Alcohol Promotes Lipogenesis in Sebocytes—Implications for Acne
Abstract
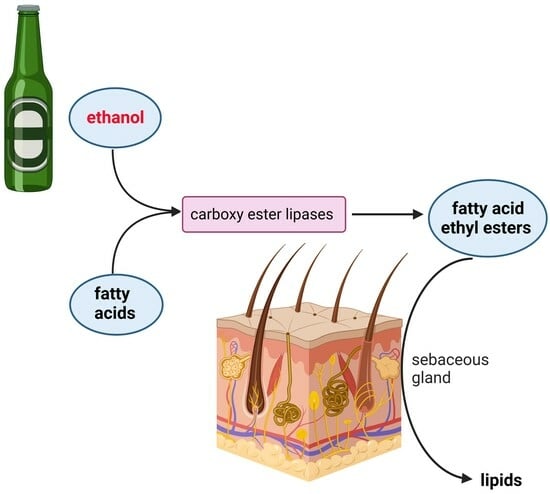
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Chemicals
2.3. Cell Culture
2.4. DNA Synthesis
2.5. Cell Lysis
2.6. Detection of Lipids
2.7. Measurement of Cellular Respiration and Mitochondrial Stress
2.8. Presentation of Data and Statistical Analysis
3. Results
3.1. Ethanol Induces Lipogenesis
3.2. Moderate Effects of Ethanol on DNA Synthesis and Membrane Integrity
3.3. Effect of Other Alcohols on Lipogenesis
3.4. Non-Oxidative Metabolism of Ethanol (NOME) Triggers Lipogenesis
3.5. Ethanol Inhibits Mitochondrial ATP Production
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carrigan, M.A.; Uryasev, O.; Frye, C.B.; Eckman, B.L.; Myers, C.R.; Hurley, T.D.; Benner, S.A. Hominids adapted to metabolize ethanol long before human-directed fermentation. Proc. Natl. Acad. Sci. USA 2015, 112, 458–463. [Google Scholar] [CrossRef] [PubMed]
- McGovern, P.E.; Zhang, J.; Tang, J.; Zhang, Z.; Hall, G.R.; Moreau, R.A.; Nuñez, A.; Butrym, E.D.; Richards, M.P.; Wang, C.S.; et al. Fermented beverages of pre- and proto-historic China. Proc. Natl. Acad. Sci. USA 2004, 101, 17593–17598. [Google Scholar] [CrossRef] [PubMed]
- Vallee, B.L. Alcohol in the western world. Sci. Am. 1998, 278, 80–85. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Status Report on Alcohol and Health 2018; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Baliunas, D.; Rehm, J.; Irving, H.; Shuper, P. Alcohol consumption and risk of incident human immunodeficiency virus infection: A meta-analysis. Int. J. Public Health 2010, 55, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Cortes, V.F.; Taveira, A.; Cruz, H.M.; Reis, A.A.; Cezar, J.S.; Silva, B.S.; D’Assunção, C.F.; Lampe, E.; Villar, L.M. Prevalence of Hepatitis B and C virus infection among alcoholic individuals: Importance of screening and vaccination. Rev. Inst. Med. Trop. Sao Paulo 2017, 59, e47. [Google Scholar] [CrossRef]
- Bagnardi, V.; Rota, M.; Botteri, E.; Tramacere, I.; Islami, F.; Fedirko, V.; Scotti, L.; Jenab, M.; Turati, F.; Pasquali, E.; et al. Alcohol consumption and site-specific cancer risk: A comprehensive dose-response meta-analysis. Br. J. Cancer 2015, 112, 580–593. [Google Scholar] [CrossRef]
- Roerecke, M.; Rehm, J. Alcohol intake revisited: Risks and benefits. Curr. Atheroscler. Rep. 2012, 14, 556–562. [Google Scholar] [CrossRef]
- Stawińska-Witoszyńska, B.; Czechowska, K.; Więckowska, B. The prevalence of Epilepsy and its co-occurrence with alcohol dependence among polish prisoners. Int. J. Equity Health 2019, 18, 102. [Google Scholar] [CrossRef]
- van de Wiel, A. Diabetes mellitus and alcohol. Diabetes Metab. Res. Rev. 2004, 20, 263–267. [Google Scholar] [CrossRef]
- Sabia, S.; Fayosse, A.; Dumurgier, J.; Dugravot, A.; Akbaraly, T.; Britton, A.; Kivimäki, M.; Singh-Manoux, A. Alcohol consumption and risk of dementia: 23 year follow-up of Whitehall II cohort study. BMJ 2018, 362, k2927. [Google Scholar] [CrossRef]
- Higgins, E.M.; du Vivier, A.W. Cutaneous disease and alcohol misuse. Br. Med. Bull. 1994, 50, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Shellow, W.V. The skin in alcoholism. Int. J. Dermatol. 1983, 22, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Parish, L.C.; Fine, E. Alcoholism and Skin Disease. Int. J. Dermatol. 1985, 24, 300–301. [Google Scholar] [CrossRef] [PubMed]
- Bhopale, K.K.; Falzon, M.; Ansari, G.A.S.; Kaphalia, B.S. Alcohol oxidizing enzymes and ethanol-induced cytotoxicity in rat pancreatic acinar AR42J cells. Vitr. Cell. Dev. Biol. Anim. 2014, 50, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Mathews, J.M.; Etheridge, A.S.; Raymer, J.H.; Black, S.R.; Pulliam, D.W., Jr.; Bucher, J.R. Selective inhibition of cytochrome P450 2E1 in vivo and in vitro with trans-1,2-dichloroethylene. Chem. Res. Toxicol. 1998, 11, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Haber, P.S.; Apte, M.V.; Moran, C.; Applegate, T.L.; Pirola, R.C.; Korsten, M.A.; McCaughan, G.W.; Wilson, J.S. Non-oxidative metabolism of ethanol by rat pancreatic acini. Pancreatol. Off. J. Int. Assoc. Pancreatol. (IAP) 2004, 4, 82–89. [Google Scholar] [CrossRef]
- Huang, W.; Booth, D.M.; Cane, M.C.; Chvanov, M.; Javed, M.A.; Elliott, V.L.; Armstrong, J.A.; Dingsdale, H.; Cash, N.; Li, Y.; et al. Fatty acid ethyl ester synthase inhibition ameliorates ethanol-induced Ca2+-dependent mitochondrial dysfunction and acute pancreatitis. Gut 2014, 63, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Bhopale, K.K.; Ansari, G.A.; Kaphalia, B.S. Ethanol-induced cytotoxicity in rat pancreatic acinar AR42J cells: Role of fatty acid ethyl esters. Alcohol Alcohol. 2008, 43, agm044. [Google Scholar] [CrossRef]
- Wróbel, A.; Seltmann, H.; Fimmel, S.; Müller-Decker, K.; Tsukada, M.; Bogdanoff, B.; Mandt, N.; Blume-Peytavi, U.; Orfanos, C.E.; Zouboulis, C.C. Differentiation and apoptosis in human immortalized sebocytes. J. Investig. Dermatol. 2003, 120, 175–181. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Seltmann, H.; Neitzel, H.; Orfanos, C.E. Establishment and characterization of an immortalized human sebaceous gland cell line (SZ95). J. Investig. Dermatol. 1999, 113, 1011–1020. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Seltmann, H.; Hiroi, N.; Chen, W.; Young, M.; Oeff, M.; Scherbaum, W.A.; Orfanos, C.E.; McCann, S.M.; Bornstein, S.R. Corticotropin-releasing hormone: An autocrine hormone that promotes lipogenesis in human sebocytes. Proc. Natl. Acad. Sci. USA 2002, 99, 7148–7153. [Google Scholar] [CrossRef] [PubMed]
- Zoller, N.; Schreiner, S.; Petry, L.; Hoffmann, S.; Steinhorst, K.; Kleemann, J.; Jager, M.; Kaufmann, R.; Meissner, M.; Kippenberger, S. Collagen I Promotes Adipocytogenesis in Adipose-Derived Stem Cells In Vitro. Cells 2019, 8, 302. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.R.; Zouboulis, C.C. Primary sebocytes and sebaceous gland cell lines for studying sebaceous lipogenesis and sebaceous gland diseases. Exp. Dermatol. 2018, 27, 484–488. [Google Scholar] [CrossRef] [PubMed]
- Tuo, J.; Wang, Q.; Zouboulis, C.C.; Liu, Y.; Ma, Y.; Ma, L.; Ying, J.; Zhang, C.; Xiang, L. ALA-PDT suppressing the cell growth and reducing the lipogenesis in human SZ95 sebocytes by mTOR signaling pathway in vitro. Photodiagnosis Photodyn. Ther. 2017, 18, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Higgins, E.; du Vivier, A. Alcohol intake and other skin disorders. Clin. Dermatol. 1999, 17, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Doyle, K.M.; Bird, D.A.; al-Salihi, S.; Hallaq, Y.; Cluette-Brown, J.E.; Goss, K.A.; Laposata, M. Fatty acid ethyl esters are present in human serum after ethanol ingestion. J. Lipid Res. 1994, 35, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yao, L.; Fu, X.; Mukherjee, R.; Xia, Q.; Jakubowska, M.A.; Ferdek, P.E.; Huang, W. Experimental Acute Pancreatitis Models: History, Current Status, and Role in Translational Research. Front. Physiol. 2020, 11, 614591. [Google Scholar] [CrossRef] [PubMed]
- Zakhari, S. Overview: How is alcohol metabolized by the body? Alcohol Res. Health J. Natl. Inst. Alcohol Abus. Alcohol. 2006, 29, 245–254. [Google Scholar]
- Cheung, C.; Smith, C.K.; Hoog, J.O.; Hotchkiss, S.A. Expression and localization of human alcohol and aldehyde dehydrogenase enzymes in skin. Biochem. Biophys. Res. Commun. 1999, 261, 100–107. [Google Scholar] [CrossRef]
- Zhai, Z.; Yamauchi, T.; Shangraw, S.; Hou, V.; Matsumoto, A.; Fujita, M. Ethanol Metabolism and Melanoma. Cancers 2023, 15, 1258. [Google Scholar] [CrossRef]
- Harjumäki, R.; Pridgeon, C.S.; Ingelman-Sundberg, M. CYP2E1 in Alcoholic and Non-Alcoholic Liver Injury. Roles of ROS, Reactive Intermediates and Lipid Overload. Int. J. Mol. Sci. 2021, 22, 8221. [Google Scholar] [CrossRef] [PubMed]
- Massart, J.; Begriche, K.; Hartman, J.H.; Fromenty, B. Role of Mitochondrial Cytochrome P450 2E1 in Healthy and Diseased Liver. Cells 2022, 11, 288. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Mukhtar, H. Cytochrome p450: A target for drug development for skin diseases. J. Investig. Dermatol. 2004, 123, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Nocchi, S.; Björklund, S.; Svensson, B.; Engblom, J.; Ruzgas, T. Electrochemical monitoring of native catalase activity in skin using skin covered oxygen electrode. Biosens. Bioelectron. 2017, 93, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Shindo, Y.; Witt, E.; Han, D.; Epstein, W.; Packer, L. Enzymic and non-enzymic antioxidants in epidermis and dermis of human skin. J. Investig. Dermatol. 1994, 102, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Manzo-Avalos, S.; Saavedra-Molina, A. Cellular and Mitochondrial Effects of Alcohol Consumption. Int. J. Environ. Res. Public Health 2010, 7, 4281–4304. [Google Scholar] [CrossRef] [PubMed]
- Rietjens, S.J.; de Lange, D.W.; Meulenbelt, J. Ethylene glycol or methanol intoxication: Which antidote should be used, fomepizole or ethanol? N. J. Med. 2014, 72, 73–79. [Google Scholar]
- Ashurst, J.V.; Nappe, T.M. Isopropanol Toxicity. In StatPearls; StatPearls Publishing Copyright© 2023; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2023. [Google Scholar]
- Slaughter, R.J.; Mason, R.W.; Beasley, D.M.; Vale, J.A.; Schep, L.J. Isopropanol poisoning. Clin. Toxicol. 2014, 52, 470–478. [Google Scholar] [CrossRef]
- Liu, L.; Xue, Y.; Chen, Y.; Pu, Y.; Zhang, Y.; Zhang, L.; Shao, X.; Chen, J.; Chen, J. Alcohol consumption and the risk of rosacea: A systematic review and meta-analysis. J. Cosmet. Dermatol. 2022, 21, 2954–2961. [Google Scholar] [CrossRef]
- Bayoumy, A.B.; Mulder, C.J.J.; Mol, J.J.; Tushuizen, M.E. Gut fermentation syndrome: A systematic review of case reports. United Eur. Gastroenterol. J. 2021, 9, 332–342. [Google Scholar] [CrossRef]
- Antoshechkin, A.G. On Intracellular Formation of Ethanol and Its Possible Role in Energy Metabolism. Alcohol Alcohol. 2001, 36, 608. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleemann, J.; Cinatl, J., Jr.; Hoffmann, S.; Zöller, N.; Özistanbullu, D.; Zouboulis, C.C.; Kaufmann, R.; Kippenberger, S. Alcohol Promotes Lipogenesis in Sebocytes—Implications for Acne. Cells 2024, 13, 328. https://doi.org/10.3390/cells13040328
Kleemann J, Cinatl J Jr., Hoffmann S, Zöller N, Özistanbullu D, Zouboulis CC, Kaufmann R, Kippenberger S. Alcohol Promotes Lipogenesis in Sebocytes—Implications for Acne. Cells. 2024; 13(4):328. https://doi.org/10.3390/cells13040328
Chicago/Turabian StyleKleemann, Johannes, Jindrich Cinatl, Jr., Stephanie Hoffmann, Nadja Zöller, Deniz Özistanbullu, Christos C. Zouboulis, Roland Kaufmann, and Stefan Kippenberger. 2024. "Alcohol Promotes Lipogenesis in Sebocytes—Implications for Acne" Cells 13, no. 4: 328. https://doi.org/10.3390/cells13040328