Loss of the Novel Myelin Protein CMTM5 in Multiple Sclerosis Lesions and Its Involvement in Oligodendroglial Stress Responses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cuprizone Intoxication
2.3. Experimental Autoimmune Encephalomyelitis Induction and Disease Scoring
2.4. Tissue Preparation
2.5. Multiple Sclerosis Tissues
2.6. Histological and Immunohistochemical Evaluation
2.7. Generation of Cmtm5 Knockdown Oli-Neu CRISPRi Cell Line
2.8. Real-Time RT-PCR
2.9. Single-Cell RNA Sequencing Analysis
2.10. Statistical Analysis
3. Results
3.1. Decreased Expression of CMTM5 Expression in the Cuprizone-Induced Animal Model of Demyelination
3.2. CMTM5-Expression Is Reduced in the Inflammatory EAE Animal Model
3.3. A Cmtm5 Knockdown Does Not Affect Oli-Neu Cell Responsiveness to ER Stress
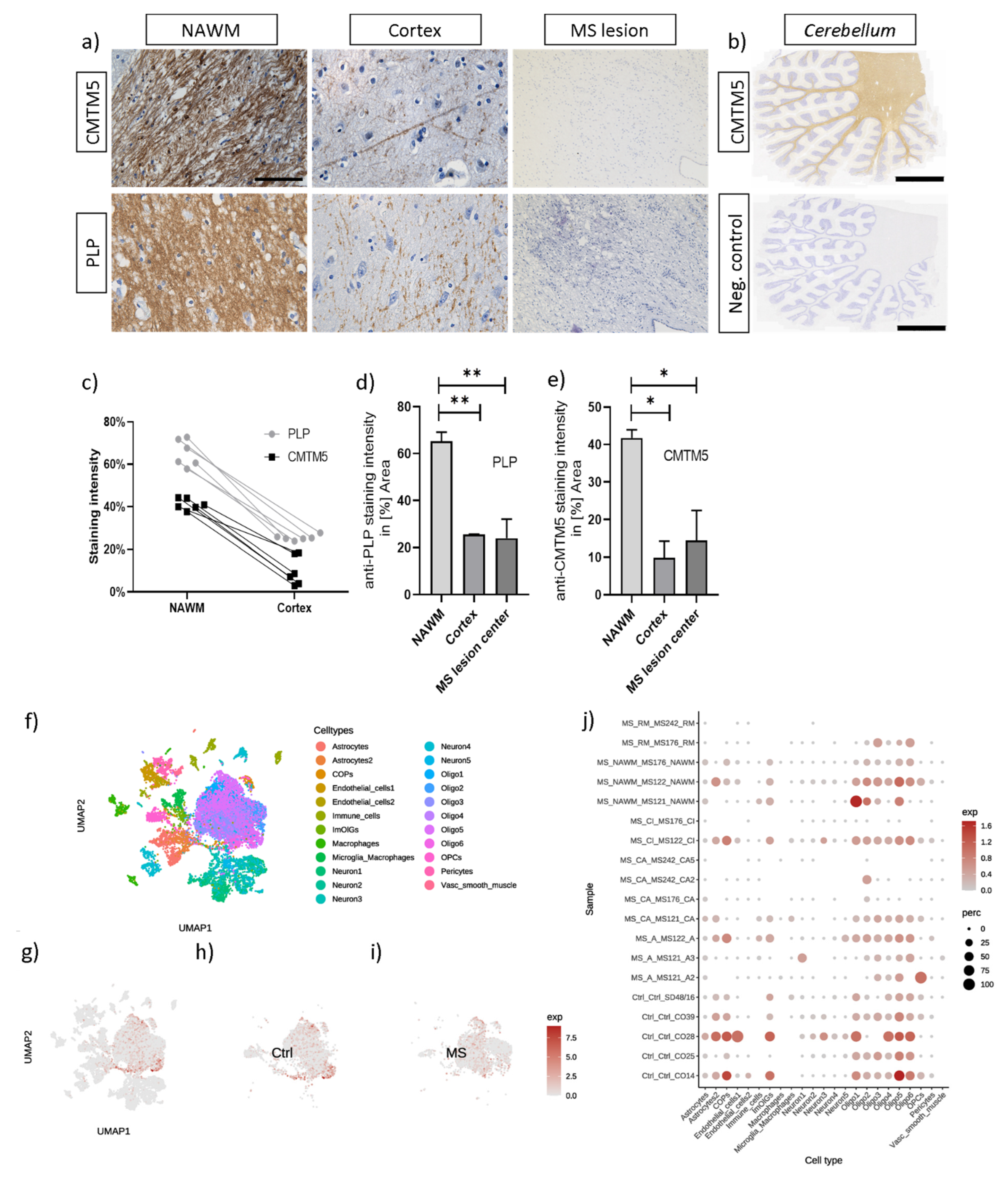
3.4. CMTM5 Protein Expression Is Reduced in Demyelinated Post Mortem MS Lesions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simkins, T.J.; Duncan, G.J.; Bourdette, D. Chronic Demyelination and Axonal Degeneration in Multiple Sclerosis: Pathogenesis and Therapeutic Implications. Curr. Neurol. Neurosci. Rep. 2021, 21, 26. [Google Scholar] [CrossRef] [PubMed]
- Nave, K.-A. Myelination and the trophic support of long axons. Nat. Rev. Neurosci. 2010, 11, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Eichel, M.A.; Gargareta, V.-I.; D’Este, E.; Fledrich, R.; Kungl, T.; Buscham, T.J.; Lüders, K.A.; Miracle, C.; Jung, R.B.; Distler, U.; et al. CMTM6 expressed on the adaxonal Schwann cell surface restricts axonal diameters in peripheral nerves. Nat. Commun. 2020, 11, 4514. [Google Scholar] [CrossRef]
- Buscham, T.J.; Eichel-Vogel, M.A.; Steyer, A.M.; Jahn, O.; Strenzke, N.; Dardawal, R.; Memhave, T.R.; Siems, S.B.; Müller, C.; Meschkat, M.; et al. Progressive axonopathy when oligodendrocytes lack the myelin protein CMTM5. eLife 2022, 11, e75523. [Google Scholar] [CrossRef] [PubMed]
- Wittekindt, M.; Kaddatz, H.; Joost, S.; Staffeld, A.; Bitar, Y.; Kipp, M.; Frintrop, L. Different Methods for Evaluating Microglial Activation Using Anti-Ionized Calcium-Binding Adaptor Protein-1 Immunohistochemistry in the Cuprizone Model. Cells 2022, 11, 1723. [Google Scholar] [CrossRef]
- Stromnes, I.M.; Goverman, J.M. Active induction of experimental allergic encephalomyelitis. Nat. Protoc. 2006, 1, 1810–1819. [Google Scholar] [CrossRef]
- Zhan, J.; Fegg, F.N.; Kaddatz, H.; Rühling, S.; Frenz, J.; Denecke, B.; Amor, S.; Ponsaerts, P.; Hochstrasser, T.; Kipp, M. Focal white matter lesions induce long-lasting axonal degeneration, neuroinflammation and behavioral deficits. Neurobiol. Dis. 2021, 155, 105371. [Google Scholar] [CrossRef]
- Zhan, J.; Yakimov, V.; Rühling, S.; Fischbach, F.; Nikolova, E.; Joost, S.; Kaddatz, H.; Greiner, T.; Frenz, J.; Holzmann, C.; et al. High Speed Ventral Plane Videography as a Convenient Tool to Quantify Motor Deficits during Pre-Clinical Experimental Autoimmune Encephalomyelitis. Cells 2019, 8, 1439. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Jung, M.; Krämer, E.; Grzenkowski, M.; Tang, K.; Blakemore, W.; Aguzzi, A.; Khazaie, K.; Chlichlia, K.; Blankenfeld, G.; Kettenmann, H.; et al. Lines of Murine Oligodendroglial Precursor Cells Immortalized by an ActivatedneuTyrosine Kinase Show Distinct Degrees of Interaction with Axons In Vitro and In Vivo. Eur. J. Neurosci. 1995, 7, 1245–1265. [Google Scholar] [CrossRef]
- Chen, J.J.; Nathaniel, D.L.; Raghavan, P.; Nelson, M.; Tian, R.; Tse, E.; Hong, J.Y.; See, S.K.; Mok, S.-A.; Hein, M.Y.; et al. Compromised function of the ESCRT pathway promotes endolysosomal escape of tau seeds and propagation of tau aggregation. J. Biol. Chem. 2019, 294, 18952–18966. [Google Scholar] [CrossRef] [PubMed]
- Horlbeck, M.A.; Gilbert, L.A.; Villalta, J.E.; Adamson, B.; Pak, R.A.; Chen, Y.; Fields, A.P.; Park, C.Y.; Corn, J.E.; Kampmann, M.; et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife 2016, 5, e19760. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, M.A.; Clark, I.C.; Tjon, E.C.; Li, Z.; Zandee, S.E.J.; Couturier, C.P.; Watson, B.R.; Scalisi, G.; Alkwai, S.; Rothhammer, V.; et al. MAFG-driven astrocytes promote CNS inflammation. Nature 2020, 578, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Jäkel, S.; Agirre, E.; Mendanha Falcão, A.; van Bruggen, D.; Lee, K.W.; Knuesel, I.; Malhotra, D.; ffrench-Constant, C.; Williams, A.; Castelo-Branco, G. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 2019, 566, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Speir, M.L.; Bhaduri, A.; Markov, N.S.; Moreno, P.; Nowakowski, T.J.; Papatheodorou, I.; Pollen, A.A.; Raney, B.J.; Seninge, L.; Kent, W.J.; et al. UCSC Cell Browser: Visualize your single-cell data. Bioinformatics 2021, 37, 4578–4580. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e3529. [Google Scholar] [CrossRef]
- Cahoy, J.D.; Emery, B.; Kaushal, A.; Foo, L.C.; Zamanian, J.L.; Christopherson, K.S.; Xing, Y.; Lubischer, J.L.; Krieg, P.A.; Krupenko, S.A.; et al. A Transcriptome Database for Astrocytes, Neurons, and Oligodendrocytes: A New Resource for Understanding Brain Development and Function. J. Neurosci. 2008, 28, 264–278. [Google Scholar] [CrossRef]
- Behrangi, N.; Heinig, L.; Frintrop, L.; Santrau, E.; Kurth, J.; Krause, B.; Atanasova, D.; Clarner, T.; Fragoulis, A.; Joksch, M.; et al. Siponimod ameliorates metabolic oligodendrocyte injury via the sphingosine-1 phosphate receptor 5. Proc. Natl. Acad. Sci. USA 2022, 119, e2204509119. [Google Scholar] [CrossRef]
- Fischbach, F.; Nedelcu, J.; Leopold, P.; Zhan, J.; Clarner, T.; Nellessen, L.; Beißel, C.; van Heuvel, Y.; Goswami, A.; Weis, J.; et al. Cuprizone-induced graded oligodendrocyte vulnerability is regulated by the transcription factor DNA damage-inducible transcript 3. Glia 2019, 67, 263–276. [Google Scholar] [CrossRef]
- Teske, N.; Liessem, A.; Fischbach, F.; Clarner, T.; Beyer, C.; Wruck, C.; Fragoulis, A.; Tauber, S.C.; Victor, M.; Kipp, M. Chemical hypoxia-induced integrated stress response activation in oligodendrocytes is mediated by the transcription factor nuclear factor (erythroid-derived 2)-like 2 (NRF2). J. Neurochem. 2018, 144, 285–301. [Google Scholar] [CrossRef]
- Way, S.W.; Podojil, J.R.; Clayton, B.L.; Zaremba, A.; Collins, T.L.; Kunjamma, R.B.; Robinson, A.P.; Brugarolas, P.; Miller, R.H.; Miller, S.D.; et al. Pharmaceutical integrated stress response enhancement protects oligodendrocytes and provides a potential multiple sclerosis therapeutic. Nat. Commun. 2015, 6, 6532. [Google Scholar] [CrossRef]
- Kipp, M.; Nyamoya, S.; Hochstrasser, T.; Amor, S. Multiple sclerosis animal models: A clinical and histopathological perspective. Brain Pathol. 2017, 27, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.; Doughty, M.; Harbaugh, C.R.; Cummins, A.; Hatten, M.E.; Heintz, N.; Gerfen, C.R. Targeting Cre Recombinase to Specific Neuron Populations with Bacterial Artificial Chromosome Constructs. J. Neurosci. 2007, 27, 9817–9823. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Yuan, Y.; Zhang, Y.; Li, J.; Liu, Z.; Zhang, X.; Sheng, Z.; Xu, T.; Wang, X. CMTM5 is reduced in prostate cancer and inhibits cancer cell growth in vitro and in vivo. Clin. Transl. Oncol. 2015, 17, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Cui, Y.; Li, H.; Liu, Y.; Zhao, H.; Wang, Y.; Zhang, Y.; Ng, K.M.; Han, W.; Ma, D. CMTM5 exhibits tumor suppressor activities and is frequently silenced by methylation in carcinoma cell lines. Clin. Cancer Res. 2007, 13, 5756–5762. [Google Scholar] [CrossRef]
- Gerber, D.; Pereira, J.A.; Gerber, J.; Tan, G.; Dimitrieva, S.; Yángüez, E.; Suter, U. Transcriptional profiling of mouse peripheral nerves to the single-cell level to build a sciatic nerve ATlas (SNAT). eLife 2021, 10, e58591. [Google Scholar] [CrossRef]
- Patzig, J.; Jahn, O.; Tenzer, S.; Wichert, S.P.; de Monasterio-Schrader, P.; Rosfa, S.; Kuharev, J.; Yan, K.; Bormuth, I.; Bremer, J. Quantitative and integrative proteome analysis of peripheral nerve myelin identifies novel myelin proteins and candidate neuropathy loci. J. Neurosci. 2011, 31, 16369–16386. [Google Scholar] [CrossRef]
- Zhan, J.; Mann, T.; Joost, S.; Behrangi, N.; Frank, M.; Kipp, M. The Cuprizone Model: Dos and Do Nots. Cells 2020, 9, 843. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Garbern, J.Y.; Cambi, F.; Tang, X.-M.; Sima, A.A.F.; Vallat, J.M.; Bosch, E.P.; Lewis, R.; Shy, M.; Sohi, J.; Kraft, G.; et al. Proteolipid Protein Is Necessary in Peripheral as Well as Central Myelin. Neuron 1997, 19, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, T.; Ding, L.; Ikenaka, K.; Inoue, Y.; Miyado, K.; Mekada, E.; Baba, H. Tetraspanin Protein CD9 Is a Novel Paranodal Component Regulating Paranodal Junctional Formation. J. Neurosci. 2004, 24, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Chernousov, M.A.; Stahl, R.C.; Carey, D.J. Tetraspanins are involved in Schwann cell-axon interaction. J. Neurosci. Res. 2013, 91, 1419–1428. [Google Scholar] [CrossRef] [PubMed]
- Bronstein, J.M. Function of tetraspan proteins in the myelin sheath. Curr. Opin. Neurobiol. 2000, 10, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Guo, X.; Shao, L.; Plate, M.; Mo, X.; Wang, Y.; Han, W. CMTM5-v1, a four-transmembrane protein, presents a secreted form released via a vesicle-mediated secretory pathway. BMB Rep. 2010, 43, 182–187. [Google Scholar] [CrossRef]
- Liu, B.; Su, Y.; Li, T.; Yuan, W.; Mo, X.; Li, H.; He, Q.; Ma, D.; Han, W. CMTM7 knockdown increases tumorigenicity of human non-small cell lung cancer cells and EGFR-AKT signaling by reducing Rab5 activation. Oncotarget 2015, 6, 41092–41107. [Google Scholar] [CrossRef]
- Yuan, W.; Liu, B.; Wang, X.; Li, T.; Xue, H.; Mo, X.; Yang, S.; Ding, S.; Han, W. CMTM3 decreases EGFR expression and EGF-mediated tumorigenicity by promoting Rab5 activity in gastric cancer. Cancer Lett. 2017, 386, 77–86. [Google Scholar] [CrossRef]
- Mezzadra, R.; Sun, C.; Jae, L.T.; Gomez-Eerland, R.; de Vries, E.; Wu, W.; Logtenberg, M.E.W.; Slagter, M.; Rozeman, E.A.; Hofland, I.; et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 2017, 549, 106–110. [Google Scholar] [CrossRef]
- Chrifi, I.; Louzao-Martinez, L.; Brandt, M.M.; van Dijk, C.G.M.; Bürgisser, P.E.; Zhu, C.; Kros, J.M.; Verhaar, M.C.; Duncker, D.J.; Cheng, C. CMTM4 regulates angiogenesis by promoting cell surface recycling of VE-cadherin to endothelial adherens junctions. Angiogenesis 2018, 22, 75–93. [Google Scholar] [CrossRef]
- Burr, M.L.; Sparbier, C.E.; Chan, Y.-C.; Williamson, J.C.; Woods, K.; Beavis, P.A.; Lam, E.Y.N.; Henderson, M.A.; Bell, C.C.; Stolzenburg, S.; et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 2017, 549, 101–105. [Google Scholar] [CrossRef]
- Emery, B.; Frühbeis, C.; Kuo-Elsner, W.P.; Müller, C.; Barth, K.; Peris, L.; Tenzer, S.; Möbius, W.; Werner, H.B.; Nave, K.-A.; et al. Oligodendrocytes support axonal transport and maintenance via exosome secretion. PLoS Biol. 2020, 18, e3000621. [Google Scholar] [CrossRef]
- Hollien, J.; Weissman, J.S. Decay of Endoplasmic Reticulum-Localized mRNAs During the Unfolded Protein Response. Science 2006, 313, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Rüther, B.J.; Scheld, M.; Dreymueller, D.; Clarner, T.; Kress, E.; Brandenburg, L.-O.; Swartenbroekx, T.; Hoornaert, C.; Ponsaerts, P.; Fallier-Becker, P.; et al. Combination of cuprizone and experimental autoimmune encephalomyelitis to study inflammatory brain lesion formation and progression. Glia 2017, 65, 1900–1913. [Google Scholar] [CrossRef] [PubMed]



| Cohort Name | Strain | Age | Sex | Experimental Group |
|---|---|---|---|---|
| Cup IHC/IF | C57BL/6 | 10 wks | ♂ | 5 wks cuprizone (n = 5) 3 wks cuprizone (n = 5) 1 wk cuprizone (n = 5) Control (n = 5) |
| 4 d Cup qPCR | C57BL/6 | 10 wks | ♂ | 4 days cuprizone (n = 6) Control (n = 7) |
| 3 wks Cup qPCR | C57BL/6 | 10 wks | ♀ | 3 wks cuprizone (n = 8) Control (n = 8) |
| EAE | C57BL/6 | 11 wks | ♀ | EAE (n = 4), Control (n = 5) |
| Sex | Age at Death in Years | MS Disease Duration in Years | Date of Birth | Date of MS Diagnosis | PMD | MS Type | Cause of Death |
|---|---|---|---|---|---|---|---|
| ♀ | 35 | 10 | 1979 | 2004 | 10:20 h | SPMS | assisted suicide |
| ♂ | 44 | 21 | 1965 | 1988 | 10:15 h | PPMS | End stage MS |
| ♀ | 66 | 32 | 1945 | 1988 | 09:35 h | PPMS | assisted suicide |
| Controls | |||||||
| ♀ | 73 | Not applicable | 1929 | Not applicable | 04:00 h | Not applicable | Lung fibrosis |
| ♀ | 60 | 1950 | 07:30 h | Infection | |||
| ♀ | 86 | 1923 | 06:30 h | Multiple myeloma | |||
| ♂ | 87 | 1918 | 06:32 h | Pneumonia | |||
| Antigen | Used for | Species | Dilution | HIER | RRID | Supplier |
|---|---|---|---|---|---|---|
| PLP | Mouse and Human | Mouse | 1:5000 | None | AB_2237198 | BioRAD, Hercules, CA, USA |
| CMTM5 | Mouse | Rabbit | 1:200 | None | Custom made | * |
| CMTM5 | Human | Rabbit | 1:200 | Citrate | AB_2928051 | Abcam, Cambridge, UK |
| HLA-DR (LN3) | Human | Mouse | 1:1500 | Citrate | AB_10979984 | Thermo Fisher, Waltham, MA, USA |
| SOX10 | Mouse | Rabbit | 1:80 | None | AB_2927464 | Abcam, Cambridge, UK |
| Synaptophysin | Mouse | Mouse | 1:300 | Citrate | AB_2198854 | Abcam, Cambridge, UK |
| VGLUT1 | Mouse | Mouse | 1:1000 | Citrate | AB_2923539 | Abcam, Cambridge, UK |
| APP | Mouse | Mouse | 1:5000 | Tris/EDTA | AB_94882 | Millipore, Burlington, MA, USA |
| OLIG2 | Mouse | Mouse | 1:200 | Tris/EDTA | AB_10807410 | Millipore, Burlington, MA, USA |
| IBA1 | Mouse | Goat | 1:200 | Tris/EDTA | AB_870576 | Abcam, Cambridge, UK |
| Gene Symbol | Sense | Antisense | Ta/°C | Bp |
|---|---|---|---|---|
| Cmtm5 | TGGTCTCCGTCTTTGCCTATG | CTCAGTGGTACTGGGCATCAG | 62 | 68 |
| Plp | TGGCGACTACAAGACCACCA | GACACACCCGCTCCAAAGAA | 60 | 116 |
| Cxcl10 | CCAAGTGCTGCCGTCATTTTC | GGCTCGCAGGGATGATTTCAA | 60 | 157 |
| Sox10 | AGGTTGCTGAACGAAAGTGAC | CCGAGGTTGGTACTTGTAGTCC | 62 | 102 |
| Atf4 | GTTGGTCAGTGCCTCAGACA | CATTCGAAACAGAGCATCGA | 60 | 109 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhan, J.; Gao, Y.; Heinig, L.; Beecken, M.; Huo, Y.; Zhang, W.; Wang, P.; Wei, T.; Tian, R.; Han, W.; et al. Loss of the Novel Myelin Protein CMTM5 in Multiple Sclerosis Lesions and Its Involvement in Oligodendroglial Stress Responses. Cells 2023, 12, 2085. https://doi.org/10.3390/cells12162085
Zhan J, Gao Y, Heinig L, Beecken M, Huo Y, Zhang W, Wang P, Wei T, Tian R, Han W, et al. Loss of the Novel Myelin Protein CMTM5 in Multiple Sclerosis Lesions and Its Involvement in Oligodendroglial Stress Responses. Cells. 2023; 12(16):2085. https://doi.org/10.3390/cells12162085
Chicago/Turabian StyleZhan, Jiangshan, Yuanxu Gao, Leo Heinig, Malena Beecken, Yangbo Huo, Wansong Zhang, Pingzhang Wang, Tianzi Wei, Ruilin Tian, Wenling Han, and et al. 2023. "Loss of the Novel Myelin Protein CMTM5 in Multiple Sclerosis Lesions and Its Involvement in Oligodendroglial Stress Responses" Cells 12, no. 16: 2085. https://doi.org/10.3390/cells12162085






