Eye Lens Organoids Made Simple: Characterization of a New Three-Dimensional Organoid Model for Lens Development and Pathology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Organoid Culture
2.3. Histology and Image Acquisition
2.4. Transparency and Light Focalization
2.5. 21EM15 2D and 3D RNA Isolation
2.6. Laser Capture Microdissection
2.7. 3′ End RNA Sequencing (RNA-Seq) and Analysis
2.8. Gene Expression Data Analysis
3. Results
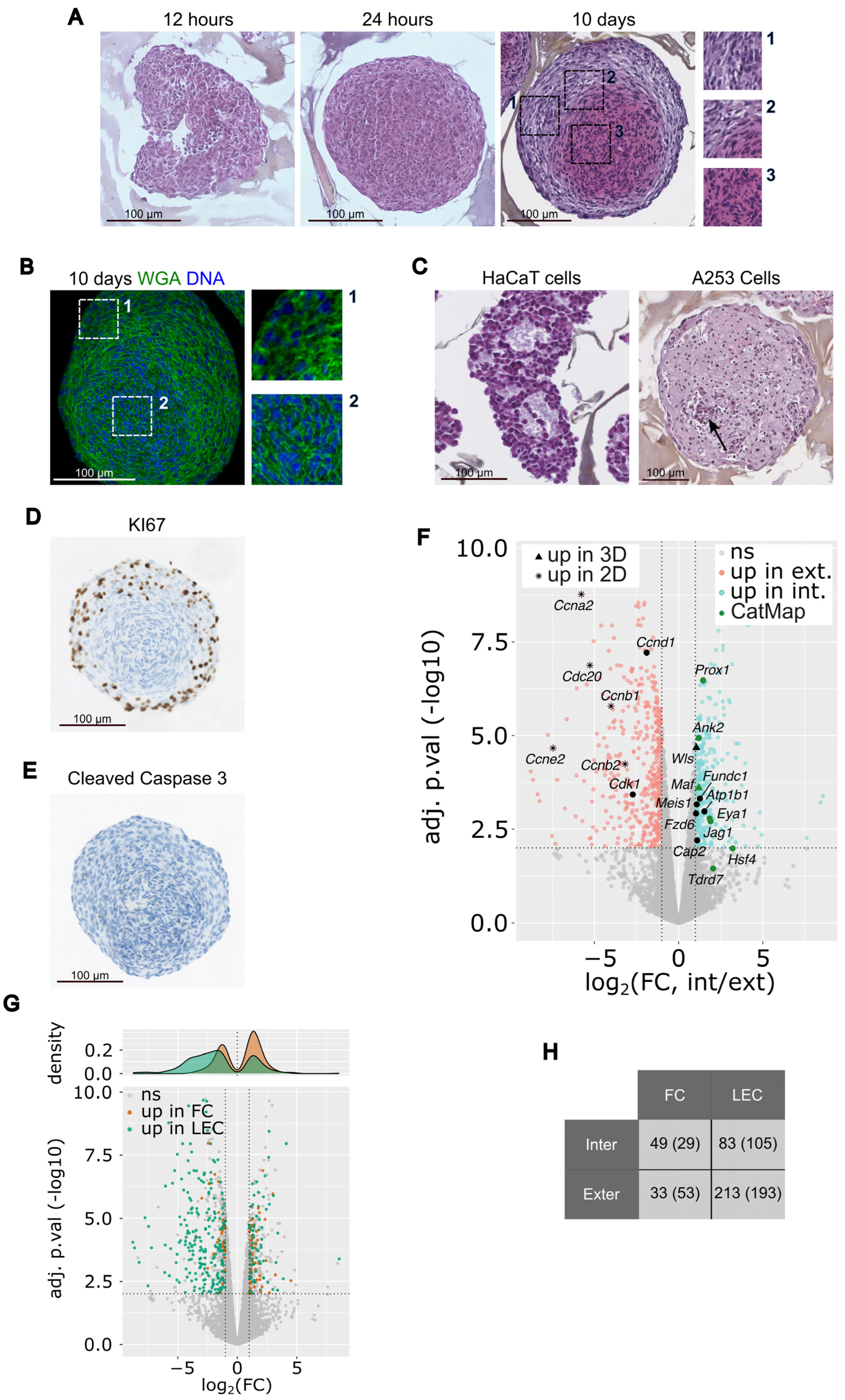
3.1. 21EM15 Spheroids Are Transparent and Have the Ability to Focus Light
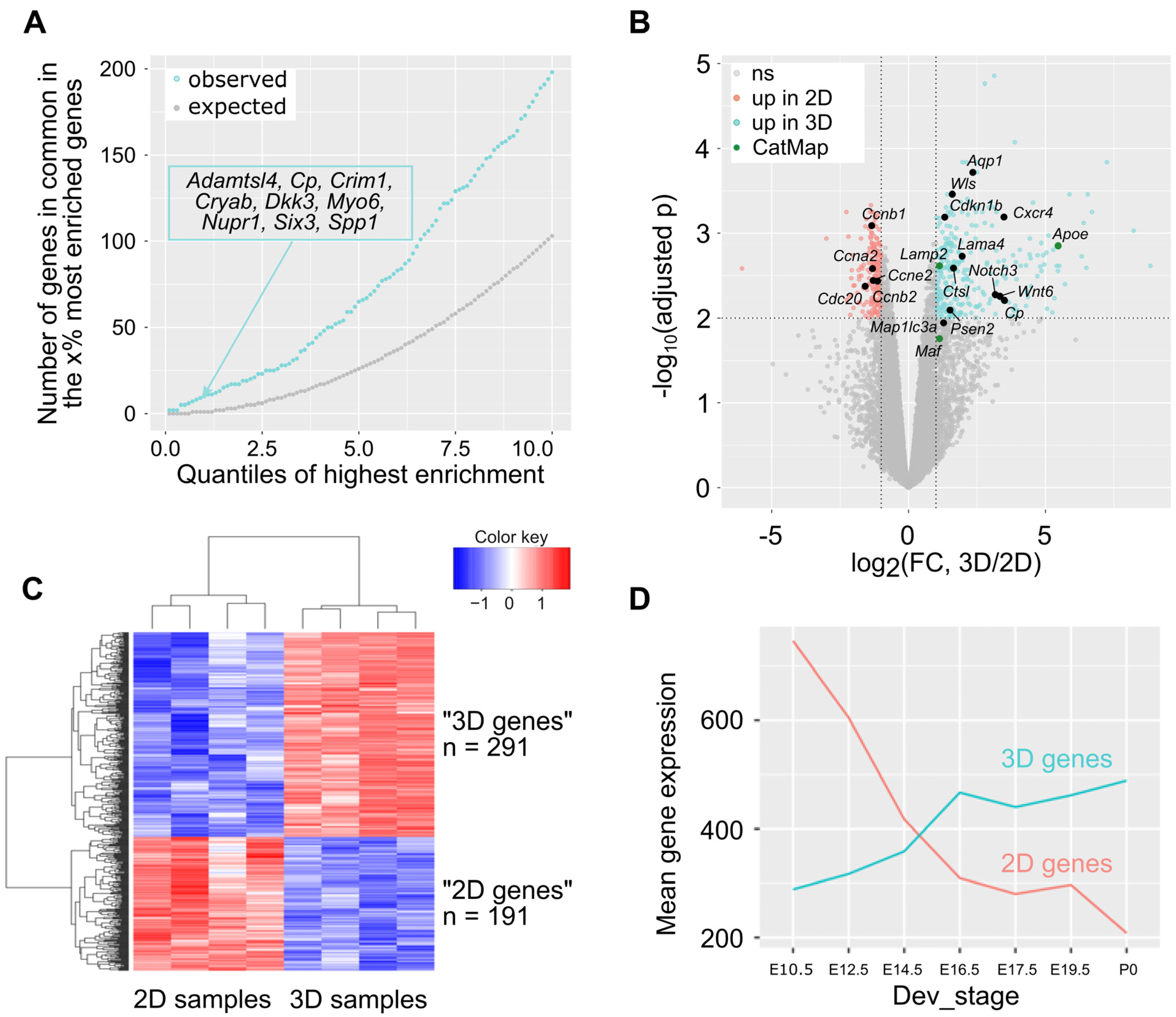
3.2. Transcriptome Analysis of 21EM15 Spheroids Reveals Strong Similarities with Lens Development
3.3. 21EM15 3D Cultured Cells form Multilayered Lens Organoids
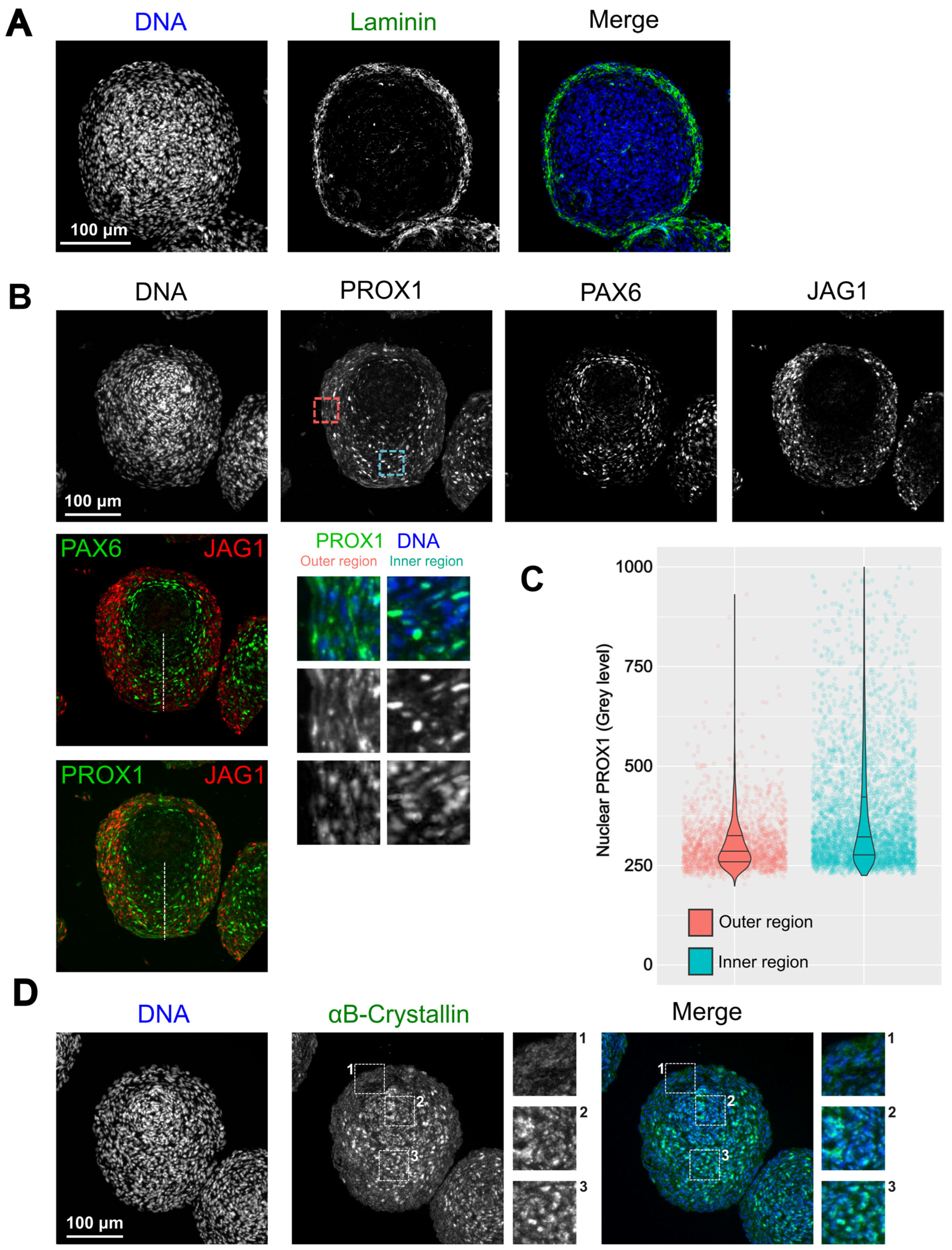
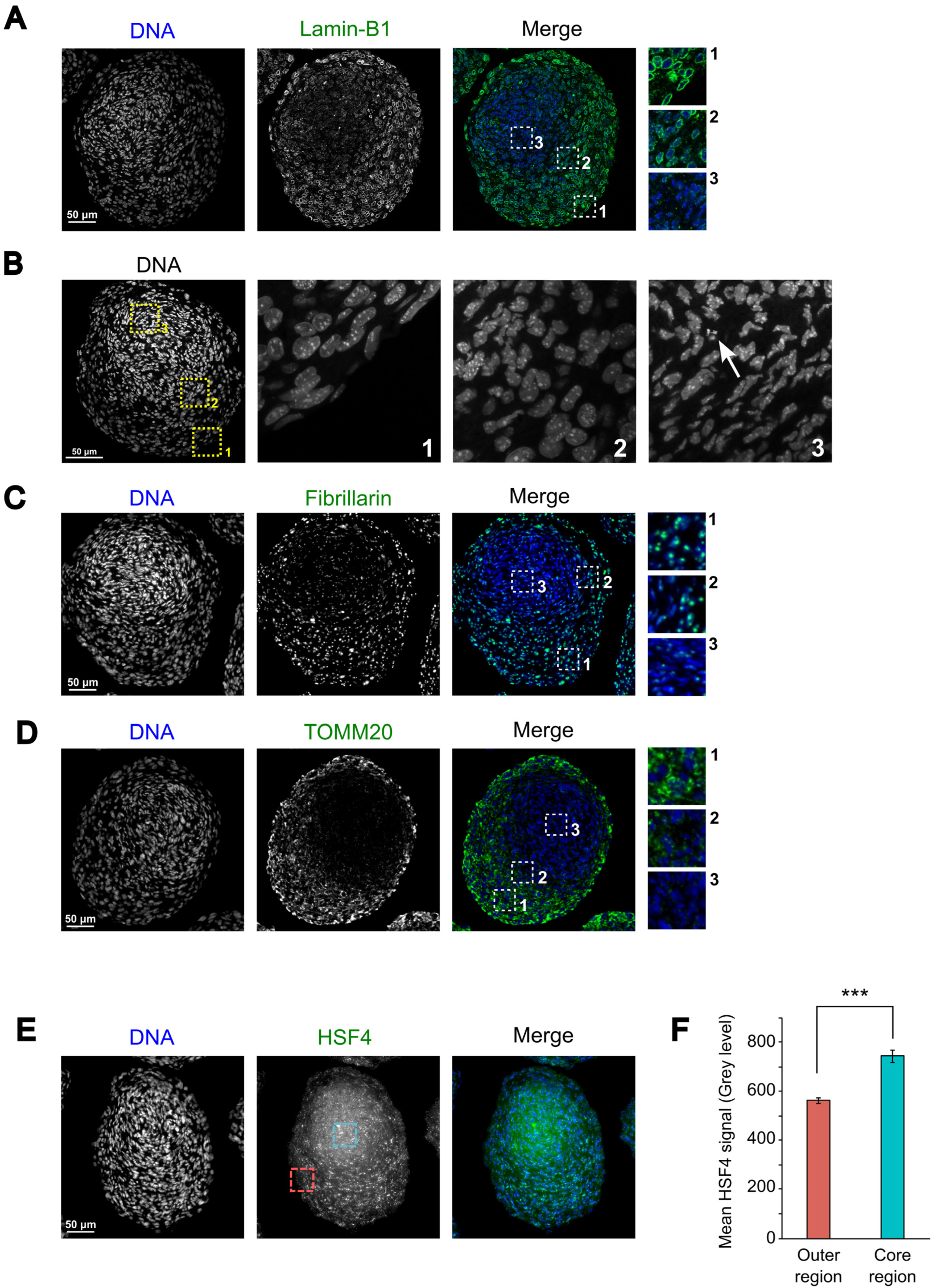
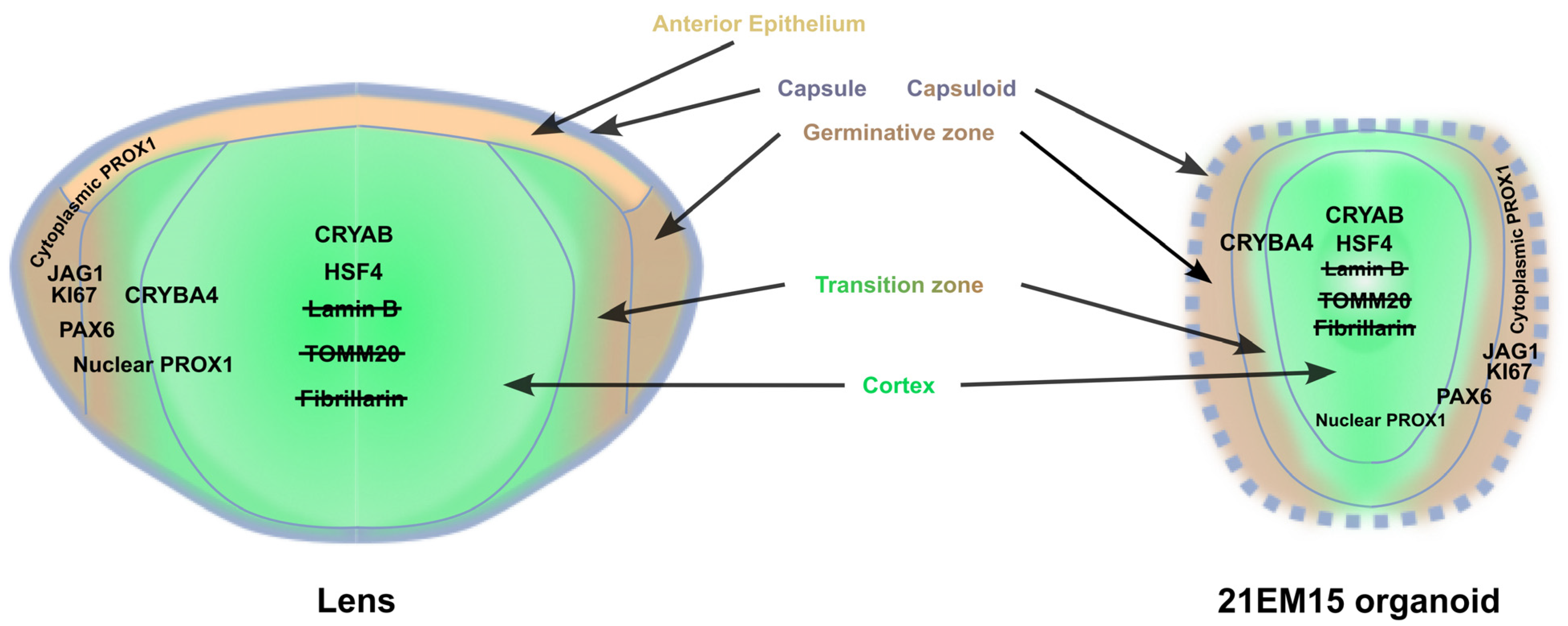
3.4. Morphological Organization of 21EM15 Organoids Partially Recapitulates Lens Patterning
3.5. 21EM15 Organoids Model Lens Cataract
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cvekl, A.; Zhang, X. Signaling and Gene Regulatory Networks in Mammalian Lens Development. Trends Genet. 2017, 33, 677–702. [Google Scholar] [CrossRef]
- Bassnett, S.; Šikić, H. The Lens Growth Process. Prog. Retin. Eye Res. 2017, 60, 181–200. [Google Scholar] [CrossRef]
- Bassnett, S.; Shi, Y.; Vrensen, G.F.J.M. Biological Glass: Structural Determinants of Eye Lens Transparency. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 1250–1264. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Huang, S.; Zheng, Y.; Zhou, T.; Hu, L.; Xiong, L.; Li, D.W.-C.; Liu, Y. The Lens Epithelium as a Major Determinant in the Development, Maintenance, and Regeneration of the Crystalline Lens. Prog. Retin. Eye Res. 2022, 92, 101112. [Google Scholar] [CrossRef] [PubMed]
- Shiels, A.; Hejtmancik, J.F. Inherited Cataracts: Genetic Mechanisms and Pathways New and Old. Exp. Eye Res. 2021, 209, 108662. [Google Scholar] [CrossRef] [PubMed]
- Shiels, A.; Hejtmancik, J.F. Biology of Inherited Cataracts and Opportunities for Treatment. Annu. Rev. Vis. Sci. 2019, 5, 123–149. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-C.; Wilkins, M.; Kim, T.; Malyugin, B.; Mehta, J.S. Cataracts. Lancet 2017, 390, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Daszynski, D.M.; Santhoshkumar, P.; Phadte, A.S.; Sharma, K.K.; Zhong, H.A.; Lou, M.F.; Kador, P.F. Failure of Oxysterols Such as Lanosterol to Restore Lens Clarity from Cataracts. Sci. Rep. 2019, 9, 8459. [Google Scholar] [CrossRef] [PubMed]
- Makley, L.N.; McMenimen, K.A.; DeVree, B.T.; Goldman, J.W.; McGlasson, B.N.; Rajagopal, P.; Dunyak, B.M.; McQuade, T.J.; Thompson, A.D.; Sunahara, R.; et al. Pharmacological Chaperone for Alpha-Crystallin Partially Restores Transparency in Cataract Models. Science 2015, 350, 674–677. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, X.J.; Zhu, J.; Xi, Y.B.; Yang, X.; Hu, L.D.; Ouyang, H.; Patel, S.H.; Jin, X.; Lin, D.; et al. Lanosterol Reverses Protein Aggregation in Cataracts. Nature 2015, 523, 607–611. [Google Scholar] [CrossRef]
- Goishi, K.; Shimizu, A.; Najarro, G.; Watanabe, S.; Rogers, R.; Zon, L.I.; Klagsbrun, M. AlphaA-Crystallin Expression Prevents Gamma-Crystallin Insolubility and Cataract Formation in the Zebrafish Cloche Mutant Lens. Development 2006, 133, 2585–2593. [Google Scholar] [CrossRef] [PubMed]
- Viet, J.; Reboutier, D.; Hardy, S.; Lachke, S.A.; Paillard, L.; Gautier-Courteille, C. Modeling Ocular Lens Disease in Xenopus. Dev. Dyn. 2019, 249, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C. Animal Models of Ophthalmic Diseases; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- Zhao, Z.; Chen, X.; Dowbaj, A.M.; Sljukic, A.; Bratlie, K.; Lin, L.; Fong, E.L.S.; Balachander, G.M.; Chen, Z.; Soragni, A.; et al. Organoids. Nat. Rev. Methods Primers 2022, 2, 1–21. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, M.D.; McAvoy, J.W. In Vitro Generation of Functional Lens-like Structures with Relevance to Age-Related Nuclear Cataract. Investig. Ophthalmol. Vis. Sci. 2007, 48, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Anchan, R.M.; Lachke, S.A.; Gerami-Naini, B.; Lindsey, J.; Ng, N.; Naber, C.; Nickerson, M.; Cavallesco, R.; Rowan, S.; Eaton, J.L.; et al. Pax6- and Six3-Mediated Induction of Lens Cell Fate in Mouse and Human ES Cells. PLoS ONE 2014, 9, e115106. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Qin, Z.; Jin, X.; Zhang, L.; Chen, Z.; He, J.; Ji, J.; Yao, K. Generation of Functional Lentoid Bodies From Human Induced Pluripotent Stem Cells Derived From Urinary Cells. Investig. Ophthalmol. Vis. Sci. 2017, 58, 517–527. [Google Scholar] [CrossRef]
- Yang, C.; Yang, Y.; Brennan, L.; Bouhassira, E.E.; Kantorow, M.; Cvekl, A. Efficient Generation of Lens Progenitor Cells and Lentoid Bodies from Human Embryonic Stem Cells in Chemically Defined Conditions. FASEB J. 2010, 24, 3274–3283. [Google Scholar] [CrossRef] [PubMed]
- Murphy, P.; Kabir, M.H.; Srivastava, T.; Mason, M.E.; Dewi, C.U.; Lim, S.; Yang, A.; Djordjevic, D.; Killingsworth, M.C.; Ho, J.W.K.; et al. Light-Focusing Human Micro-Lenses Generated from Pluripotent Stem Cells Model Lens Development and Drug-Induced Cataract in Vitro. Development 2018, 145, dev155838. [Google Scholar] [CrossRef]
- Plüss, C.J.; Kustermann, S. A Human Three-Dimensional In Vitro Model of Lens Epithelial Cells as a Model to Study Mechanisms of Drug-Induced Posterior Subcapsular Cataracts. J. Ocul. Pharmacol. Ther. 2019, 36, 56–64. [Google Scholar] [CrossRef]
- Cvekl, A.; Camerino, M.J. Generation of Lens Progenitor Cells and Lentoid Bodies from Pluripotent Stem Cells: Novel Tools for Human Lens Development and Ocular Disease Etiology. Cells 2022, 11, 3516. [Google Scholar] [CrossRef]
- Lin, H.; Ouyang, H.; Zhu, J.; Huang, S.; Liu, Z.; Chen, S.; Cao, G.; Li, G.; Signer, R.A.J.; Xu, Y.; et al. Lens Regeneration Using Endogenous Stem Cells with Gain of Visual Function. Nature 2016, 531, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Terrell, A.M.; Anand, D.; Smith, S.F.; Dang, C.A.; Waters, S.M.; Pathania, M.; Beebe, D.C.; Lachke, S.A. Molecular Characterization of Mouse Lens Epithelial Cell Lines and Their Suitability to Study RNA Granules and Cataract Associated Genes. Exp. Eye Res. 2015, 131, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Weatherbee, B.A.T.; Barton, J.R.; Siddam, A.D.; Anand, D.; Lachke, S.A. Molecular Characterization of the Human Lens Epithelium-Derived Cell Line SRA01/04. Exp. Eye Res. 2019, 188, 107787. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From Reads to Genes to Pathways: Differential Expression Analysis of RNA-Seq Experiments Using Rsubread and the edgeR Quasi-Likelihood Pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef] [PubMed]
- Kakrana, A.; Yang, A.; Anand, D.; Djordjevic, D.; Ramachandruni, D.; Singh, A.; Huang, H.; Ho, J.W.K.; Lachke, S.A. iSyTE 2.0: A Database for Expression-Based Gene Discovery in the Eye. Nucleic Acids Res. 2018, 46, D875–D885. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wilmarth, P.A.; Cheng, C.; Limi, S.; Fowler, V.M.; Zheng, D.; David, L.L.; Cvekl, A. Proteome-Transcriptome Analysis and Proteome Remodeling in Mouse Lens Epithelium and Fibers. Exp. Eye Res. 2019, 179, 32–46. [Google Scholar] [CrossRef]
- Zhao, Y.; Zheng, D.; Cvekl, A. A Comprehensive Spatial-Temporal Transcriptomic Analysis of Differentiating Nascent Mouse Lens Epithelial and Fiber Cells. Exp. Eye Res. 2018, 175, 56–72. [Google Scholar] [CrossRef]
- Anand, G.M.; Megale, H.C.; Murphy, S.H.; Weis, T.; Lin, Z.; He, Y.; Wang, X.; Liu, J.; Ramanathan, S. Controlling Organoid Symmetry Breaking Uncovers an Excitable System Underlying Human Axial Elongation. Cell 2023, 186, 497–512.e23. [Google Scholar] [CrossRef]
- Brassard, J.A.; Lutolf, M.P. Engineering Stem Cell Self-Organization to Build Better Organoids. Cell Stem Cell 2019, 24, 860–876. [Google Scholar] [CrossRef] [PubMed]
- Serra, D.; Mayr, U.; Boni, A.; Lukonin, I.; Rempfler, M.; Challet Meylan, L.; Stadler, M.B.; Strnad, P.; Papasaikas, P.; Vischi, D.; et al. Self-Organization and Symmetry Breaking in Intestinal Organoid Development. Nature 2019, 569, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Cvekl, A.; McGreal, R.; Liu, W. Lens Development and Crystallin Gene Expression. Prog. Mol. Biol. Transl. Sci. 2015, 134, 129–167. [Google Scholar] [CrossRef]
- DeDreu, J.; Walker, J.L.; Menko, A.S. Dynamics of the Lens Basement Membrane Capsule and Its Interaction with Connective Tissue-like Extracapsular Matrix Proteins. Matrix Biol. 2021, 96, 18–46. [Google Scholar] [CrossRef] [PubMed]
- Duncan, M.K.; Cui, W.; Oh, D.-J.; Tomarev, S.I. Prox1 Is Differentially Localized during Lens Development. Mech. Dev. 2002, 112, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Hannan, A.; Wang, Q.; Makrides, N.; Zhong, J.; Li, H.; Yoon, S.; Mao, Y.; Zhang, X. Etv Transcription Factors Functionally Diverge from Their Upstream FGF Signaling in Lens Development. Elife 2020, 9, e51915. [Google Scholar] [CrossRef] [PubMed]
- Brennan, L.; Disatham, J.; Kantorow, M. Mechanisms of Organelle Elimination for Lens Development and Differentiation. Exp. Eye Res. 2021, 209, 108682. [Google Scholar] [CrossRef] [PubMed]
- Costello, M.J.; Brennan, L.A.; Basu, S.; Chauss, D.; Mohamed, A.; Gilliland, K.O.; Johnsen, S.; Menko, S.; Kantorow, M. Autophagy and Mitophagy Participate in Ocular Lens Organelle Degradation. Exp. Eye Res. 2013, 116, 141–150. [Google Scholar] [CrossRef]
- Cui, X.; Liu, H.; Li, J.; Guo, K.; Han, W.; Dong, Y.; Wan, S.; Wang, X.; Jia, P.; Li, S.; et al. Heat Shock Factor 4 Regulates Lens Epithelial Cell Homeostasis by Working with Lysosome and Anti-Apoptosis Pathways. Int. J. Biochem. Cell Biol. 2016, 79, 118–127. [Google Scholar] [CrossRef]
- Imai, F.; Yoshizawa, A.; Fujimori-Tonou, N.; Kawakami, K.; Masai, I. The Ubiquitin Proteasome System Is Required for Cell Proliferation of the Lens Epithelium and for Differentiation of Lens Fiber Cells in Zebrafish. Development 2010, 137, 3257–3268. [Google Scholar] [CrossRef]
- Morishita, H.; Eguchi, S.; Kimura, H.; Sasaki, J.; Sakamaki, Y.; Robinson, M.L.; Sasaki, T.; Mizushima, N. Deletion of Autophagy-Related 5 (Atg5) and Pik3c3 Genes in the Lens Causes Cataract Independent of Programmed Organelle Degradation. J. Biol. Chem. 2013, 288, 11436–11447. [Google Scholar] [CrossRef] [PubMed]
- Sandilands, A.; Hutcheson, A.M.; Long, H.A.; Prescott, A.R.; Vrensen, G.; Löster, J.; Klopp, N.; Lutz, R.B.; Graw, J.; Masaki, S.; et al. Altered Aggregation Properties of Mutant Gamma-Crystallins Cause Inherited Cataract. EMBO J. 2002, 21, 6005–6014. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Wang, L.; Zhang, J.; Du, R.; Liao, S.; Li, D.; Li, C.; Ke, T.; Li, D.W.-C.; Huang, H.; et al. HSF4 Regulates DLAD Expression and Promotes Lens De-Nucleation. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2013, 1832, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Ruiss, M.; Kronschläger, M.; Schlatter, A.; Dechat, T.; Findl, O. Comparison of Methods to Experimentally Induce Opacification and Elasticity Change in Ex Vivo Porcine Lenses. Sci. Rep. 2021, 11, 23406. [Google Scholar] [CrossRef] [PubMed]
- Siddam, A.D.; Gautier-Courteille, C.; Perez-Campos, L.; Anand, D.; Kakrana, A.; Dang, C.A.; Legagneux, V.; Méreau, A.; Viet, J.; Gross, J.M.; et al. The RNA-Binding Protein Celf1 Post-Transcriptionally Regulates p27Kip1 and Dnase2b to Control Fiber Cell Nuclear Degradation in Lens Development. PLoS Genet. 2018, 14, e1007278. [Google Scholar] [CrossRef] [PubMed]
- Aryal, S.; Viet, J.; Weatherbee, B.A.T.; Siddam, A.D.; Hernandez, F.G.; Gautier-Courteille, C.; Paillard, L.; Lachke, S.A. The Cataract-Linked RNA-Binding Protein Celf1 Post-Transcriptionally Controls the Spatiotemporal Expression of the Key Homeodomain Transcription Factors Pax6 and Prox1 in Lens Development. Hum. Genet. 2020, 139, 1541–1554. [Google Scholar] [CrossRef]
- Young, L.K.; Jarrin, M.; Saunter, C.D.; Quinlan, R.A.; Girkin, J.M. Non-Invasive in Vivo Quantification of the Developing Optical Properties and Graded Index of the Embryonic Eye Lens Using SPIM. Biomed. Opt. Express 2018, 9, 2176–2188. [Google Scholar] [CrossRef]
- Khago, D.; Bierma, J.C.; Roskamp, K.W.; Kozlyuk, N.; Martin, R.W. Protein Refractive Index Increment Is Determined by Conformation as Well as Composition. J. Phys. Condens. Matter 2018, 30, 435101. [Google Scholar] [CrossRef]
- Mahendiran, K.; Elie, C.; Nebel, J.-C.; Ryan, A.; Pierscionek, B.K. Primary Sequence Contribution to the Optical Function of the Eye Lens. Sci. Rep. 2014, 4, 5195. [Google Scholar] [CrossRef]
- Wistow, G. The Human Crystallin Gene Families. Hum. Genom. 2012, 6, 26. [Google Scholar] [CrossRef]
- Clark, A.R.; Lubsen, N.H.; Slingsby, C. sHSP in the Eye Lens: Crystallin Mutations, Cataract and Proteostasis. Int. J. Biochem. Cell Biol. 2012, 44, 1687–1697. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.I. Functional Sequences in Human alphaB Crystallin. Biochim. Biophys. Acta 2016, 1860, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Nourazaran, M.; Yousefi, R.; Moosavi-Movahedi, F.; Panahi, F.; Hong, J.; Moosavi-Movahedi, A.A. The Structural and Functional Consequences of Melatonin and Serotonin on Human αB-Crystallin and Their Dual Role in the Eye Lens Transparency. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2023, 1871, 140928. [Google Scholar] [CrossRef] [PubMed]
- Boelens, W.C. Cell Biological Roles of αB-Crystallin. Prog. Biophys. Mol. Biol. 2014, 115, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Sprague-Piercy, M.A.; Rocha, M.A.; Kwok, A.O.; Martin, R.W. α-Crystallins in the Vertebrate Eye Lens: Complex Oligomers and Molecular Chaperones. Annu. Rev. Phys. Chem. 2021, 72, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Hejtmancik, J.F.; Riazuddin, S.A.; McGreal, R.; Liu, W.; Cvekl, A.; Shiels, A. Lens Biology and Biochemistry. Prog. Mol. Biol. Transl. Sci. 2015, 134, 169–201. [Google Scholar] [CrossRef]
- Quinlan, R. Cytoskeletal Competence Requires Protein Chaperones. Prog. Mol. Subcell. Biol. 2002, 28, 219–233. [Google Scholar] [CrossRef]
- Ho, D.W.H.; Yap, M.K.H.; Ng, P.W.; Fung, W.Y.; Yip, S.P. Association of High Myopia with Crystallin Beta A4 (CRYBA4) Gene Polymorphisms in the Linkage-Identified MYP6 Locus. PLoS ONE 2012, 7, e40238. [Google Scholar] [CrossRef]
- Oliver, G.; Loosli, F.; Köster, R.; Wittbrodt, J.; Gruss, P. Ectopic Lens Induction in Fish in Response to the Murine Homeobox Gene Six3. Mech. Dev. 1996, 60, 233–239. [Google Scholar] [CrossRef]
- Wang, Y.; Mahesh, P.; Wang, Y.; Novo, S.G.; Shihan, M.H.; Hayward-Piatkovskyi, B.; Duncan, M.K. Spatiotemporal Dynamics of Canonical Wnt Signaling during Embryonic Eye Development and Posterior Capsular Opacification (PCO). Exp. Eye Res. 2018, 175, 148–158. [Google Scholar] [CrossRef]
- Harned, J.; Fleisher, L.N.; McGahan, M.C. Lens Epithelial Cells Synthesize and Secrete Ceruloplasmin: Effects of Ceruloplasmin and Transferrin on Iron Efflux and Intracellular Iron Dynamics. Exp. Eye Res. 2006, 83, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Pontoriero, G.F.; Smith, A.N.; Miller, L.-A.D.; Radice, G.L.; West-Mays, J.A.; Lang, R.A. Co-Operative Roles for E-Cadherin and N-Cadherin during Lens Vesicle Separation and Lens Epithelial Cell Survival. Dev. Biol. 2009, 326, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Landgren, H.; Blixt, A.; Carlsson, P. Persistent FoxE3 Expression Blocks Cytoskeletal Remodeling and Organelle Degradation during Lens Fiber Differentiation. Investig. Ophthalmol. Vis. Sci. 2008, 49, 4269–4277. [Google Scholar] [CrossRef] [PubMed]
- Bassnett, S. On the Mechanism of Organelle Degradation in the Vertebrate Lens. Exp. Eye Res. 2009, 88, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Rowan, S.; Conley, K.W.; Le, T.T.; Donner, A.L.; Maas, R.L.; Brown, N.L. Notch Signaling Regulates Growth and Differentiation in the Mammalian Lens. Dev. Biol. 2008, 321, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Saravanamuthu, S.S.; Gao, C.Y.; Zelenka, P.S. Notch Signaling Is Required for Lateral Induction of Jagged1 during FGF-Induced Lens Fiber Differentiation. Dev. Biol. 2009, 332, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Shaham, O.; Smith, A.N.; Robinson, M.L.; Taketo, M.M.; Lang, R.A.; Ashery-Padan, R. Pax6 Is Essential for Lens Fiber Cell Differentiation. Development 2009, 136, 2567–2578. [Google Scholar] [CrossRef]
- McAvoy, J.W.; Richardson, N.A. Nuclear Pyknosis during Lens Fibre Differentiation in Epithelial Explants. Curr. Eye Res. 1986, 5, 711–715. [Google Scholar] [CrossRef]
- Mochizuki, T.; Masai, I. The Lens Equator: A Platform for Molecular Machinery That Regulates the Switch from Cell Proliferation to Differentiation in the Vertebrate Lens. Dev. Growth Differ. 2014, 56, 387–401. [Google Scholar] [CrossRef]
- Shaham, O.; Menuchin, Y.; Farhy, C.; Ashery-Padan, R. Pax6: A Multi-Level Regulator of Ocular Development. Prog. Retin. Eye Res. 2012, 31, 351–376. [Google Scholar] [CrossRef]
- Cvekl, A.; Ashery-Padan, R. The Cellular and Molecular Mechanisms of Vertebrate Lens Development. Development 2014, 141, 4432–4447. [Google Scholar] [CrossRef] [PubMed]
- Wigle, J.T.; Chowdhury, K.; Gruss, P.; Oliver, G. Prox1 Function Is Crucial for Mouse Lens-Fibre Elongation. Nat. Genet. 1999, 21, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Audette, D.S.; Anand, D.; So, T.; Rubenstein, T.B.; Lachke, S.A.; Lovicu, F.J.; Duncan, M.K. Prox1 and Fibroblast Growth Factor Receptors Form a Novel Regulatory Loop Controlling Lens Fiber Differentiation and Gene Expression. Development 2016, 143, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Richardson, R.; Tracey-White, D.; Webster, A.; Moosajee, M. The Zebrafish Eye-a Paradigm for Investigating Human Ocular Genetics. Eye 2017, 31, 68–86. [Google Scholar] [CrossRef] [PubMed]
- Spector, A.; Wang, G.M.; Wang, R.R.; Garner, W.H.; Moll, H. The Prevention of Cataract Caused by Oxidative Stress in Cultured Rat Lenses. I. H2O2 and Photochemically Induced Cataract. Curr. Eye Res. 1993, 12, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Spector, A. Oxidative Stress-Induced Cataract: Mechanism of Action. FASEB J. 1995, 9, 1173–1182. [Google Scholar] [CrossRef]
- Hernebring, M.; Adelöf, J.; Wiseman, J.; Petersen, A.; Zetterberg, M. H2O2-Induced Cataract as a Model of Age-Related Cataract: Lessons Learned from Overexpressing the Proteasome Activator PA28αβ in Mouse Eye Lens. Exp. Eye Res. 2021, 203, 108395. [Google Scholar] [CrossRef]
- Gu, Y.; Xu, B.; Feng, C.; Ni, Y.; Hong, N.; Wang, J.; Jiang, B. Topical Use of NaCl Solution with Different Concentration Affects Lens Transparency in Anesthetized Mice. Curr. Eye Res. 2016, 41, 943–950. [Google Scholar] [CrossRef]
- Butler, P.A. Reversible Cataracts in Diabetes Mellitus. J. Am. Optom. Assoc. 1994, 65, 559–563. [Google Scholar]
- Obrosova, I.G.; Chung, S.S.M.; Kador, P.F. Diabetic Cataracts: Mechanisms and Management. Diabetes Metab. Res. Rev. 2010, 26, 172–180. [Google Scholar] [CrossRef]
- Dasgupta, T.; Ladd, A.N. The Importance of CELF Control: Molecular and Biological Roles of the CUG-BP, Elav-like Family of RNA-Binding Proteins. Wiley Interdiscip. Rev. RNA 2012, 3, 104–121. [Google Scholar] [CrossRef]
- Siddam, A.D.; Duot, M.; Coomson, S.Y.; Anand, D.; Aryal, S.; Weatherbee, B.A.T.; Audic, Y.; Paillard, L.; Lachke, S.A. High-Throughput Transcriptomics of Celf1 Conditional Knockout Lens Identifies Downstream Networks Linked to Cataract Pathology. Cells 2023, 12, 1070. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duot, M.; Viel, R.; Viet, J.; Le Goff-Gaillard, C.; Paillard, L.; Lachke, S.A.; Gautier-Courteille, C.; Reboutier, D. Eye Lens Organoids Made Simple: Characterization of a New Three-Dimensional Organoid Model for Lens Development and Pathology. Cells 2023, 12, 2478. https://doi.org/10.3390/cells12202478
Duot M, Viel R, Viet J, Le Goff-Gaillard C, Paillard L, Lachke SA, Gautier-Courteille C, Reboutier D. Eye Lens Organoids Made Simple: Characterization of a New Three-Dimensional Organoid Model for Lens Development and Pathology. Cells. 2023; 12(20):2478. https://doi.org/10.3390/cells12202478
Chicago/Turabian StyleDuot, Matthieu, Roselyne Viel, Justine Viet, Catherine Le Goff-Gaillard, Luc Paillard, Salil A. Lachke, Carole Gautier-Courteille, and David Reboutier. 2023. "Eye Lens Organoids Made Simple: Characterization of a New Three-Dimensional Organoid Model for Lens Development and Pathology" Cells 12, no. 20: 2478. https://doi.org/10.3390/cells12202478





