USP10 Contributes to Colon Carcinogenesis via mTOR/S6K Mediated HIF-1α but Not HIF-2α Protein Synthesis
Abstract
:1. Introduction
2. Materials and Methods
3. Results
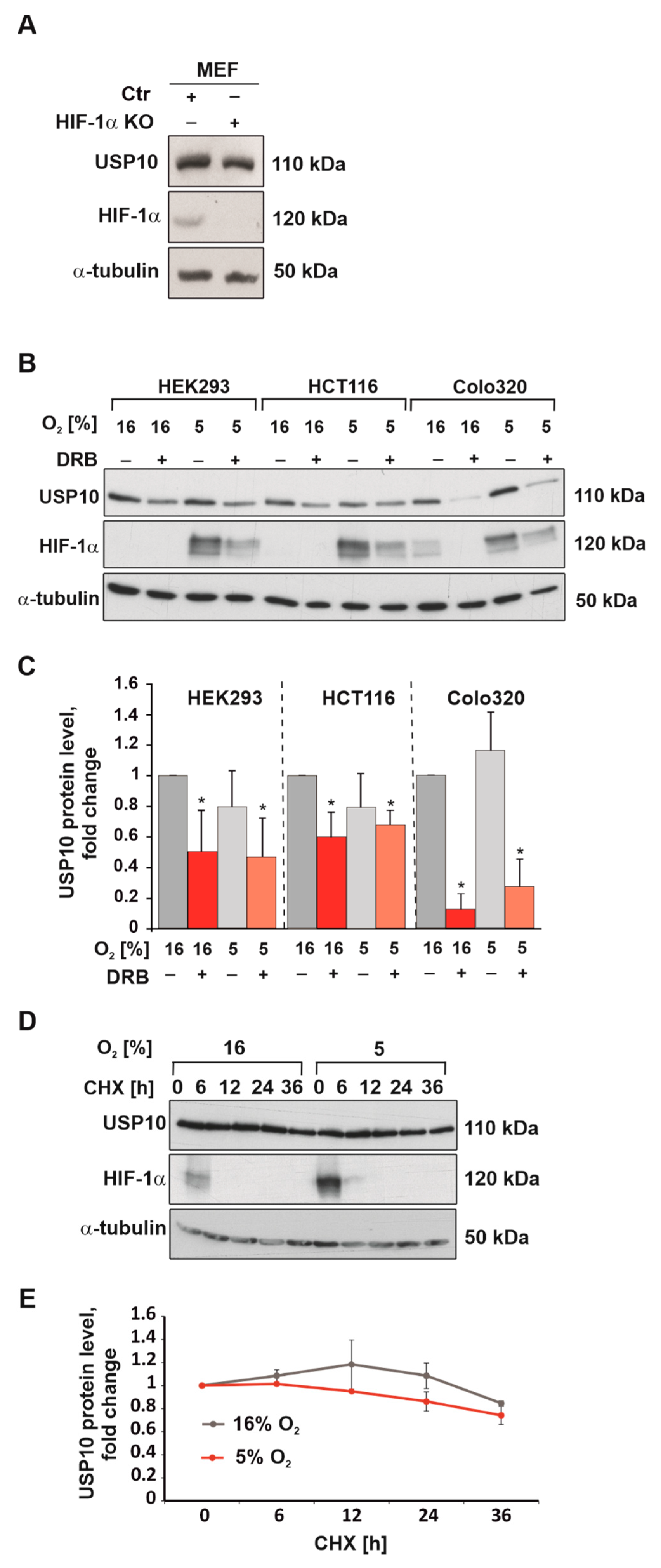
3.1. Hypoxia Reduces USP10 mRNA and Protein Levels
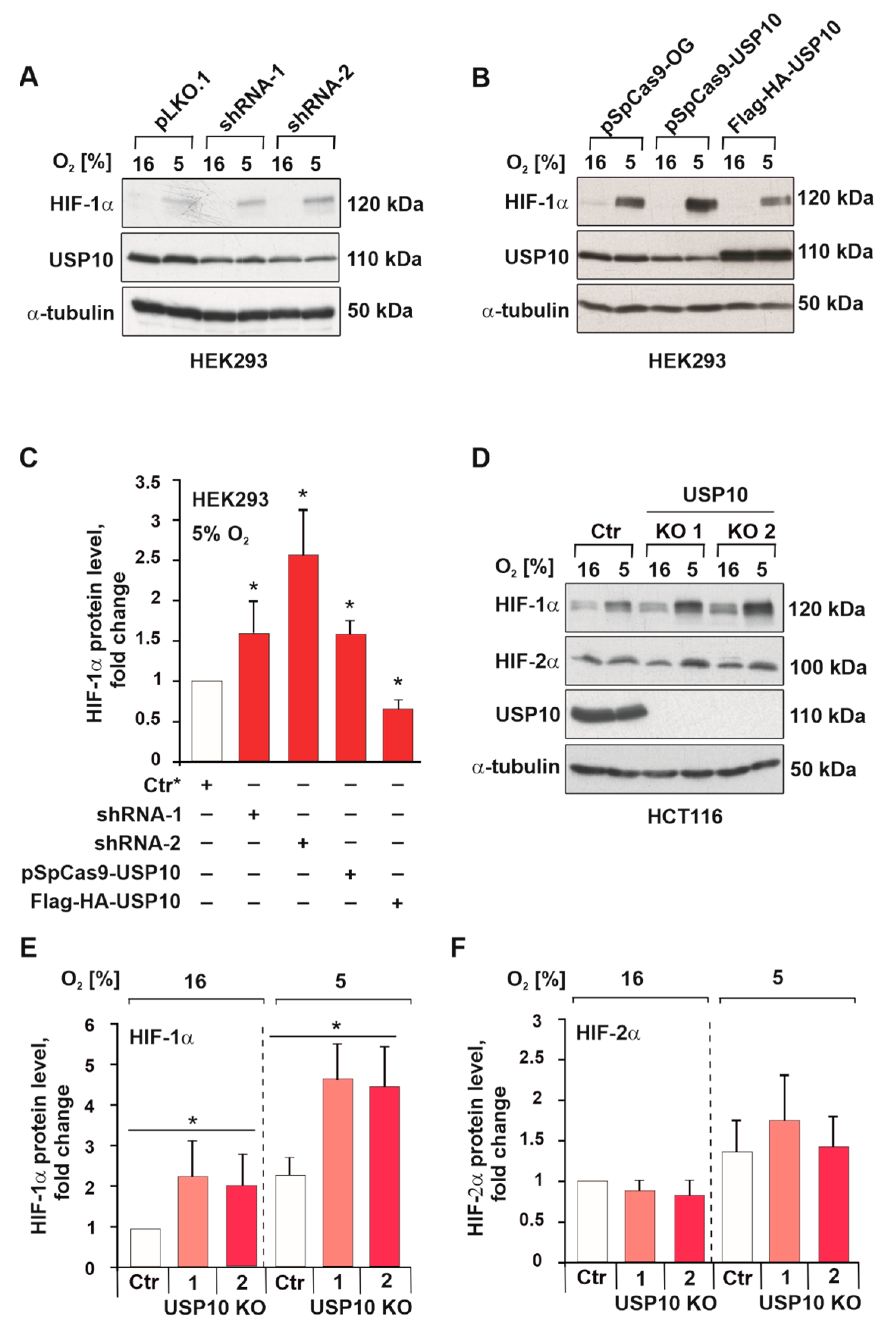
3.2. USP10 Knockout Elevates HIF-1α but Not HIF-2α Protein Levels
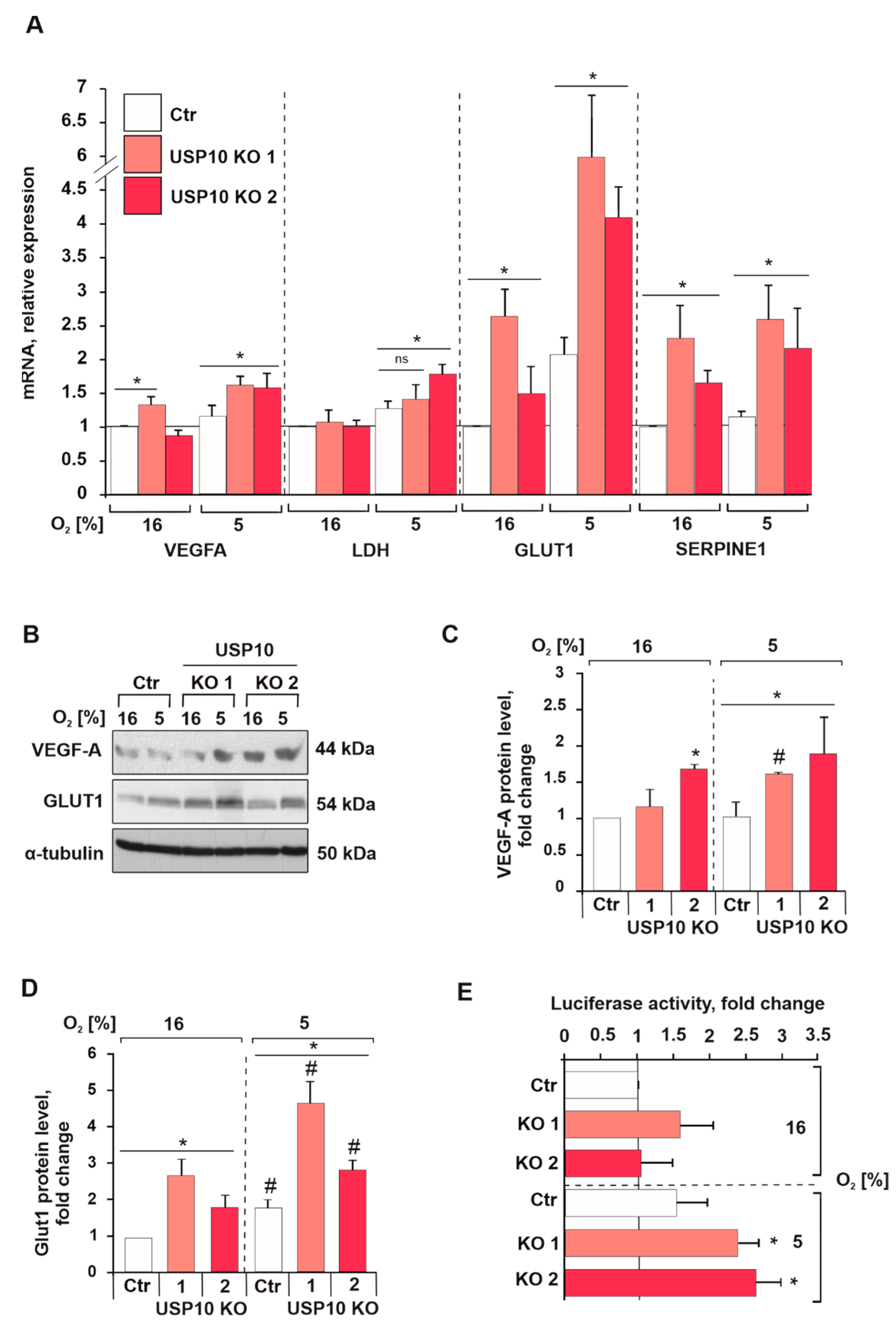
3.3. USP10 Deficiency Affects HIF-1α Signaling and Transcriptional Activity
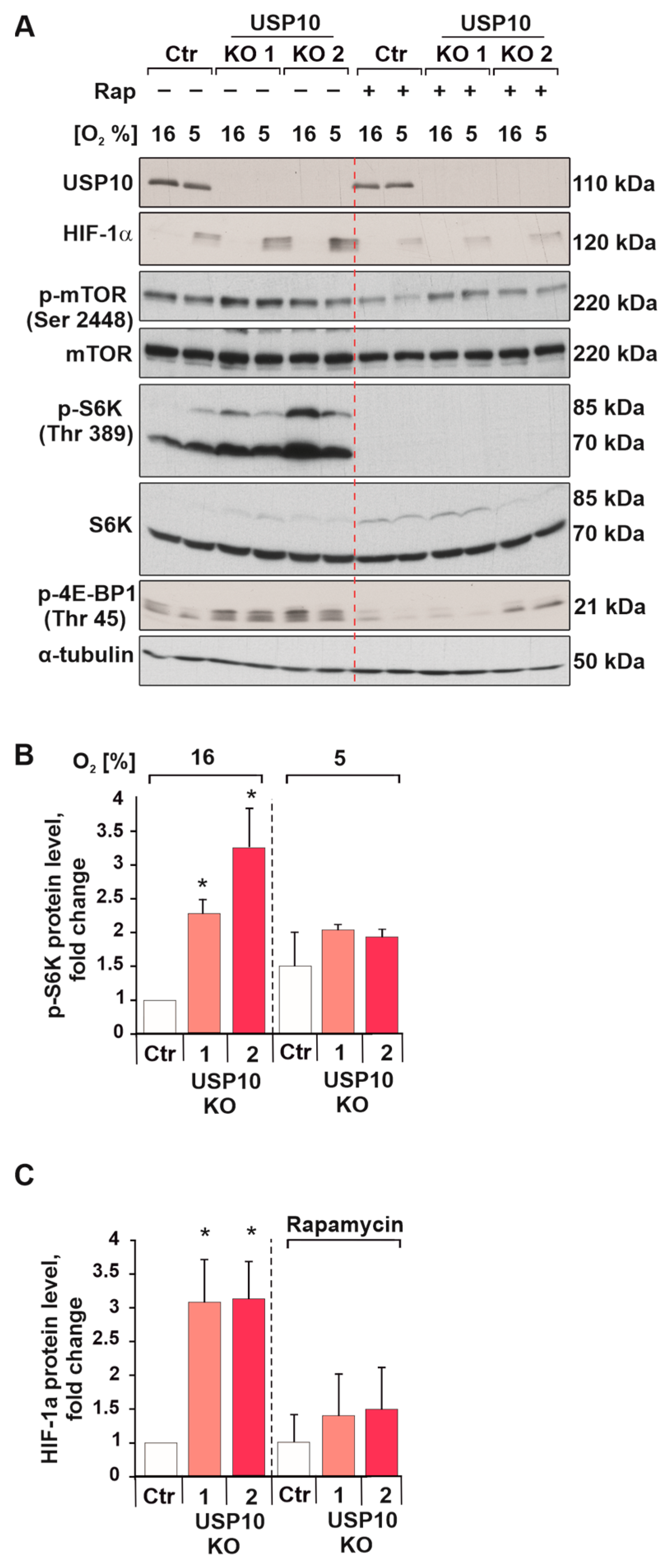
3.4. USP10 Deficiency Increases S6 Kinase Activity and HIF-1α Synthesis
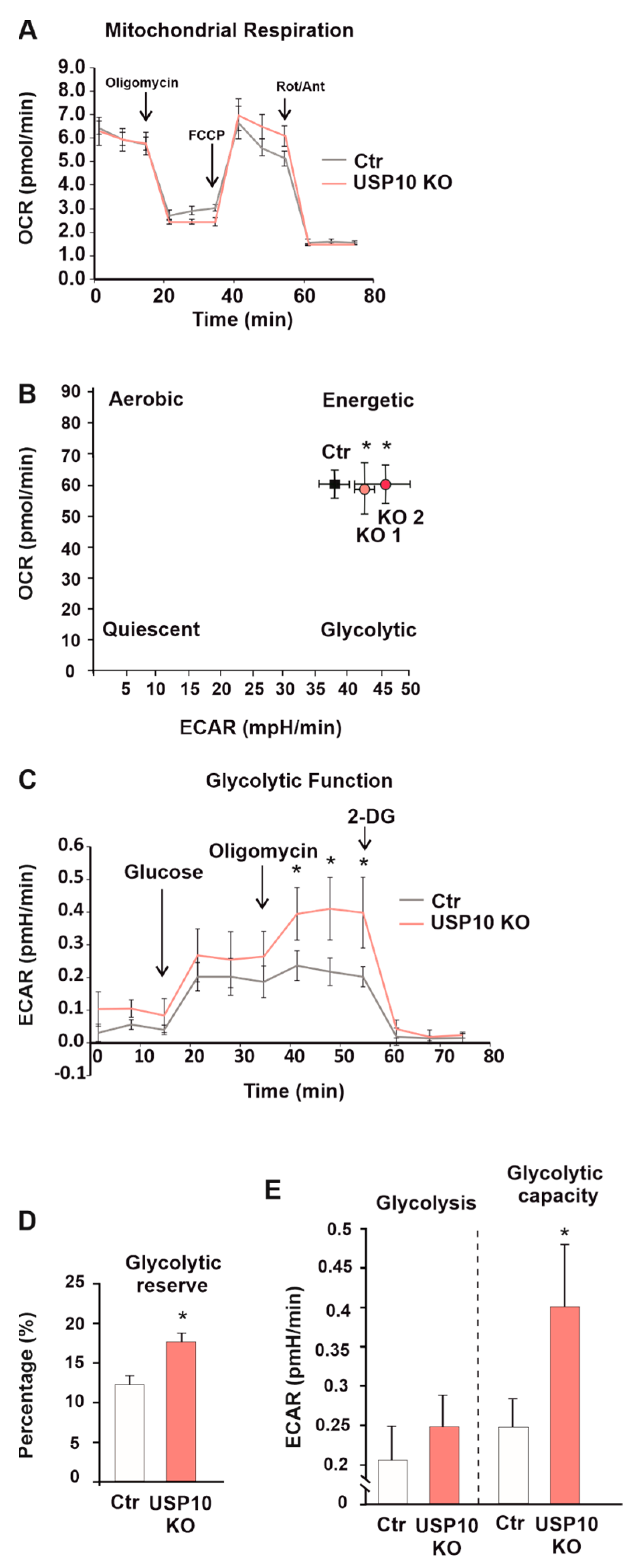
3.5. USP10 Knockout Affects the Cellular Energy Phenotype and Induces Glycolysis
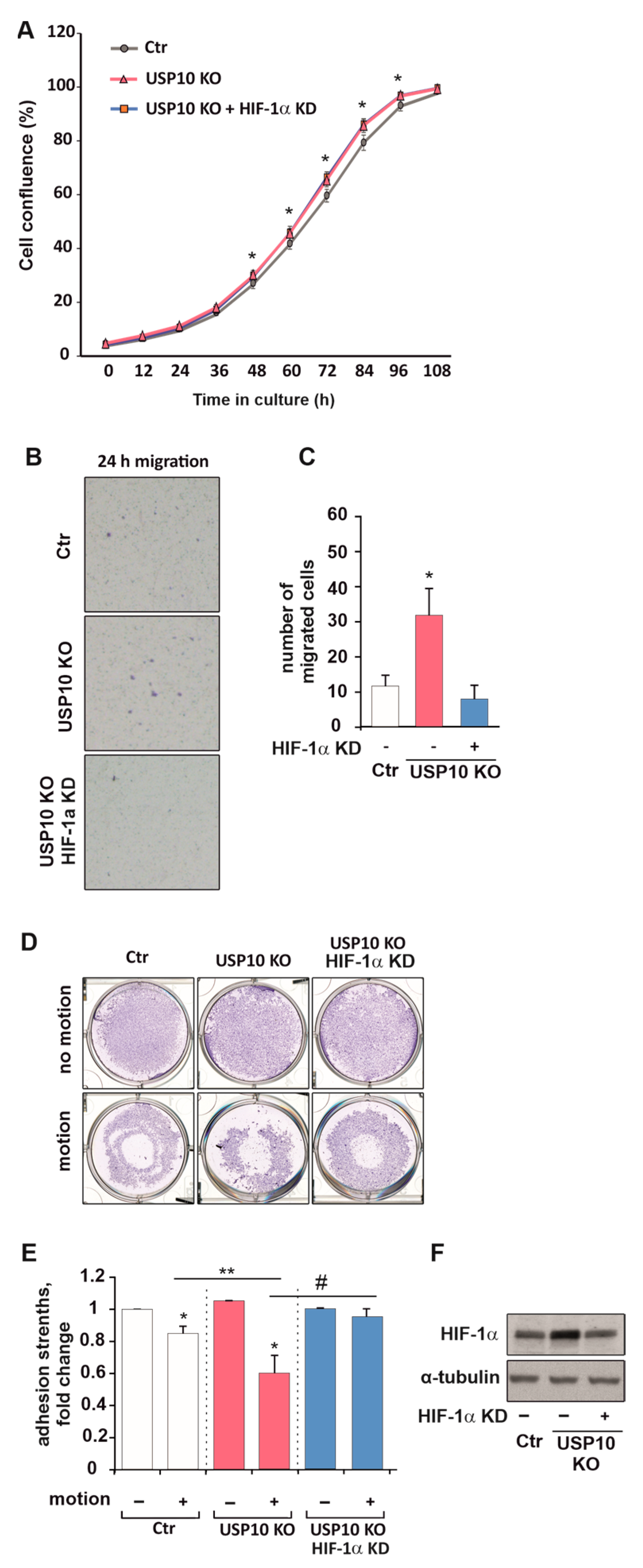
3.6. HIF Knockdown Rescues USP10-Dependent Migration and Adhesion but Not Proliferation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, J.; Ding, Z.; Peng, Y.; Pan, F.; Li, J.; Zou, L.; Zhang, Y.; Liang, H. HIF-1α Inhibition Reverses Multidrug Resistance in Colon Cancer Cells via Downregulation of MDR1/P-Glycoprotein. PLoS ONE 2014, 9, e98882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, T.T.; Lin, Y.T.; Tang, S.P.; Luo, C.K.; Tsai, C.T.; Shun, C.T.; Chen, C.C. Metabolic Targeting of HIF-1α Potentiates the Therapeutic Efficacy of Oxaliplatin in Colorectal Cancer. Oncogene 2020, 39, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, K.; Fidler, I.J. Hypoxia Increases Resistance of Human Pancreatic Cancer Cells to Apoptosis Induced by Gemcitabine. Clin. Cancer Res. 2004, 10, 2299–2306. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Ning, X.; Sun, L.; Zhang, H.; Shi, Y.; Guo, C.; Han, S.; Liu, J.; Sun, S.; Han, Z.; et al. Hypoxia-Inducible Factor-1α Contributes to Hypoxia-Induced Chemoresistance in Gastric Cancer. Cancer Sci. 2007, 99, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Hussein, D.; Estlin, E.J.; Dive, C.; Makin, G.W.J. Chronic Hypoxia Promotes Hypoxia-Inducible Factor-1α–Dependent Resistance to Etoposide and Vincristine in Neuroblastoma Cells. Mol. Cancer Ther. 2006, 5, 2241–2250. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Liu, X.; Chi, W.; Liu, Y.; Wei, L.; Wang, X.; Yu, J. Hypoxia-Induced Resistance to Cisplatin and Doxorubicin in Non-Small Cell Lung Cancer Is Inhibited by Silencing of HIF-1α Gene. Cancer Chemother. Pharmacol. 2006, 58, 776–784. [Google Scholar] [CrossRef]
- Wang, G.L.; Semenza, G.L. Purification and Characterization of Hypoxia-Inducible Factor. J. Biol. Chem. 1995, 270, 1230–1237. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.-Z.; Moran, S.M.; Hogenesch, J.B.; Wartman, L.; Bradfield, C.A. Molecular Characterization and Chromosomal Localization of a Third α-Class Hypoxia Inducible Factor Subunit, HIF3α. Gene Expr. 1998, 7, 205–213. [Google Scholar]
- Tian, H.; McKnight, S.L.; Russell, D.W. Endothelial PAS Domain Protein 1 (EPAS1), a Transcription Factor Selectively Expressed in Endothelial Cells. Genes Dev. 1997, 11, 72–82. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.E.; Gu, J.; Schau, M.; Bunn, H.F. Regulation of Hypoxia-Inducible Factor 1α Is Mediated by an O2-Dependent Degradation Domain via the Ubiquitin-Proteasome Pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 7987–7992. [Google Scholar] [CrossRef] [Green Version]
- Metzen, E.; Stiehl, D.P.; Doege, K.; Marxsen, J.H.; Hellwig-Bürgel, T.; Jelkmann, W. Regulation of the Prolyl Hydroxylase Domain Protein 2 (Phd2/Egln-1) Gene: Identification of a Functional Hypoxia-Responsive Element. Biochem. J. 2005, 387, 711–717. [Google Scholar] [CrossRef] [Green Version]
- Fong, G.-H.; Takeda, K. Role and Regulation of Prolyl Hydroxylase Domain Proteins. Cell Death Differ. 2008, 15, 635–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Günter, J.; Ruiz-Serrano, A.; Pickel, C.; Wenger, R.H.; Scholz, C.C. The Functional Interplay between the HIF Pathway and the Ubiquitin System—More than a One-Way Road. Exp. Cell Res. 2017, 356, 152–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubaichuk, K.; Kietzmann, T. Involvement of E3 Ligases and Deubiquitinases in the Control of Hif-α Subunit Abundance. Cells 2019, 8, 596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the Chains: Structure and Function of the Deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef]
- Mevissen, T.E.T.; Komander, D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 2017, 86, 159–192. [Google Scholar] [CrossRef] [Green Version]
- Flügel, D.; Görlach, A.; Kietzmann, T. GSK-3β Regulates Cell Growth, Migration, and Angiogenesis via Fbw7 and USP28-Dependent Degradation of HIF-1α. Blood 2012, 119, 1292–1301. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Oliveira, R.I.; Guedes, R.A.; Salvador, J.A.R. Highlights in USP7 Inhibitors for Cancer Treatment. Front. Chem. 2022, 10, 1005727. [Google Scholar] [CrossRef]
- Tao, L.; Liu, X.; Jiang, X.; Zhang, K.; Wang, Y.; Li, X.; Jiang, S.; Han, T. USP10 as a Potential Therapeutic Target in Human Cancers. Genes 2022, 13, 831. [Google Scholar] [CrossRef]
- Guo, F.; Zhang, C.; Wang, F.; Zhang, W.; Shi, X.; Zhu, Y.; Fang, Z.; Yang, B.; Sun, Y. Deubiquitinating Enzyme USP33 Restrains Docetaxel-Induced Apoptosis via Stabilising the Phosphatase DUSP1 in Prostate Cancer. Cell Death Differ. 2020, 27, 1938–1951. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, Q.; Li, K.; Gong, Z.; Liu, Z.; Xu, Y.; Swaney, M.H.; Xiao, K.; Chen, Y. Prognostic Significance of USP33 in Advanced Colorectal Cancer Patients: New Insights into β-Arrestin-Dependent ERK Signaling. Oncotarget 2016, 7, 81223–81240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.-H.; Wang, H.; Ji, X.-Y.; Zhang, F.-X.; Gao, B.-L.; Hu, J.-A.; Zheng, J. A Detrimental Mutation on USP40 Unlocks the Tumorigenesis in a Rare Case of Lung Cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 740–749. [Google Scholar] [PubMed]
- Li, K.; Wang, Q.; Bian, H.; Chen, Z.; He, H.; Zhao, X.; Gong, P. Comprehensive Analysis Reveals USP45 as a Novel Putative Oncogene in Pan-Cancer. Front. Mol. Biosci. 2022, 9, 886904. [Google Scholar] [CrossRef] [PubMed]
- Kiran, S.; Dar, A.; Singh, S.K.; Lee, K.Y.; Dutta, A. The Deubiquitinase USP46 Is Essential for Proliferation and Tumor Growth of HPV-Transformed Cancers. Mol. Cell 2018, 72, 823–835.e5. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Stevens, P.D.; Yang, H.; Gulhati, P.; Wang, W.; Evers, B.M.; Gao, T. The Deubiquitination Enzyme USP46 Functions as a Tumor Suppressor by Controlling PHLPP-Dependent Attenuation of Akt Signaling in Colon Cancer. Oncogene 2013, 32, 471–478. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zhao, W.; Li, Y.; Li, Y.; Cheng, H.; Zheng, L.; Sun, X.; Liu, H.; Shao, R. YOD1 Serves as a Potential Prognostic Biomarker for Pancreatic Cancer. Cancer Cell Int. 2022, 22, 203. [Google Scholar] [CrossRef]
- Wu, X.; Luo, Q.; Zhao, P.; Chang, W.; Wang, Y.; Shu, T.; Ding, F.; Li, B.; Liu, Z. JOSD1 Inhibits Mitochondrial Apoptotic Signalling to Drive Acquired Chemoresistance in Gynaecological Cancer by Stabilizing MCL1. Cell Death Differ. 2020, 27, 55–70. [Google Scholar] [CrossRef]
- Jing, C.; Liu, D.; Lai, Q.; Li, L.; Zhou, M.; Ye, B.; Wu, Y.; Li, H.; Yue, K.; Wu, Y.; et al. JOSD1 Promotes Proliferation and Chemoresistance of Head and Neck Squamous Cell Carcinoma under the Epigenetic Regulation of BRD4. Cancer Cell Int. 2021, 21, 375. [Google Scholar] [CrossRef]
- Li, B.; Qi, Z.-P.; He, D.-L.; Chen, Z.-H.; Liu, J.-Y.; Wong, M.-W.; Zhang, J.-W.; Xu, E.-P.; Shi, Q.; Cai, S.-L.; et al. NLRP7 Deubiquitination by USP10 Promotes Tumor Progression and Tumor-Associated Macrophage Polarization in Colorectal Cancer. J. Exp. Clin. Cancer Res. 2021, 40, 126. [Google Scholar] [CrossRef]
- Novellasdemunt, L.; Foglizzo, V.; Cuadrado, L.; Antas, P.; Kucharska, A.; Encheva, V.; Snijders, A.P.; Li, V.S.W. USP7 Is a Tumor-Specific WNT Activator for APC -Mutated Colorectal Cancer by Mediating β-Catenin Deubiquitination. Cell Rep. 2017, 21, 612–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Huh, T.; Park, Y.; Koo, D.-H.; Kim, H.; Hwang, I.; Choi, C.H.; Yi, J.M.; Chung, J.-Y. Prognostic Significance of USP10 and P14ARF Expression in Patients with Colorectal Cancer. Pathol. Res. Pract. 2020, 216, 152988. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Yang, H.; Tan, C.; Li, J.; Liu, Z.; Quan, Q.; Kong, S.; Ye, J.; Gao, B.; Fang, D. USP10 Antagonizes C-Myc Transcriptional Activation through SIRT6 Stabilization to Suppress Tumor Formation. Cell Rep. 2013, 5, 1639–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziello, J.E.; Jovin, I.S.; Huang, Y. Hypoxia-Inducible Factor (HIF)-1 Regulatory Pathway and Its Potential for Therapeutic Intervention in Malignancy and Ischemia. Yale J. Biol. Med. 2007, 80, 51–60. [Google Scholar]
- Dodd, K.M.; Yang, J.; Shen, M.H.; Sampson, J.R.; Tee, A.R. MTORC1 Drives HIF-1α and VEGF-A Signalling via Multiple Mechanisms Involving 4E-BP1, S6K1 and STAT3. Oncogene 2015, 34, 2239–2250. [Google Scholar] [CrossRef] [Green Version]
- Denko, N.C. Hypoxia, HIF1 and Glucose Metabolism in the Solid Tumour. Nat. Rev. Cancer 2008, 8, 705–713. [Google Scholar] [CrossRef]
- Karar, J.; Maity, A. PI3K/AKT/MTOR Pathway in Angiogenesis. Front. Mol. Neurosci. 2011, 4, 51. [Google Scholar] [CrossRef] [Green Version]
- Hubbi, M.E.; Semenza, G.L. Regulation of Cell Proliferation by Hypoxia-Inducible Factors. Am. J. Physiol. Cell Physiol. 2015, 309, C775–C782. [Google Scholar] [CrossRef] [Green Version]
- Dang, C.V. The Interplay Between MYC and HIF in the Warburg Effect. Ernst Scher. Found. Symp. Proc. 2007, 4, 35–53. [Google Scholar] [CrossRef]
- Carrera, S.; Senra, J.; Acosta, M.I.; Althubiti, M.; Hammond, E.M.; de Verdier, P.J.; Macip, S. The Role of the HIF-1α Transcription Factor in Increased Cell Division at Physiological Oxygen Tensions. PLoS ONE 2014, 9, e97938. [Google Scholar] [CrossRef] [Green Version]
- Tam, S.Y.; Wu, V.W.C.; Law, H.K.W. Hypoxia-Induced Epithelial-Mesenchymal Transition in Cancers: HIF-1α and Beyond. Front. Oncol. 2020, 10, 486. [Google Scholar] [CrossRef] [PubMed]
- Ouchida, A.T.; Kacal, M.; Zheng, A.; Ambroise, G.; Zhang, B.; Norberg, E.; Vakifahmetoglu-Norberg, H. USP10 Regulates the Stability of the EMT-Transcription Factor Slug/SNAI2. Biochem. Biophys. Res. Commun. 2018, 502, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubaichuk, K.; Kietzmann, T. USP10 Contributes to Colon Carcinogenesis via mTOR/S6K Mediated HIF-1α but Not HIF-2α Protein Synthesis. Cells 2023, 12, 1585. https://doi.org/10.3390/cells12121585
Kubaichuk K, Kietzmann T. USP10 Contributes to Colon Carcinogenesis via mTOR/S6K Mediated HIF-1α but Not HIF-2α Protein Synthesis. Cells. 2023; 12(12):1585. https://doi.org/10.3390/cells12121585
Chicago/Turabian StyleKubaichuk, Kateryna, and Thomas Kietzmann. 2023. "USP10 Contributes to Colon Carcinogenesis via mTOR/S6K Mediated HIF-1α but Not HIF-2α Protein Synthesis" Cells 12, no. 12: 1585. https://doi.org/10.3390/cells12121585





