Synaptopodin-2 Isoforms Have Specific Binding Partners and Display Distinct, Muscle Cell Type-Specific Expression Patterns
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Human Tissue Specimens
2.2. RT-PCR
2.3. Preparation of Tissue Extracts and Western Blotting
2.4. Immunolocalization in Tissue Sections
2.5. Primary and Secondary Antibodies, and Other Reagents
2.6. Yeast Two-Hybrid Assays
2.7. Protein Expression and Purification
2.8. Co-Immunoprecipitation Assays and Western Blot Overlays
2.9. Culture and Transfection of A7r5 Smooth Muscle Cells
2.10. Densitometry
3. Results
3.1. Tissue-Specific Expression of SYNPO2 Isoforms at the RNA Level
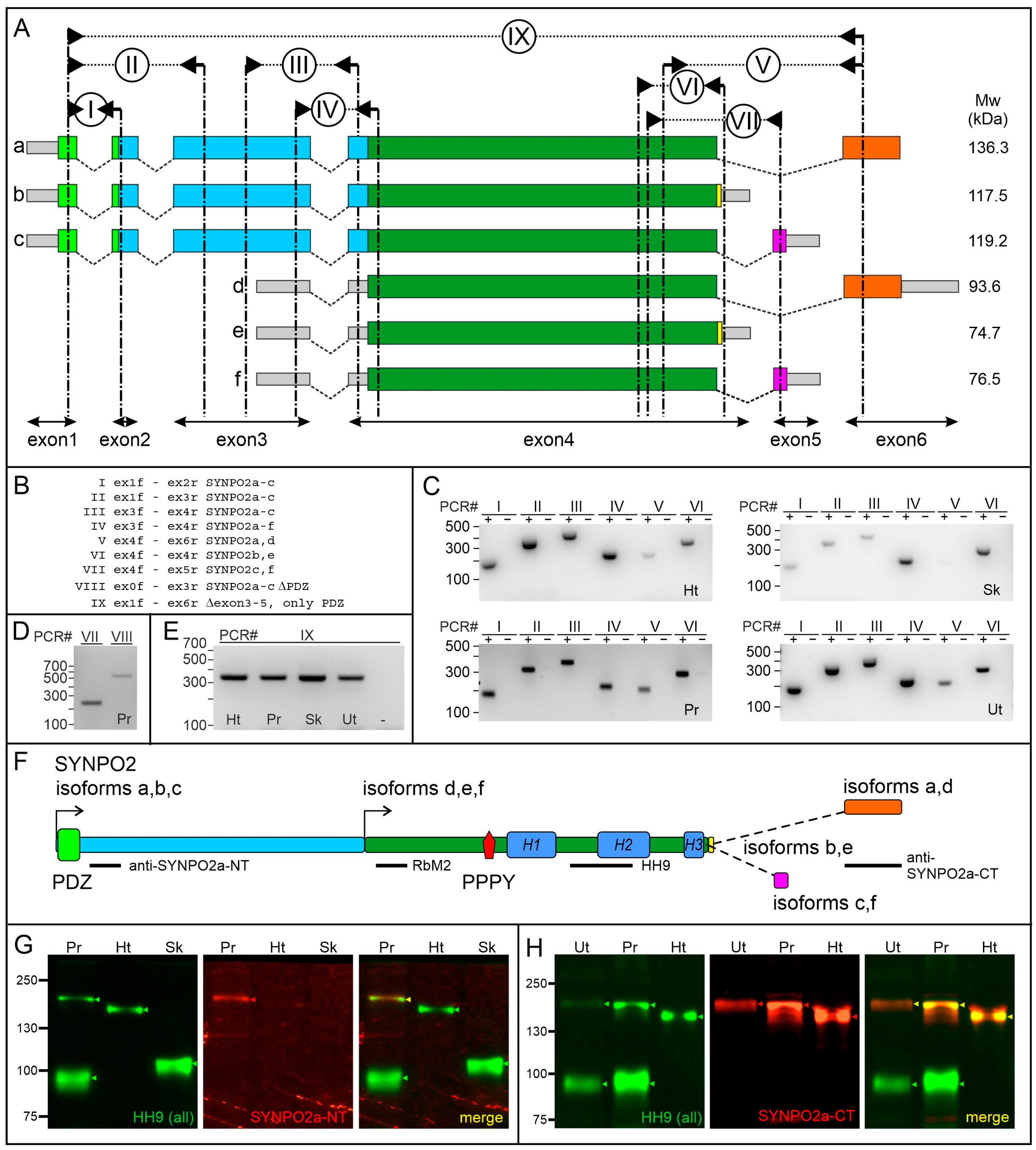
3.2. Tissue-Specific Expression of SYNPO2 Protein Isoforms
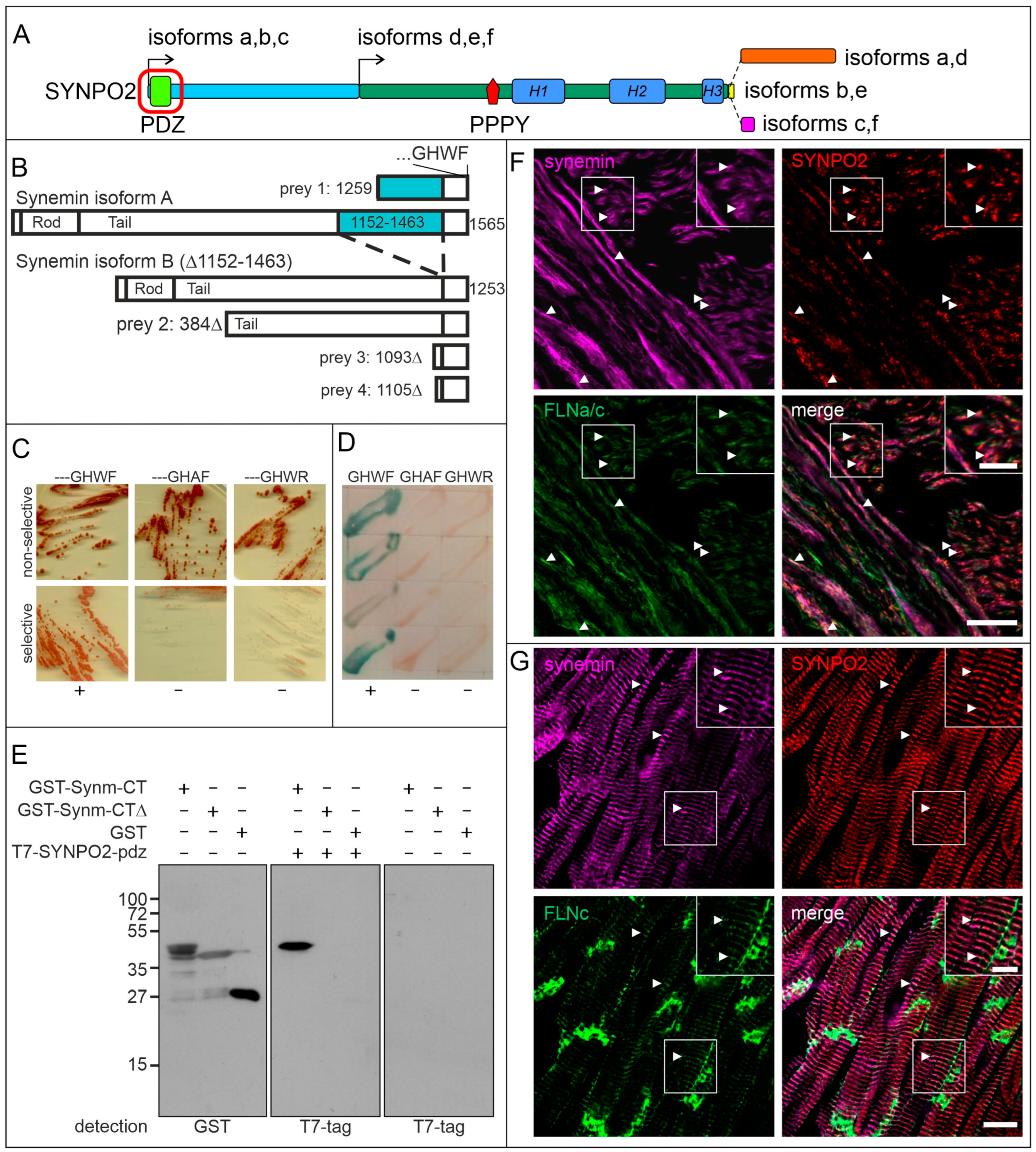
3.3. The PDZ-Domain of SYNPO2 Interacts with Synemin
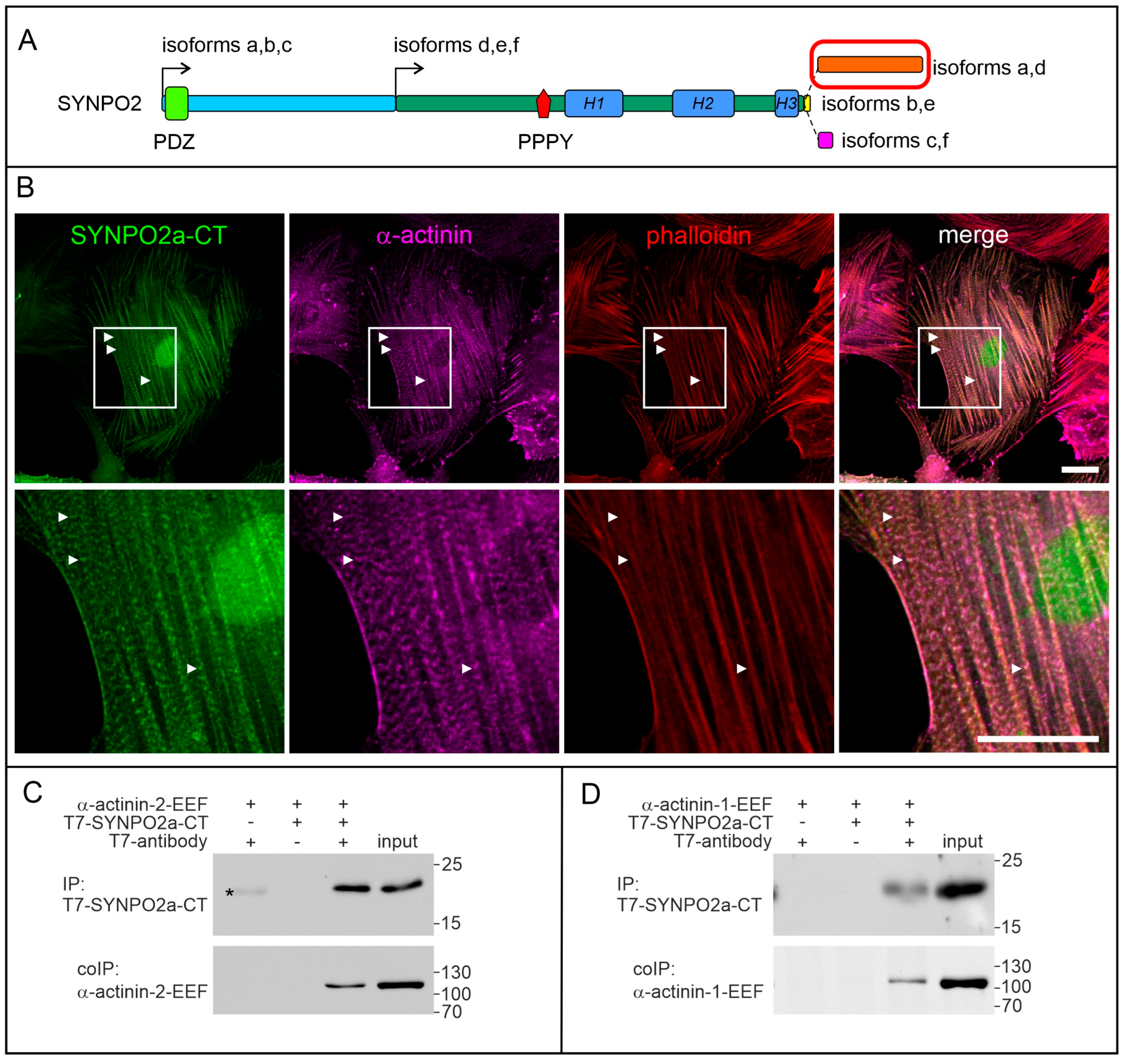
3.4. The Isoform-Specific Part of SYNPO2 Encoded by Exon 6 Binds α-Actinin
3.5. SYNPO2 Is Expressed in Uterus Smooth Muscle Cells
3.6. SYNPO2 Is Mainly Localized in Smooth Muscle Cells and Not in Epithelial Cells of the Human Prostate
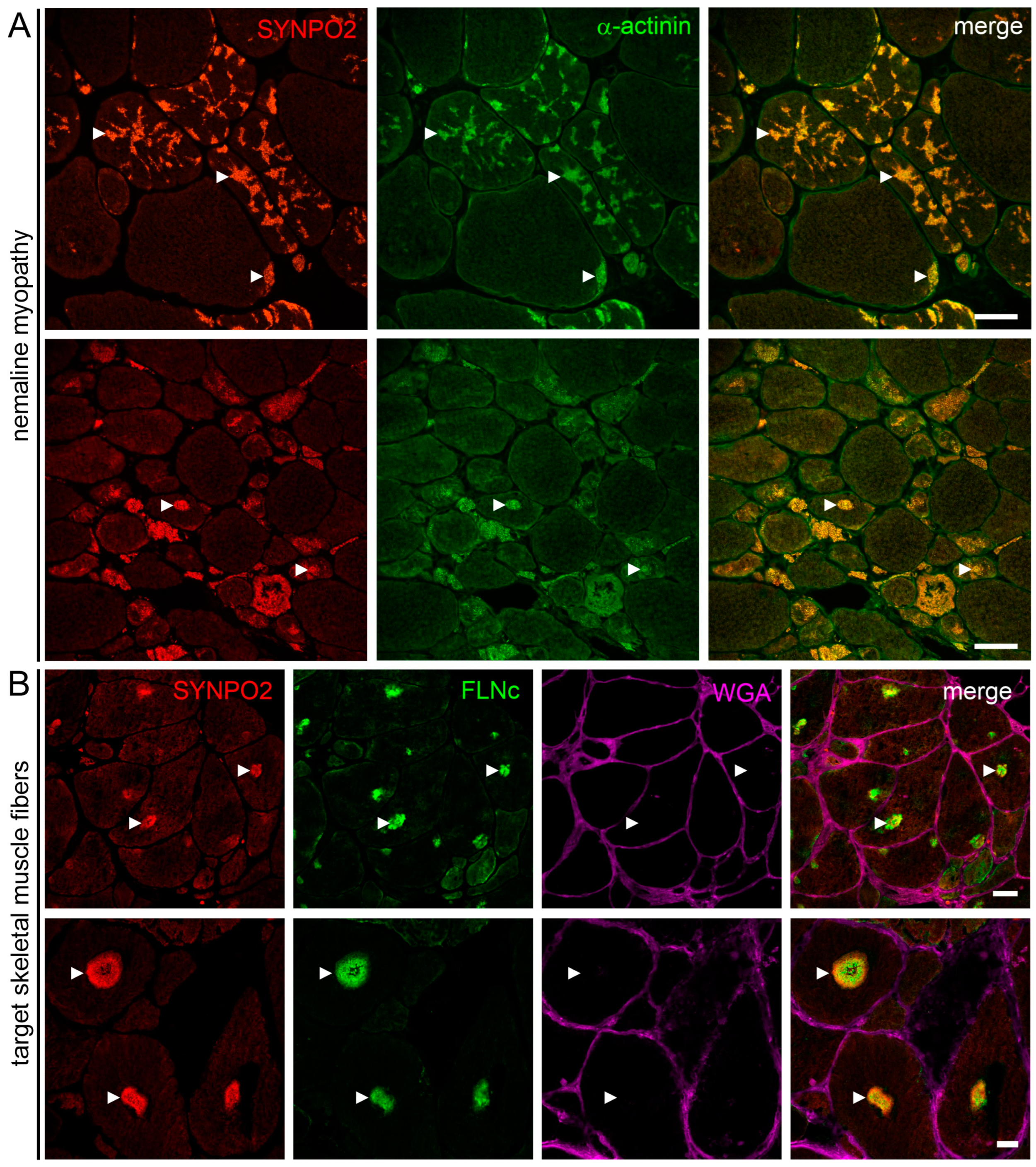
3.7. SYNPO2 Is a Component of Nemaline Rods and the Central and Intermediate Zone of Target Fibers
4. Discussion
4.1. SYNPO2 Isoform Expression in Striated Muscles
4.2. The PDZ Domain of SYNPO2 Isoforms a–c Also Interacts with Synemin: An Additional Novel Link between Z-Discs and the Surrounding Intermediate Filaments
4.3. SYNPO2 in Smooth Muscles
4.4. The 5’ UTR of Individual SYNPO2 mRNAs Is Extremely Long in Smooth Muscle Cells
4.5. Further Predicted Isoforms
4.6. SYNPO2 in Skeletal Muscle Pathology
4.7. SYNPO2 in Cancer
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leinweber, B.D.; Fredricksen, R.S.; Hoffman, D.R.; Chalovich, J.M. Fesselin: A novel synaptopodin-like actin binding protein from muscle tissue. J. Muscle Res. Cell. Motil. 1999, 20, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Schroeter, M.M.; Beall, B.; Heid, H.W.; Chalovich, J.M. The actin binding protein, fesselin, is a member of the synaptopodin family. Biochem. Biophys. Res. Commun. 2008, 371, 582–586. [Google Scholar] [CrossRef] [PubMed]
- Tkatchenko, A.V.; Pietu, G.; Cros, N.; Gannoun-Zaki, L.; Auffray, C.; Léger, J.J.; Dechesne, C.A. Identification of altered gene expression in skeletal muscles from Duchenne muscular dystrophy patients. Neuromuscul. Disord. 2001, 11, 269–277. [Google Scholar] [CrossRef]
- Weins, A.; Schwarz, K.; Faul, C.; Barisoni, L.; Linke, W.A.; Mundel, P. Differentiation- and stress-dependent nuclear cytoplasmic redistribution of myopodin, a novel actin-bundling protein. J. Cell Biol. 2001, 155, 393–404. [Google Scholar] [CrossRef]
- Renegar, R.H.; Chalovich, J.M.; Leinweber, B.D.; Zary, J.T.; Schroeter, M.M. Localization of the actin-binding protein fesselin in chicken smooth muscle. Histochem. Cell Biol. 2009, 131, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Faul, C.; Hüttelmaier, S.; Oh, J.; Hachet, V.; Singer, R.H.; Mundel, P. Promotion of importin a-mediated nuclear import by the phosphorylation-dependent binding of cargo protein to 14-3-3. J. Cell Biol. 2005, 169, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Faul, C.; Dhume, A.; Schecter, A.D.; Mundel, P. Protein kinase A, Ca2+/calmodulin-dependent kinase II, and calcineurin regulate the intracellular trafficking of myopodin between the Z-disc and the nucleus of cardiac myocytes. Mol. Cell Biol. 2007, 27, 8215–8227. [Google Scholar] [CrossRef]
- De Ganck, A.; de Corte, V.; Staes, A.; Gevaert, K.; Vandekerckhove, J.; Gettemans, J. Multiple isoforms of the tumor suppressor myopodin are simultaneously transcribed in cancer cells. Biochem. Biophys. Res. Commun. 2008, 370, 269–273. [Google Scholar] [CrossRef]
- Chalovich, J.M.; Schroeter, M.M. Synaptopodin family of natively unfolded, actin binding proteins: Physical properties and potential biological functions. Biophys. Rev. 2010, 2, 181–189. [Google Scholar] [CrossRef]
- Linnemann, A.; van der Ven, P.F.M.; Vakeel, P.; Albinus, B.; Simonis, D.; Bendas, G.; Schenk, J.A.; Micheel, B.; Kley, R.A.; Fürst, D.O. The sarcomeric Z-disc component myopodin is a multiadapter protein that interacts with filamin and α-actinin. Eur. J. Cell Biol. 2010, 89, 681–692. [Google Scholar] [CrossRef]
- Kai, F.; Tanner, K.; King, C.; Duncan, R. Myopodin isoforms alter the chemokinetic response of PC3 cells in response to different migration stimuli via differential effects on Rho-ROCK signaling pathways. Carcinogenesis 2012, 33, 2100–2107. [Google Scholar] [CrossRef] [PubMed]
- Lohanadan, K.; Molt, S.; Dierck, F.; van der Ven, P.F.M.; Frey, N.; Höhfeld, J.; Fürst, D.O. Isoform-specific functions of synaptopodin-2 variants in cytoskeleton stabilization and autophagy regulation in muscle under mechanical stress. Exp. Cell Res. 2021, 408, 112865. [Google Scholar] [CrossRef] [PubMed]
- Ulbricht, A.; Eppler, F.J.; Tapia, V.E.; van der Ven, P.F.M.; Hampe, N.; Hersch, N.; Vakeel, P.; Stadel, D.; Haas, A.; Saftig, P.; et al. Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr. Biol. 2013, 23, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Linnemann, A.; Vakeel, P.; Bezerra, E.; Orfanos, Z.; Djinović-Carugo, K.; van der Ven, P.F.M.; Kirfel, G.; Fürst, D.O. Myopodin is an F-actin bundling protein with multiple independent actin-binding regions. J. Muscle Res. Cell. Motil. 2012, 34, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, M.E.; Flenniken, A.M.; Ji, X.; Teboul, L.; Wong, M.D.; White, J.K.; Meehan, T.F.; Weninger, W.J.; Westerberg, H.; Adissu, H.; et al. High-throughput discovery of novel developmental phenotypes. Nature 2016, 537, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Schneider, R.; van der Ven, P.F.M.; Assent, M.; Lohanadan, K.; Klämbt, V.; Buerger, F.; Kitzler, T.M.; Deutsch, K.; Nakayama, M.; et al. Recessive mutations in SYNPO2 as a candidate of monogenic nephrotic syndrome. Kidney Int. Rep. 2020, 6, 472–483. [Google Scholar] [CrossRef] [PubMed]
- Elshafey, S.A.; Thabet, M.A.E.H.; Abo Elwafa, R.A.H.; Schneider, R.; Shril, S.; Buerger, F.; Hildebrandt, F.; Fathy, H.M. Genetic stratification reveals COL4A variants and spontaneous remission in Egyptian children with proteinuria in the first 2 years of life. Acta Paediatr. 2023, 112, 1324–1332. [Google Scholar] [CrossRef]
- Zheng, Z.; Song, Y. Synaptopodin-2: A potential tumor suppressor. Cancer Cell Int. 2023, 23, 158. [Google Scholar] [CrossRef]
- De Ganck, A.; de Corte, V.; Bruyneel, E.; Bracke, M.; Vandekerckhove, J.; Gettemans, J. Down-regulation of myopodin expression reduces invasion and motility of PC-3 prostate cancer cells. Int. J. Oncol. 2009, 34, 1403–1409. [Google Scholar] [CrossRef]
- Krause, K.; Eggers, B.; Uszkoreit, J.; Eulitz, S.; Rehmann, R.; Güttsches, A.K.; Schreiner, A.; van der Ven, P.F.M.; Fürst, D.O.; Marcus, K.; et al. Target formation in muscle fibres indicates reinnervation—A proteomic study in muscle samples from peripheral neuropathies. Neuropathol. Appl. Neurobiol. 2023, 49, e12853. [Google Scholar] [CrossRef]
- Gomori, G. A rapid one-step trichrome stain. Am. J. Clin. Pathol. 1950, 20, 661–664. [Google Scholar] [CrossRef] [PubMed]
- Schuld, J.; Orfanos, Z.; Chevessier, F.; Eggers, B.; Heil, L.; Uszkoreit, J.; Unger, A.; Kirfel, G.; van der Ven, P.F.M.; Marcus, K.; et al. Homozygous expression of the myofibrillar myopathy-associated p.W2710X filamin C variant reveals major pathomechanisms of sarcomeric lesion formation. Acta Neuropathol. Comm. 2020, 8, 154. [Google Scholar] [CrossRef] [PubMed]
- Van der Ven, P.F.M.; Obermann, W.M.J.; Lemke, B.; Gautel, M.; Weber, K.; Fürst, D.O. Characterization of muscle filamin isoforms suggests a possible role of γ-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil. Cytoskelet. 2000, 45, 149–162. [Google Scholar] [CrossRef]
- Wehland, J.; Willingham, M.C.; Sandoval, I.V. A rat monoclonal antibody reacting specifically with the tyrosylated form of alpha-tubulin. I. Biochemical characterization, effects on microtubule polymerization in vitro, and microtubule polymerization and organization in vivo. J. Cell Biol. 1983, 97, 1467–1475. [Google Scholar] [CrossRef]
- Van der Ven, P.F.M.; Wiesner, S.; Salmikangas, P.; Auerbach, D.; Himmel, M.; Kempa, S.; Hayeß, K.; Pacholsky, D.; Taivainen, A.; Schröder, R.; et al. Indications for a novel muscular dystrophy pathway. γ-filamin, the muscle-specific filamin isoform, interacts with myotilin. J. Cell Biol. 2000, 151, 235–248. [Google Scholar] [CrossRef]
- Obermann, W.M.J.; van der Ven, P.F.M.; Steiner, F.; Weber, K.; Fürst, D.O. Mapping of a myosin-binding domain and a regulatory phosphorylation site in M-protein, a structural protein of the sarcomeric M band. Mol. Biol. Cell 1998, 9, 829–840. [Google Scholar] [CrossRef]
- Reimann, L.; Wiese, H.; Leber, Y.; Schwäble, A.N.; Fricke, A.L.; Rohland, A.; Knapp, B.; Peikert, C.D.; Drepper, F.; van der Ven, P.F.M.; et al. Myofibrillar Z-discs are a protein phosphorylation hot spot with protein kinase C (PKCα) modulating protein dynamics. Mol. Cell Proteom. 2017, 16, 346–367. [Google Scholar] [CrossRef]
- Reimann, L.; Schwäble, A.N.; Fricke, A.L.; Mühlhäuser, W.W.D.; Leber, Y.; Lohanadan, K.; Puchinger, M.G.; Schäuble, S.; Faessler, E.; Wiese, H.; et al. Phosphoproteomics identifies dual-site phosphorylation in an extended basophilic motif regulating FILIP1-mediated degradation of filamin-C. Commun. Biol. 2020, 3, 253. [Google Scholar] [CrossRef]
- Lin, F.; Yu, Y.P.; Woods, J.; Cieply, K.; Gooding, B.; Finkelstein, P.; Dhir, R.; Krill, D.; Becich, M.J.; Michalopoulos, G.; et al. Myopodin, a synaptopodin homologue, is frequently deleted in invasive prostate cancers. Am. J. Pathol. 2001, 159, 1603–1612. [Google Scholar] [CrossRef]
- Murgia, M.; Nogara, L.; Baraldo, M.; Reggiani, C.; Mann, M.; Schiaffino, S. Protein profile of fiber types in human skeletal muscle: A single-fiber proteomics study. Skelet. Muscle 2021, 11, 24. [Google Scholar] [CrossRef]
- Eggers, B.; Schork, K.; Turewicz, M.; Barkovits, K.; Eisenacher, M.; Schröder, R.; Clemen, C.S.; Marcus, K. Advanced Fiber Type-Specific Protein Profiles Derived from Adult Murine Skeletal Muscle. Proteomes 2021, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Granger, B.L.; Lazarides, E. Synemin: A new high molecular weight protein associated with desmin and vimentin filaments in muscle. Cell 1980, 22, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Lund, L.M.; Kerr, J.P.; Lupinetti, J.; Zhang, Y.; Russell, M.A.; Bloch, R.J.; Bond, M. Synemin isoforms differentially organize cell junctions and desmin filaments in neonatal cardiomyocytes. FASEB J. 2012, 26, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Bellin, R.M.; Huiatt, T.W.; Critchley, D.R.; Robson, R.M. Synemin may function to directly link muscle cell intermediate filaments to both myofibrillar Z-lines and costameres. J. Biol. Chem. 2001, 276, 32330–32337. [Google Scholar] [CrossRef] [PubMed]
- Bellin, R.M.; Sernett, S.W.; Becker, B.; Ip, W.; Huiatt, T.W.; Robson, R.M. Molecular Characteristics and Interactions of the Intermediate Filament Protein Synemin. Interactions with alpha-actinin may anchor synemin-containing heterofilaments. J. Biol. Chem. 1999, 274, 29493–29499. [Google Scholar] [CrossRef] [PubMed]
- Winter, L.; Staszewska-Daca, I.; Zittrich, S.; Elhamine, F.; Zrelski, M.M.; Schmidt, K.; Fischer, I.; Jüngst, C.; Schauss, A.; Goldmann, W.H.; et al. Z-disk-associated plectin (isoform 1d): Spatial arrangement, interaction partners, and role in filamin C homeostasis. Cells 2023, 12, 1259. [Google Scholar] [CrossRef] [PubMed]
- Small, J.V.; Fürst, D.O.; Mey, J.D. Localization of filamin in smooth muscle. J. Cell Biol. 1986, 102, 210–220. [Google Scholar] [CrossRef]
- Swärd, K.; Krawczyk, K.K.; Morén, B.; Zhu, B.; Matic, L.; Holmberg, J.; Hedin, U.; Uvelius, B.; Stenkula, K.; Rippe, C. Identification of the intermediate filament protein synemin/SYNM as a target of myocardin family coactivators. Am. J. Physiol. Cell Physiol. 2019, 317, C1128–C1142. [Google Scholar] [CrossRef]
- Perisic Matic, L.; Rykaczewska, U.; Razuvaev, A.; Sabater-Lleal, M.; Lengquist, M.; Miller, C.L.; Ericsson, I.; Röhl, S.; Kronqvist, M.; Aldi, S.; et al. Phenotypic modulation of smooth muscle cells in atherosclerosis is associated with downregulation of LMOD1, SYNPO2, PDLIM7, PLN, and SYNM. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 1947–1961. [Google Scholar] [CrossRef]
- Brosens, E.; Felix, J.F.; Boerema-de Munck, A.; Jong, E.M.D.; Lodder, E.M.; Swagemakers, S.; Buscop-van Kempen, M.; Krijger, R.R.D.; Wijnen, R.M.H.; van IJcken, W.F.J.; et al. Histological, immunohistochemical and transcriptomic characterization of human tracheoesophageal fistulas. PLoS ONE 2020, 15, e0242167. [Google Scholar] [CrossRef]
- Green, I.D.; Liu, R.; Wong, J.J.L. The Expanding Role of Alternative Splicing in Vascular Smooth Muscle Cell Plasticity. Int. J. Mol. Sci. 2021, 22, 10213. [Google Scholar] [CrossRef] [PubMed]
- Turczyńska, K.M.; Swärd, K.; Hien, T.T.; Wohlfahrt, J.; Mattisson, I.Y.; Ekman, M.; Nilsson, J.; Sjögren, J.; Murugesan, V.; Hultgårdh-Nilsson, A.; et al. Regulation of smooth muscle dystrophin and synaptopodin 2 expression by actin polymerization and vascular injury. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1489–1497. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C. What Are 3′ UTRs Doing? Cold Spring Harb. Perspect. Biol. 2019, 11, a034728. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Jeong, S. 3′UTR Diversity: Expanding Repertoire of RNA Alterations in Human mRNAs. Mol. Cells 2023, 46, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Claeys, K.G.; van der Ven, P.F.M.; Behin, A.; Stojkovic, T.; Eymard, B.; Dubourg, O.; Laforet, P.; Faulkner, G.; Richard, P.; Vicart, P.; et al. Differential involvement of sarcomeric proteins in myofibrillar myopathies: A morphological and immunohistochemical study. Acta Neuropathol. 2009, 117, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Kley, R.A.; Maerkens, A.; Leber, Y.; Theis, V.; Schreiner, A.; van der Ven, P.F.M.; Uszkoreit, J.; Stephan, C.; Eulitz, S.; Euler, N.; et al. A combined laser microdissection and mass spectrometry approach reveals new disease relevant proteins accumulating in aggregates of filaminopathy patients. Mol. Cell Proteom. 2013, 12, 215–227. [Google Scholar] [CrossRef]
- Kley, R.A.; Leber, Y.; Schrank, B.; Zhuge, H.; Orfanos, Z.; Kostan, J.; Onipe, A.; Sellung, D.; Güttsches, A.K.; Eggers, B.; et al. FLNC-Associated Myofibrillar Myopathy: New Clinical, Functional, and Proteomic Data. Neurol. Genet. 2021, 7, e590. [Google Scholar] [CrossRef]
- Nicolau, S.; Dasgupta, A.; Dasari, S.; Charlesworth, M.C.; Johnson, K.L.; Pandey, A.; Doles, J.D.; Milone, M. Molecular signatures of inherited and acquired sporadic late onset nemaline myopathies. Acta Neuropathol. Comm. 2023, 11, 20. [Google Scholar] [CrossRef]
- Jing, L.; Liu, L.; Yu, Y.P.; Dhir, R.; Acquafondada, M.; Landsittel, D.; Cieply, K.; Wells, A.; Luo, J.H. Expression of myopodin induces suppression of tumor growth and metastasis. Am. J. Pathol. 2004, 164, 1799–1806. [Google Scholar] [CrossRef]
- Esteban, S.; Moya, P.; Fernandez-Suarez, A.; Vidaurreta, M.; Gonzalez-Peramato, P.; Sanchez-Carbayo, M. Diagnostic and prognostic utility of methylation and protein expression patterns of myopodin in colon cancer. Tumour. Biol. 2012, 33, 337–346. [Google Scholar] [CrossRef]
- Sanchez-Carbayo, M.; Schwarz, K.; Charytonowicz, E.; Cordon-Cardo, C.; Mundel, P. Tumor suppressor role for myopodin in bladder cancer: Loss of nuclear expression of myopodin is cell-cycle dependent and predicts clinical outcome. Oncogene 2003, 22, 5298–5305. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Björling, E.; Agaton, C.; Szigyarto, C.A.-K.; Amini, B.; Andersen, E.; Andersson, A.-C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.; Zhang, C.; Méar, L.; Zhong, W.; Digre, A.; Katona, B.; Sjöstedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single-cell type transcriptomics map of human tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef]


| RT-PCR | Forward Primer | Reverse Primer |
|---|---|---|
| I | ATGACTGGAGGGGCGCCCTG exon1 | AGCTTGATGACTTCAGGGTA exon2 |
| II | ATGACTGGAGGGGCGCCCTG exon1 | CTGCAGGGTGGTACTTTCCA exon3 |
| III | TGTCCACATAAATTCGATCC exon3, 5′ | ACTTTCACTCCTCCTGAGCC exon4 |
| IV | AGCAGACCTCACAAGCACCG exon3, 3′ | CCTCTCGCTCAAGCTCGCCA exon 4 |
| V | TCAGTCAAGGTCAATTCAGCC exon4 | TCATCCACTAGACCGAGAGG exon 6 |
| VI | TTCTCAGCCAAGAAAAGTGG exon4 | TCCACAACAGATGGTTTCCA exon 4 |
| VII (exon 5) | GCAGGCTTCGTCAGTGTACT exon4 | AGGCTGGGAAGTAGGTCCTT exon 5 |
| VIII (+ex0/Δex1) | GAATTCGGCAACTCGGAAGC exon0 | CTTTCAAGCTGCCTGCACAG exon 3 |
| IX (Δex2,3,4) | ATGACTGGAGGGGCGCCCTG exon1 | TCATCCACTAGACCGAGAGG exon 6 |
| Antibody/Reagent | Target | Species | Source and/or Reference | Dilution |
|---|---|---|---|---|
| HH9 | aa 803-916; SYNPO2a-f | mouse IgG1 | [10] | IF 1:2; WB 1:50 |
| RbM2 | aa 428-534; SYNPO2a-f | rabbit | [10] | IF 1:1000 |
| anti-SYNPO2a-CT | aa 1085-1262; SYNPO2a,d | rabbit | custom made, BioGenes, Berlin, Germany | WB 1:1000 |
| anti-SYNPO2a-NT | aa 66-151; SYNPO2a,b,c | rabbit | Atlas antibodies, Bromma Sweden, HPA049707 | WB 1:500 |
| BM75.2 | α-actinin | mouse IgM | Sigma-Aldrich Chemie, Taufkirchen, Germany A5044 | IF 1:75 |
| RR90 | filamin A/C | mouse IgA | [23] | IF 1:50 |
| anti-SYNM | synemin | rabbit | Atlas antibodies HPA040066 | IF 1:200 |
| C11 | cytokeratins (pan) | mouse IgG1 | Sigma-Aldrich Chemie, C2931 | IF 1:400 |
| 1A4 | smooth muscle actin | mouse IgG2a | Sigma A2547 | IF 1:600 |
| YL1/2 | EEF-tag | rat IgG2a | ThermoFisher Scientific, Dreieich, Germany, MA1-80017 [24] | WB 1:700 |
| T7-tag | T7-tag | mouse IgG2b | Sigma-Aldrich Chemie 69522 | WB 1:10,000 |
| GST-tag | GST | mouse IgG1 | Sigma-Aldrich Chemie, 71097 | WB 1:10,000 |
| GAM IgA AF488 | mouse IgA | goat | SouthernBiotech, Birmingham, AL, USA, 1040-30 | IF 1:40 |
| GAM IgG2a AF594 | mouse IgG2a | goat | ThermoFisher Scientific A-21135 | IF 1:500 |
| GAM IgG1 AF594 | mouse IgG1 | goat | Jackson ImmunoResearch, Ely, UK, 115-585-205 | IF 1:300 |
| GAM IgM AF546 | mouse IgM | goat | ThermoFisher Scientific A-21045 | IF 1:100 |
| GAM IgM Cy5 | mouse IgM | goat | Jackson ImmunoResearch 115-175-075 | IF 1:100 |
| GAR Cy3 | rabbit Ig | goat | Jackson ImmunoResearch 111-165-045 | IF 1:800 |
| GAR AF647 | rabbit Ig | goat | ThermoFisher Scientific A-21245 | IF 1:100 |
| GAR-HRP | rabbit IgG | goat | Jackson ImmunoResearch 111-035-144 | WB 1:10,000 |
| GAM-HRP | mouse IgG + IgM | goat | Jackson ImmunoResearch 115-035-068 | WB 1:10,000 |
| GARat-HRP | rat IgG + IgM | goat | Jackson ImmunoResearch 112-035-068 | WB 1:10,000 |
| GAR IRdye800CW | rabbit | goat | LI-COR Biosciences, Bad Homburg, Germany 926-32211 | WB 1:10,000 |
| CoraLite594- phalloidin | actin filaments | - | Proteintech, Planegg-Martinsried, Germany, PF00003 | 1:100 |
| WGA Oregon Green 488 | glycoproteins, cell membrane | - | ThermoFisher Scientific W6748 | 1:250 |
| DAPI | nuclei | - | Sigma-Aldrich Chemie, D9542 | 1:40,000 |
| RNA (RT-PCR) | Protein (Western Blotting) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SYNPO2 Isoforms | Skeletal Muscle | Cardiac Muscle | Prostate | Uterus | SYNPO2 Isoform | Skeletal Muscle | Cardiac Muscle | Prostate | Uterus # |
| a–c | + | +++ | +++ | ++ | a | - | - | + | ± |
| a–f | +++ | +++ | +++ | +++ | b | - | ++ | - | - |
| a,d | ± | + | ++ | + | c * | ||||
| b,e | +++ | ++ | +++ | ++ | d | - | - | - | - |
| c,f | - | - | ± | - | e | +++ | - | ++ | + |
| f * | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lohanadan, K.; Assent, M.; Linnemann, A.; Schuld, J.; Heukamp, L.C.; Krause, K.; Vorgerd, M.; Reimann, J.; Schänzer, A.; Kirfel, G.; et al. Synaptopodin-2 Isoforms Have Specific Binding Partners and Display Distinct, Muscle Cell Type-Specific Expression Patterns. Cells 2024, 13, 85. https://doi.org/10.3390/cells13010085
Lohanadan K, Assent M, Linnemann A, Schuld J, Heukamp LC, Krause K, Vorgerd M, Reimann J, Schänzer A, Kirfel G, et al. Synaptopodin-2 Isoforms Have Specific Binding Partners and Display Distinct, Muscle Cell Type-Specific Expression Patterns. Cells. 2024; 13(1):85. https://doi.org/10.3390/cells13010085
Chicago/Turabian StyleLohanadan, Keerthika, Marvin Assent, Anja Linnemann, Julia Schuld, Lukas C. Heukamp, Karsten Krause, Matthias Vorgerd, Jens Reimann, Anne Schänzer, Gregor Kirfel, and et al. 2024. "Synaptopodin-2 Isoforms Have Specific Binding Partners and Display Distinct, Muscle Cell Type-Specific Expression Patterns" Cells 13, no. 1: 85. https://doi.org/10.3390/cells13010085





