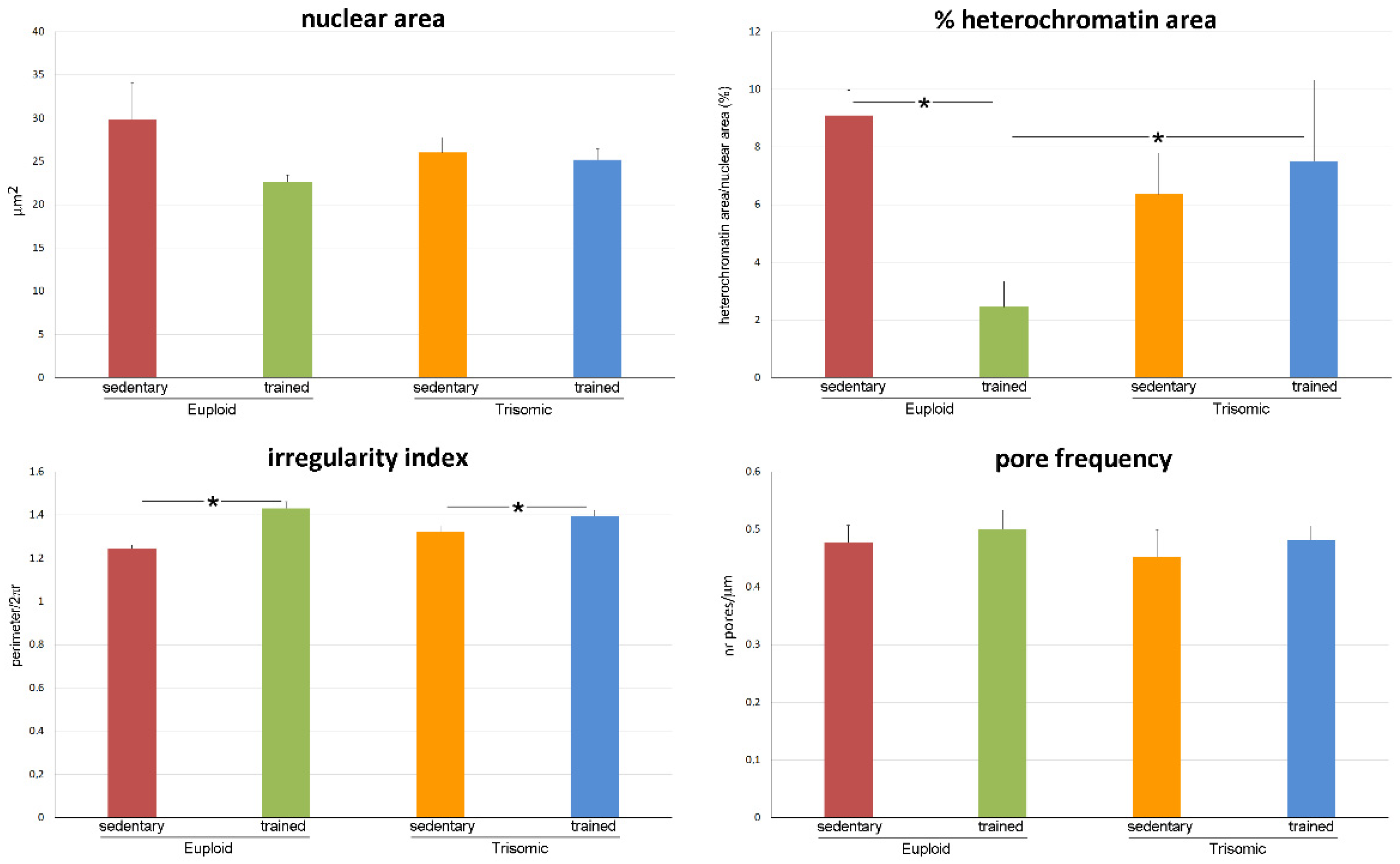
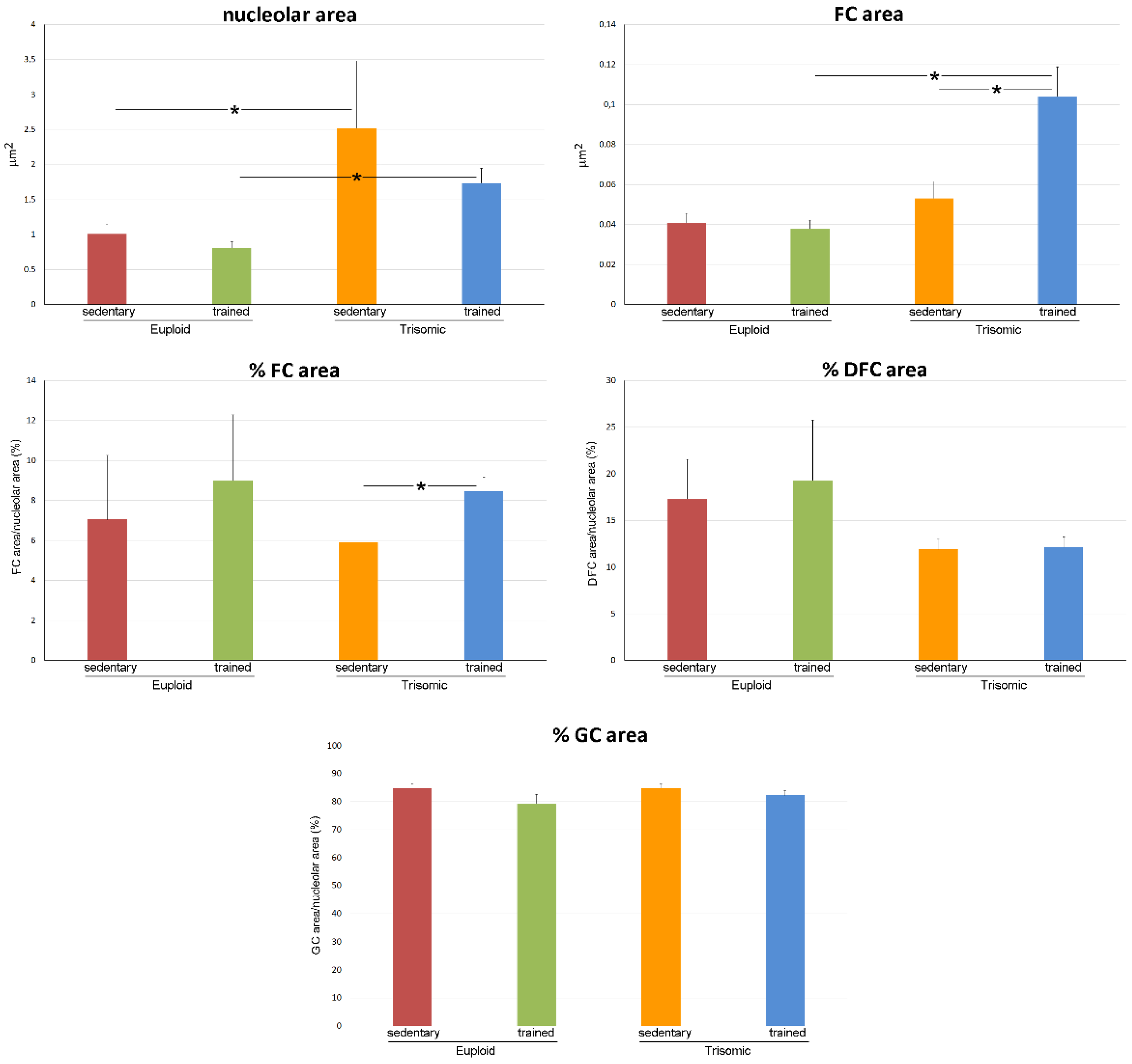
3.2. Ultrastructural Morphometry
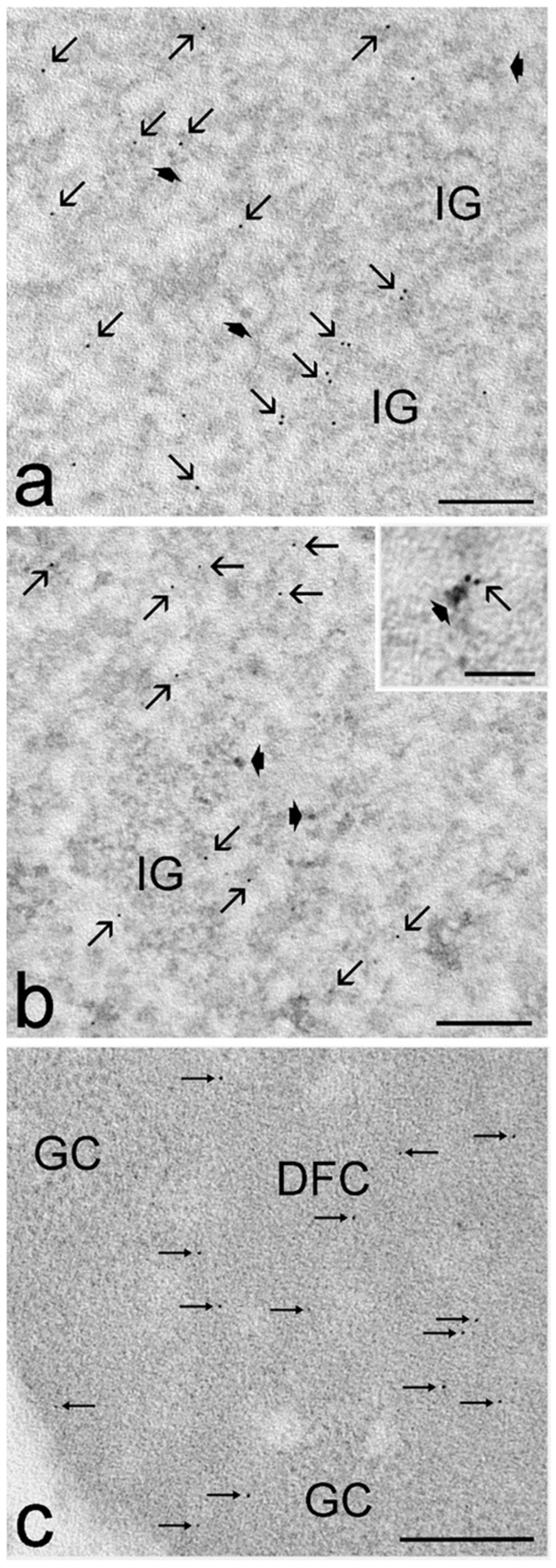
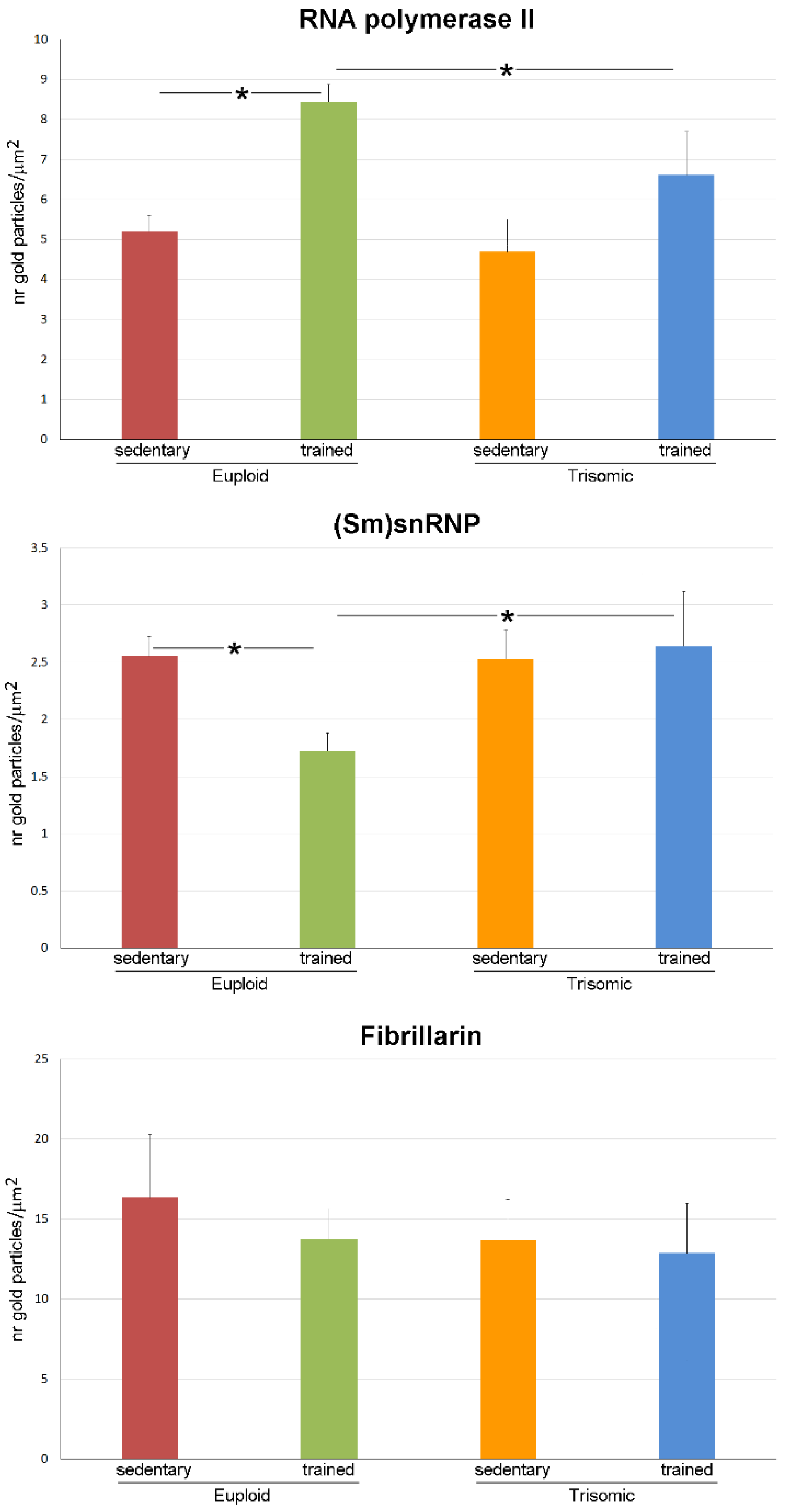
3.3. Ultrastructural Immunocytochemistry
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parker, S.E.; Mai, C.T.; Canfield, M.A.; Rickard, R.; Wang, Y.; Meyer, R.E.; Anderson, P.; Mason, C.A.; Collins, J.S.; Kirby, R.S.; et al. Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defects Res. A Clin. Mol. Teratol. 2010, 88, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Latash, M.L.; Kang, N.; Patterson, D. Finger coordination in persons with Down syndrome: Atypical patterns of coordination and the effects of practice. Exp. Brain Res. 2002, 146, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Vimercati, S.L.; Galli, M.; Stella, G.; Caiazzo, G.; Ancillao, A.; Albertini, G. Clumsiness in fine motor tasks: Evidence from the quantitative drawing evaluation of children with Down Syndrome. J. Intellect. Disabil. Res. 2015, 59, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.F.; Vaughan, S.E.; Vaccaro, P. Measurements of neuromuscular tone and strength in Down’s syndrome children. J. Ment. Defic. Res. 1982, 26, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Korenberg, J.R.; Chen, X.N.; Schipper, R.; Sun, Z.; Gonsky, R.; Gerwehr, S.; Carpenter, N.; Daumer, C.; Dignan, P.; Disteche, C.; et al. Down syndrome phenotypes: The consequences of chromosomal imbalance. Proc. Natl. Acad. Sci. USA 1994, 91, 4997–5001. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.A.; Stergiou, N.; Ulrich, B.D. Patterns of gait variability across the lifespan in persons with and without down syndrome. J. Neurol. Phys. Ther. 2011, 35, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Roth, G.M.; Sun, B.; Greensite, F.S.; Lott, I.T.; Dietrich, R.B. Premature aging in persons with Down syndrome: MR findings. AJNR Am. J. Neuroradiol. 1996, 17, 1283–1289. [Google Scholar] [PubMed]
- Nakamura, E.; Tanaka, S. Biological ages of adult men and women with Down’s syndrome and its changes with aging. Mech. Ageing Dev. 1998, 105, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jentoft, A.J.; Sayer, A.A. Sarcopenia. Lancet 2019, 393, 2636–2646. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, D.A.; Ulrich, B.D.; Angulo-Kinzler, R.M.; Yun, J. Treadmill training of infants with Down syndrome: Evidence-based developmental outcomes. Pediatrics 2001, 108, E84. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Reif, M.; Field, T.; Largie, S.; Mora, D.; Bornstein, J.; Waldman, R. Children with Down syndrome improved in motor functioning and muscle tone following massage therapy. Early Child Dev. Care 2006, 176, 395–410. [Google Scholar] [CrossRef]
- Smith, B.A.; Kubo, M.; Black, D.P.; Holt, K.G.; Ulrich, B.D. Effect of practice on a novel task--walking on a treadmill: Preadolescents with and without Down syndrome. Phys. Ther. 2007, 87, 766–777. [Google Scholar] [CrossRef] [PubMed]
- Ordonez, F.J.; Rosety, I.; Rosety, M.A.; Camacho-Molina, A.; Fornieles, G.; Rosety, M.; Rosety-Rodriguez, M. Aerobic training at moderate intensity reduced protein oxidation in adolescents with Down syndrome. Scand. J. Med. Sci. Sports 2012, 22, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Espinosa, R.M.; Molina Vila, M.D.; Reig García-Galbis, M. Evidences from Clinical Trials in Down Syndrome: Diet, Exercise and Body Composition. Int. J. Environ. Res. Public Health 2020, 17, 4294. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.F.; Scariot, E.L.; da Rosa, L.H.T. The effect of different exercise programs on sarcopenia criteria in older people: A systematic review of systematic reviews with meta-analysis. Arch. Gerontol. Geriatr. 2023, 105, 104868. [Google Scholar] [CrossRef] [PubMed]
- Cassilhas, R.C.; Tufik, S.; de Mello, M.T. Physical exercise, neuroplasticity, spatial learning and memory. Cell. Mol. Life Sci. 2016, 73, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Dhanasekaran, A.R.; Gardiner, K.J. Mouse models of Down syndrome: Gene content and consequences. Mamm. Genome 2016, 27, 538–555. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R.H.; Irving, N.G.; Moran, T.H.; Wohn, A.; Kitt, C.; Sisodia, S.S.; Schmidt, C.; Bronson, R.T.; Davisson, M.T. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat. Genet. 1995, 11, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.C.; Walsh, K.; Davisson, M.T. Motor dysfunction in a mouse model for Down syndrome. Physiol. Behav. 1999, 68, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Cowley, P.M.; Keslacy, S.; Middleton, F.A.; DeRuisseau, L.R.; Fernhall, B.; Kanaley, J.A.; DeRuisseau, K.C. Functional and biochemical characterization of soleus muscle in Down syndrome mice: Insight into the muscle dysfunction seen in the human condition. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 303, R1251–R1260. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.C.; Stasko, M.R.; Schmidt, C.; Davisson, M.T. Behavioral validation of the Ts65Dn mouse model for Down syndrome of a genetic background free of the retinal degeneration mutation Pde6b(rd1). Behav. Brain Res. 2010, 206, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Holtzman, D.M.; Santucci, D.; Kilbridge, J.; Chua-Couzens, J.; Fontana, D.J.; Daniels, S.E.; Johnson, R.M.; Chen, K.; Sun, Y.; Carlson, E.; et al. Developmental abnormalities and age-related neurodegeneration in a mouse model of Down syndrome. Proc. Natl. Acad. Sci. USA 1996, 93, 13333–13338. [Google Scholar] [CrossRef] [PubMed]
- Cisterna, B.; Sobolev, A.P.; Costanzo, M.; Malatesta, M.; Zancanaro, C. Combined Microscopic and Metabolomic Approach to Characterize the Skeletal Muscle Fiber of the Ts65Dn Mouse, A Model of Down Syndrome. Microsc. Microanal. 2020, 26, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Cisterna, B.; Costanzo, M.; Scherini, E.; Zancanaro, C.; Malatesta, M. Ultrastructural features of skeletal muscle in adult and aging Ts65Dn mice, a murine model of Down syndrome. Muscles Ligaments Tendons J. 2013, 3, 287–294. [Google Scholar] [PubMed]
- Llorens-Martín, M.V.; Rueda, N.; Tejeda, G.S.; Flórez, J.; Trejo, J.L.; Martínez-Cué, C. Effects of voluntary physical exercise on adult hippocampal neurogenesis and behavior of Ts65Dn mice, a model of Down syndrome. Neuroscience 2010, 171, 1228–1240. [Google Scholar] [CrossRef]
- Kida, E.; Walus, M.; Albertini, G.; Golabek, A.A. Long-term voluntary running modifies the levels of proteins of the excitatory/inhibitory system and reduces reactive astrogliosis in the brain of Ts65Dn mouse model for Down syndrome. Brain Res. 2021, 1766, 147535. [Google Scholar] [CrossRef] [PubMed]
- Parrini, M.; Ghezzi, D.; Deidda, G.; Medrihan, L.; Castroflorio, E.; Alberti, M.; Baldelli, P.; Cancedda, L.; Contestabile, A. Aerobic exercise and a BDNF-mimetic therapy rescue learning and memory in a mouse model of Down syndrome. Sci. Rep. 2017, 7, 16825. [Google Scholar] [CrossRef]
- Cisterna, B.; Bontempi, P.; Sobolev, A.P.; Costanzo, M.; Malatesta, M.; Zancanaro, C. Quantitative magnetic resonance characterization of the effect of physical training on skeletal muscle of the Ts65Dn mice, a model of Down syndrome. Quant. Imaging Med. Surg. 2022, 12, 2066–2074. [Google Scholar] [CrossRef]
- Cisterna, B.; Boschi, F.; Lacavalla, M.A.; Vattemi, G.N.A.; Zancanaro, C.; Malatesta, M. Physical training promotes remodeling of the skeletal muscle extracellular matrix: An ultrastructural study in a murine model of the Down syndrome. Micr. Res. Tech.
- Puente-Bedia, A.; Berciano, M.T.; Tapia, O.; Martínez-Cué, C.; Lafarga, M.; Rueda, N. Nuclear Reorganization in Hippocampal Granule Cell Neurons from a Mouse Model of Down Syndrome: Changes in Chromatin Configuration, Nucleoli and Cajal Bodies. Int. J. Mol. Sci. 2021, 22, 1259. [Google Scholar] [CrossRef] [PubMed]
- Jordan, E.G. Nucleolar nomenclature. J. Cell Sci. 1984, 67, 217–220. [Google Scholar] [CrossRef]
- Schwarzacher, H.G.; Wachtler, F. The nucleolus. Anat. Embryol. 1993, 188, 515–536. [Google Scholar] [CrossRef] [PubMed]
- Biggiogera, M.; Cisterna, B.; Spedito, A.; Vecchio, L.; Malatesta, M. Perichromatin fibrils as early markers of transcriptional alterations. Differentiation 2008, 76, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Rouquette, J.; Cremer, C.; Cremer, T.; Fakan, S. Functional nuclear architecture studied by microscopy: Present and future. Int. Rev. Cell Mol. Biol. 2010, 282, 1–90. [Google Scholar] [CrossRef] [PubMed]
- Cisterna, B.; Biggiogera, M. Ribosome biogenesis: From structure to dynamics. Int. Rev. Cell Mol. Biol. 2010, 284, 67–111. [Google Scholar] [CrossRef] [PubMed]
- Reinholdt, L.G.; Ding, Y.; Gilbert, G.J.; Czechanski, A.; Solzak, J.P.; Roper, R.J.; Johnson, M.T.; Donahue, L.R.; Lutz, C.; Davisson, M.T. Molecular characterization of the translocation breakpoints in the Down syndrome mouse model Ts65Dn. Mamm. Genome 2011, 22, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Malatesta, M.; Perdoni, F.; Muller, S.; Zancanaro, C.; Pellicciari, C. Nuclei of aged myofibres undergo structural and functional changes suggesting impairment in RNA processing. Eur. J. Histochem. 2009, 53, e12. [Google Scholar] [CrossRef]
- Malatesta, M.; Costanzo, M.; Cisterna, B.; Zancanaro, C. Satellite Cells in Skeletal Muscle of the Hibernating Dormouse, a Natural Model of Quiescence and Re-Activation: Focus on the Cell Nucleus. Cells 2020, 9, 1050. [Google Scholar] [CrossRef] [PubMed]
- Lührmann, R.; Kastner, B.; Bach, M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim. Biophys. Acta 1990, 1087, 265–292. [Google Scholar] [CrossRef] [PubMed]
- Kass, S.; Tyc, K.; Steitz, J.A.; Sollner-Webb, B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell 1990, 60, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Lacavalla, M.A.; Cisterna, B. Uranyl-Free Staining as a Suitable Contrasting Technique for Nuclear Structures at Transmission Electron Microscopy. Methods Mol. Biol. 2023, 2566, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Kurt, M.A.; Davies, D.C.; Kidd, M.; Dierssen, M.; Flórez, J. Synaptic deficit in the temporal cortex of partial trisomy 16 (Ts65Dn) mice. Brain Res. 2000, 858, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Guidi, S.; Ciani, E.; Bonasoni, P.; Santini, D.; Bartesaghi, R. Widespread proliferation impairment and hypocellularity in the cerebellum of fetuses with down syndrome. Brain Pathol. 2011, 21, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Necchi, D.; Lomoio, S.; Scherini, E. Axonal abnormalities in cerebellar Purkinje cells of the Ts65Dn mouse. Brain Res. 2008, 1238, 181–188. [Google Scholar] [CrossRef]
- Necchi, D.; Lomoio, S.; Scherini, E. Dysfunction of the ubiquitin-proteasome system in the cerebellum of aging Ts65Dn mice. Exp. Neurol. 2011, 232, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Fakan, S. Ultrastructural cytochemical analyses of nuclear functional architecture. Eur. J. Histochem. 2004, 48, 5–14. [Google Scholar]
- Kemeny, S.; Tatout, C.; Salaun, G.; Pebrel-Richard, C.; Goumy, C.; Ollier, N.; Maurin, E.; Pereira, B.; Vago, P.; Gouas, L. Spatial organization of chromosome territories in the interphase nucleus of trisomy 21 cells. Chromosoma 2018, 127, 247–259. [Google Scholar] [CrossRef]
- Kahlem, P.; Sultan, M.; Herwig, R.; Steinfath, M.; Balzereit, D.; Eppens, B.; Saran, N.G.; Pletcher, M.T.; South, S.T.; Stetten, G.; et al. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of down syndrome. Genome Res. 2004, 14, 1258–1267. [Google Scholar] [CrossRef]
- Lyle, R.; Gehrig, C.; Neergaard-Henrichsen, C.; Deutsch, S.; Antonarakis, S.E. Gene expression from the aneuploid chromosome in a trisomy mouse model of down syndrome. Genome Res. 2004, 14, 1268–1274. [Google Scholar] [CrossRef]
- Malatesta, M.; Fattoretti, P.; Giagnacovo, M.; Pellicciari, C.; Zancanaro, C. Physical training modulates structural and functional features of cell nuclei in type II myofibers of old mice. Rejuvenation Res. 2011, 14, 543–552. [Google Scholar] [CrossRef]
- Cisterna, B.; Giagnacovo, M.; Costanzo, M.; Fattoretti, P.; Zancanaro, C.; Pellicciari, C.; Malatesta, M. Adapted physical exercise enhances activation and differentiation potential of satellite cells in the skeletal muscle of old mice. J. Anat. 2016, 228, 771–783. [Google Scholar] [CrossRef]
- Puvion-Dutilleul, F.; Puvion, E. Relationship between chromatin and perichromatin granules in cadmium-treated isolated hepatocytes. J. Ultrastruct. Res. 1981, 74, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Nin, G.H.; Echeverria, O.M.; Fakan, S.; Leser, G.; Martin, T.E. Immunoelectron microscope localization of snRNPs in the polytene nucleus of salivary glands ofChironomus thummi. Chromosoma 1990, 99, 44–51. [Google Scholar] [CrossRef]
- Spedito, A.; Cisterna, B.; Malatesta, M.; Biggiogera, M. Use of halogenated precursors to define a transcription time window after treatment with hypometabolizing molecules. Histochem. Cell Biol. 2014, 141, 243–249. [Google Scholar] [CrossRef]
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb. Perspect Biol. 2011, 3, e16210. [Google Scholar] [CrossRef] [PubMed]
- Aziz, D.C.; Barathur, R.B. Quantitation and morphometric analysis of tumors by image analysis. J. Cell Biochem. 1994, 19, 120–125. [Google Scholar]
- Malatesta, M.; Boraldi, F.; Annovi, G.; Baldelli, B.; Battistelli, S.; Biggiogera, M.; Quaglino, D. A long-term study on female mice fed on a genetically modified soybean: Effects on liver ageing. Histochem. Cell Biol. 2008, 130, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Biggiogera, M.; Malatesta, M.; Abolhassani-Dadras, S.; Amalric, F.; Rothblum, L.I.; Fakan, S. Revealing the unseen: The organizer region of the nucleolus. J. Cell Sci. 2001, 114, 3199–3205. [Google Scholar] [CrossRef]
- Sanders, N.C.; Williams, D.K.; Wenger, G.R. Does the learning deficit observed under an incremental repeated acquisition schedule of reinforcement in Ts65Dn mice, a model for Down syndrome, change as they age? Behav. Brain Res. 2009, 203, 137–142. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inguscio, C.R.; Lacavalla, M.A.; Cisterna, B.; Zancanaro, C.; Malatesta, M. Physical Training Chronically Stimulates the Motor Neuron Cell Nucleus in the Ts65Dn Mouse, a Model of Down Syndrome. Cells 2023, 12, 1488. https://doi.org/10.3390/cells12111488
Inguscio CR, Lacavalla MA, Cisterna B, Zancanaro C, Malatesta M. Physical Training Chronically Stimulates the Motor Neuron Cell Nucleus in the Ts65Dn Mouse, a Model of Down Syndrome. Cells. 2023; 12(11):1488. https://doi.org/10.3390/cells12111488
Chicago/Turabian StyleInguscio, Chiara Rita, Maria Assunta Lacavalla, Barbara Cisterna, Carlo Zancanaro, and Manuela Malatesta. 2023. "Physical Training Chronically Stimulates the Motor Neuron Cell Nucleus in the Ts65Dn Mouse, a Model of Down Syndrome" Cells 12, no. 11: 1488. https://doi.org/10.3390/cells12111488




